Mutations in the Vicinity of the IRAK3 Guanylate Cyclase Center Impact Its Subcellular Localization and Ability to Modulate Inflammatory Signaling in Immortalized Cell Lines
Abstract
1. Introduction
2. Results
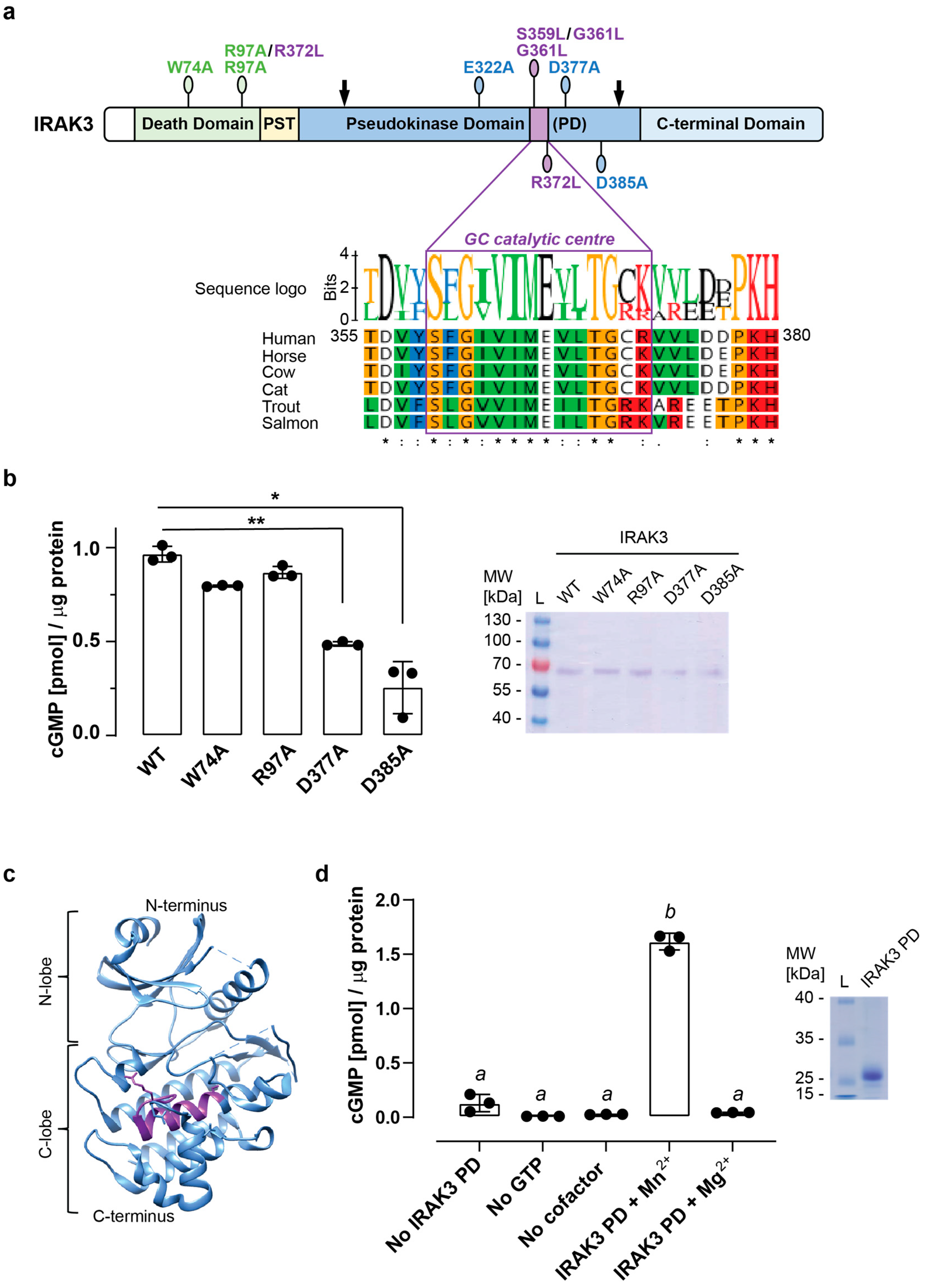
2.1. Mutations in the Vicinity of the Guanylate Cyclase Center Modulate IRAK3 Capacity to Generate cGMP
2.2. Mutations in IRAK3 Pseudokinase Domain Modify LPS-Induced NFκB Activity
2.3. Wild Type and Mutant IRAK3 Differentially Affect Cytokine Production
2.4. Mutations in the Pseudokinase Domain of IRAK3 Alter Its Subcellular Localization
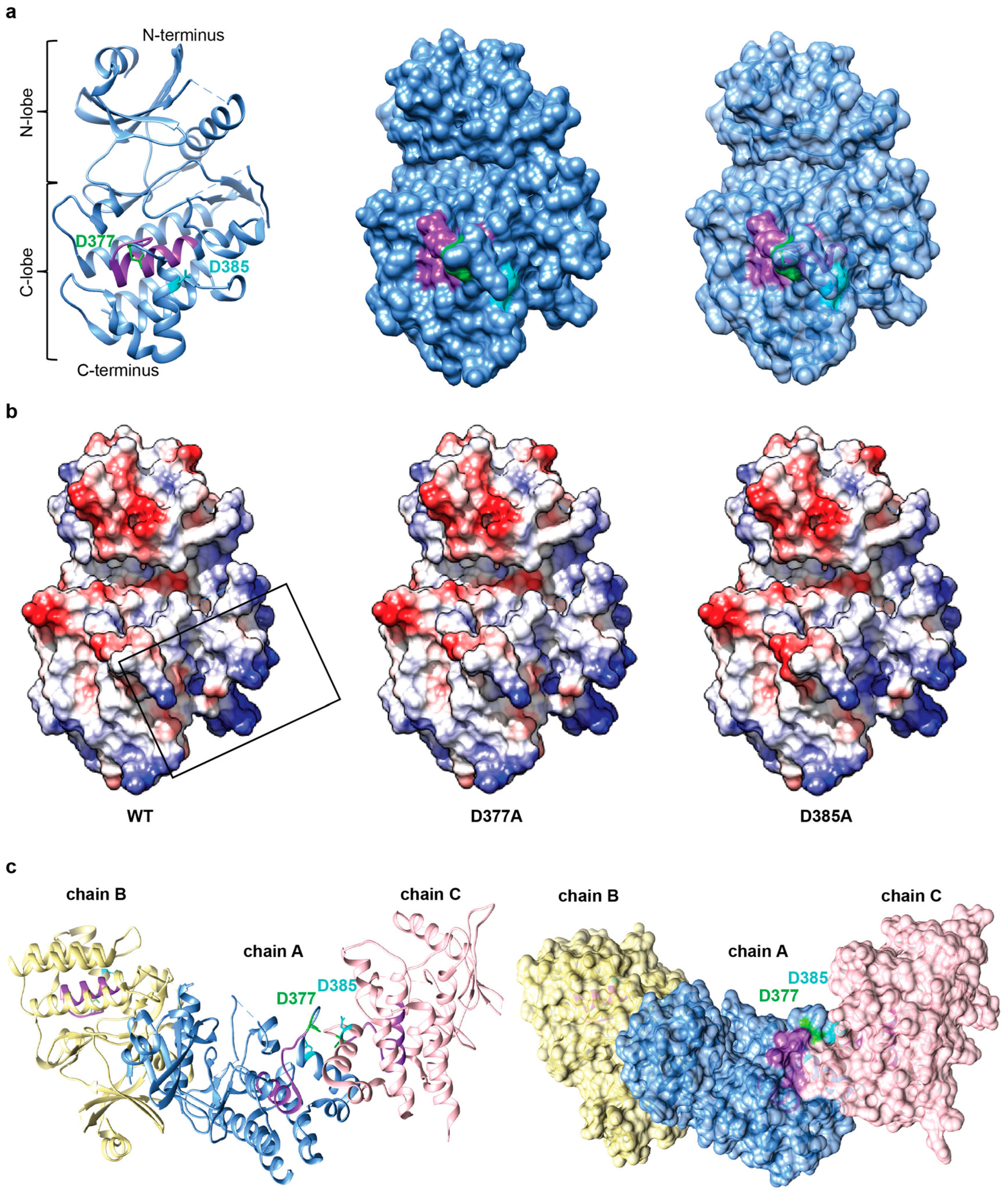
2.5. Negatively Charged Patch in the Conserved Region of IRAK3 C-Lobe May Be Required for Cofactor Binding and Is Predicted to Mediate Protein-Protein Interactions
3. Discussion
4. Materials and Methods
4.1. Amino Acid Sequence Alignment
4.2. Generation of the Truncated IRAK3 (IRAK3 PD) and Mutated Full-Length IRAK3 Constructs Using Gateway Recombination Cloning Technology
4.3. Expression and Purification of Recombinant Proteins in Bacteria
4.4. Detection and Quantification of cGMP Generated In Vitro
4.5. Protein Expression in the Baculovirus/Insect Cell Expression System
4.6. Mammalian Cell Culture, Transient Transfection and Imaging
4.7. HEK-Blue hTLR4 Cell Treatments for Secreted Embryonic Alkaline Phosphatase (SEAP) Assay
4.8. HEK293T Cell Treatments for cGMP Generation and Quantification
4.9. Complementation of IRAK3 Knockout Cell Lines and Measurement of Cytokine Levels
4.10. Prediction of Protein Associations and Protein—GTP Docking
4.11. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Medzhitov, R. Origin and physiological roles of inflammation. Nature 2008, 454, 07201. [Google Scholar] [CrossRef]
- Pandey, S.; Kawai, T.; Akira, S. Microbial sensing by Toll-like receptors and intracellular nucleic acid sensors. Cold Spring Harb. Perspect. Biol. 2015, 7, a016246. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, K.; Takeuchi, O.; Kawai, T.; Sanjo, H.; Ogawa, T.; Takeda, Y.; Takeda, K.; Akira, S. Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: Evidence for TLR4 as the LPS gene product. J. Immunol. 1999, 162, 3749–3752. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Lee, J.-O. Recognition of lipopolysaccharide pattern by TLR4 complexes. Exp. Mol. Med. 2013, 45, e66. [Google Scholar] [CrossRef] [PubMed]
- Park, B.S.; Song, D.H.; Kim, H.M.; Choi, B.-S.; Lee, H.; Lee, J.-O. The structural basis of lipopolysaccharide recognition by the TLR4–MD-2 complex. Nature 2009, 458, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Wesche, H.; Gao, X.; Li, X.; Kirschning, C.J.; Stark, G.R.; Cao, Z. IRAK-M s a novel member of the Pelle/Interleukin-1 Receptor-Associated Kinase (IRAK) family. J. Biol. Chem. 1999, 274, 19403–19410. [Google Scholar] [CrossRef] [PubMed]
- Medzhitov, R.; Preston-Hurlburt, P.; Kopp, E.; Stadlen, A.; Chen, C.; Ghosh, S.; Janeway, C.A. MyD88 is an adaptor protein in the HToll/IL-1 receptor family signaling pathways. Mol. Cell. 1998, 2, 253–258. [Google Scholar] [CrossRef]
- Motshwene, P.G.; Moncrieffe, M.C.; Grossmann, J.G.; Kao, C.; Ayaluru, M.; Sandercock, A.M.; Robinson, C.V.; Latz, E.; Gay, N.J. An oligomeric signaling platform formed by the Toll-like receptor signal transducers MyD88 and IRAK-4. J. Biol. Chem. 2009, 284, 25404–25411. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-C.; Lo, Y.-C.; Wu, H. Helical assembly in the MyD88–IRAK4–IRAK2 complex in TLR/IL-1R signalling. Nature 2010, 465, 885–890. [Google Scholar] [CrossRef] [PubMed]
- Sen, R.; Baltimore, D. Inducibility of K immunoglobulin enhancer-binding protein NFκB by a posttranslational mechanism. Cell 1966, 47, 921–926. [Google Scholar] [CrossRef]
- Hambleton, J.; Weinsteint, S.L.; Lemt, L.; Defrancots, A.L. Activation of C-Jun N-terminal kinase in bacterial lipopolysaccharide-stimulated macrophages (c-Jun N-terminal kinase/endotoxin/macrophages). Immunology 1996, 93, 2774–2778. [Google Scholar]
- van der Bruggen, T.; Nijenhuis, S.; van Raaij, E.; Verhoef, J.; Sweder Van Asbeck, A.B. Lipopolysaccharide-induced tumor necrosis factor alpha production by human monocytes involves the Raf-1/MEK1-MEK2/ERK1-ERK2 Pathway. Infect. Immun. 1999, 67, 3824–3829. [Google Scholar] [CrossRef] [PubMed]
- Casey, J.R.; Petranka, J.G.; Kottra, J.; Fleenor, D.E.; Rosse, W.F. The structure of the urokinase-type plasminogen activator receptor gene. Blood 1994, 84, 1151–1156. [Google Scholar] [CrossRef] [PubMed]
- Fitzgerald, K.A.; Kagan, J.C. Toll-like receptors and the control of immunity. Cell 2020, 180, 1044–1066. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Kim, T.W.; Qin, J.; Jiang, Z.; Qian, Y.; Xiao, H.; Lu, Y.; Qian, W.; Gulen, M.F.; Sizemore, N.; et al. Interleukin-1 (IL-1)-induced TAK1-dependent versus MEKK3-dependent NFκB activation pathways bifurcate at IL-1 receptor-associated kinase modification. J. Biol. Chem. 2007, 282, 6075–6089. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Xiong, Y.; Li, Q.; Yang, H. Inhibition of Toll-like receptor signaling as a promising therapy for inflammatory diseases: A journey from molecular to nano therapeutics. Front. Physiol. 2017, 8, 508. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, K.; Hernandez, L.D.; Galan, J.E.; Janeway, C.A.; Medzhitov, R.; Flavell, R.A. IRAK-M is a negative regulator of Toll-like receptor signaling. Cell 2002, 110, 191–202. [Google Scholar] [CrossRef]
- Liu, Z.-J.; Yan, L.-N.; Li, X.-H.; Xu, F.-L.; Chen, X.-F.; You, H.-B.; Gong, J.-P. Up-regulation of IRAK-M is essential for endotoxin tolerance induced by a low dose of lipopolysaccharide in Kupffer cells. J. Surg. Res. 2008, 150, 34–39. [Google Scholar] [CrossRef]
- Zhou, H.; Yu, M.; Fukuda, K.; Im, J.; Yao, P.; Cui, W.; Bulek, K.; Zepp, J.; Wan, Y.; Kim, T.W.; et al. IRAK-M mediates Toll-like receptor/IL-1R-induced NFκB activation and cytokine production. EMBO J. 2013, 32, 583–596. [Google Scholar] [CrossRef]
- Su, J.; Zhang, T.; Tyson, J.; Li, L. Innate immunity the interleukin-1 receptor-associated kinase M selectively inhibits the alternative, instead of the classical NFκB pathway. J. Innate Immun. 2009, 1, 164–174. [Google Scholar] [CrossRef]
- Kollewe, C.; Mackensen, A.-C.; Neumann, D.; Knop, J.; Cao, P.; Li, S.; Wesche, H.; Martin, M.U. Sequential autophosphorylation steps in the interleukin-1 receptor-associated kinase-1 regulate its availability as an adapter in interleukin-1 signaling. J. Biol. Chem. 2004, 279, 5227–5236. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Arron, J.R.; Lamothe, B.; Cirilli, M.; Kobayashi, T.; Shevde, N.K.; Segal, D.; Dzivenu, O.K.; Vologodskaia, M.; Yim, M.; et al. Distinct molecular mechanism for initiating TRAF6 signalling. Nature 2002, 418, 443–447. [Google Scholar] [CrossRef] [PubMed]
- Geng, D.; Ciavattone, N.; Lasola, J.J.; Shrestha, R.; Sanchez, A.; Guo, J.; Vlk, A.; Younis, R.; Wang, L.; Brown, A.J.; et al. Induction of IRAK-M in melanoma induces caspase-3 dependent apoptosis by reducing TRAF6 and calpastatin levels. Commun. Biol. 2020, 3, 306. [Google Scholar] [CrossRef]
- Turek, I.; Irving, H. Moonlighting proteins shine new light on molecular signaling niches. Int. J. Mol. Sci. 2021, 22, 1367. [Google Scholar] [CrossRef] [PubMed]
- Freihat, L.; Muleya, V.; Manallack, D.T.; Wheeler, J.I.; Irving, H.R. Comparison of moonlighting guanylate cyclases: Roles in signal direction? Biochem. Soc. Trans. 2014, 42, 1773–1779. [Google Scholar] [CrossRef] [PubMed]
- Freihat, L.A.; Wheeler, J.I.; Wong, A.; Turek, I.; Manallack, D.T.; Irving, H.R. IRAK3 modulates downstream innate immune signalling through its guanylate cyclase activity. Sci. Rep. 2019, 9, 15468. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Axell, A.; Turek, I.; Wright, B.; Meehan-Andrews, T.; Irving, H.R. Modulation of inflammatory cytokine production in human monocytes by cGMP and IRAK3. Int. J. Mol. Sci. 2022, 23, 2552. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.H.; Turek, I.; Meehan-Andrews, T.; Zacharias, A.; Irving, H. Analysis of interleukin-1 receptor associated kinase-3 (IRAK3) function in modulating expression of inflammatory markers in cell culture models: A systematic review and meta-analysis. PLoS ONE 2020, 15, e0244570. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Turek, I.; Meehan-Andrews, T.; Zacharias, A.; Irving, H.R. A systematic review and meta-analyses of interleukin-1 receptor associated kinase 3 (IRAK3) action on inflammation in in vivo models for the study of sepsis. PLoS ONE 2022, 17, e0263968. [Google Scholar] [CrossRef]
- Hulsmans, M.; Geeraert, B.; De Keyzer, D.; Mertens, A.; Lannoo, M.; Vanaudenaerde, B.; Hoylaerts, M.; Benhabilès, N.; Tsatsanis, C.; Mathieu, C.; et al. Interleukin-1 Receptor-Associated Kinase-3 Is a Key Inhibitor of Inflammation in Obesity and Metabolic Syndrome. PLoS ONE 2012, 7, e30414. [Google Scholar] [CrossRef]
- da Silva, I.I.F.G.; Barbosa, A.D.; Souto, F.O.; Maia, M.D.M.D.; Crovella, S.; de Souza, P.R.E.; Sandrin-Garcia, P. MYD88, IRAK3 and Rheumatoid Arthritis pathogenesis: Analysis of differential gene expression in CD14+ monocytes and the inflammatory cytokine levels. Immunobiology 2021, 226, 152152. [Google Scholar] [CrossRef] [PubMed]
- del Fresno, C.; Otero, K.; Gómez-García, L.; González-León, M.C.; Soler-Ranger, L.; Fuentes-Prior, P.; Escoll, P.; Baos, R.; Caveda, L.; García, F.; et al. Tumor Cells Deactivate Human Monocytes by Up-Regulating IL-1 Receptor Associated Kinase-M Expression via CD44 and TLR4. J. Immunol. 2005, 174, 3032–3040. [Google Scholar] [CrossRef]
- Tunalı, G.; Rúbies Bedós, M.; Nagarajan, D.; Fridh, P.; Papakyriacou, I.; Mao, Y. Interleukin-1 receptor-associated kinase-3 acts as an immune checkpoint in myeloid cells to limit cancer immunotherapy. J. Clin. Investig. 2023, 133, e161084. [Google Scholar] [CrossRef] [PubMed]
- Shen, P.; Li, Q.; Ma, J.; Tian, M.; Hong, F.; Zhai, X.; Li, J.; Huang, H.; Shi, C. IRAK-M alters the polarity of macrophages to facilitate the survival of Mycobacterium tuberculosis. BMC Microbiol. 2017, 17, 185. [Google Scholar] [CrossRef]
- Pantazi, I.; Al-Qahtani, A.A.; Alhamlan, F.S.; Alothaid, H.; Matou-Nasri, S.; Sourvinos, G.; Vergadi, E.; Tsatsanis, C. SARS-CoV-2/ACE2 interaction suppresses IRAK-M expression and promotes pro-inflammatory cytokine production in macrophages. Front. Immunol. 2021, 12, 683800. [Google Scholar] [CrossRef] [PubMed]
- Ko, Y.; Chan, Y.-H.; Liu, C.-H.; Liang, J.-J.; Chuang, T.-H.; Hsueh, Y.-P.; Lin, Y.-L.; Lin, K.-I. Blimp-1-mediated pathway promotes type I IFN production in plasmacytoid dendritic cells by targeting to interleukin-1 receptor-associated kinase M. Front. Immunol. 2018, 9, 1828. [Google Scholar] [CrossRef] [PubMed]
- Lange, S.M.; Nelen, M.I.; Cohen, P.; Kulathu, Y. Dimeric structure of the pseudokinase IRAK3 suggests an allosteric mechanism for negative regulation. Structure 2021, 29, 238–251.e4. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.; Chan, S.L.; Chan, S.S.L.; Fiscus, R.R.; Tsang, B.K. Regulation of p53 and suppression of apoptosis by the soluble guanylyl cyclase/cGMP pathway in human ovarian cancer cells. Oncogene 2006, 25, 2203–2212. [Google Scholar] [CrossRef] [PubMed]
- Piazza, G.A.; Chen, X.; Ward, A.; Coley, A.; Zhou, G.; Buchsbaum, D.J.; Maxuitenko, Y.; Keeton, A.B. Targeting cGMP/PKG signaling for the treatment or prevention of colorectal cancer with novel sulindac derivatives lacking cyclooxygenase inhibitory activity. Oncol. Signal. 2020, 3, 1–6. [Google Scholar] [CrossRef]
- Gilmore, T.D. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein V-Rel. Oncogene 1999, 18, 6925–6937. [Google Scholar] [CrossRef]
- Karin, M. NFκB as a critical link between inflammation and cancer. Cold Spring Harb. Perspect. Biol. 2009, 1, a000141. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Zhang, L.; Joo, D.; Sun, S.-C. NF-κB signaling in inflammation. Signal. Transduct. Target. Ther. 2017, 2, e17023. [Google Scholar] [CrossRef] [PubMed]
- Du, J.; Nicolaes, G.A.F.; Kruijswijk, D.; Versloot, M.; van der Poll, T.; van’t Veer, C. The structure function of the death domain of human IRAK-M. Cell. Commun. Signal. 2014, 12, 77. [Google Scholar] [CrossRef] [PubMed]
- Nechama, M.; Kwon, J.; Wei, S.; Kyi, A.T.; Welner, R.S.; Ben-Dov, I.Z.; Arredouani, M.S.; Asara, J.M.; Chen, C.H.; Tsai, C.Y.; et al. The IL-33-PIN1-IRAK-M axis is critical for type 2 immunity in IL-33-induced allergic airway inflammation. Nat. Commun. 2018, 9, 1603. [Google Scholar] [CrossRef]
- Liu, W.; Zhou, H.; Wang, H.; Zhang, Q.; Zhang, R.; Willard, B.; Liu, C.; Kang, Z.; Li, X.; Li, X. IL-1R-IRAKM-Slc25a1 signaling axis reprograms lipogenesis in adipocytes to promote diet-induced obesity in mice. Nat. Commun. 2022, 13, 2748. [Google Scholar] [CrossRef]
- DeFelice, M.M.; Clark, H.R.; Hughey, J.J.; Maayan, I.; Kudo, T.; Gutschow, M.V.; Covert, M.W.; Regot, S. NF-κB signaling dynamics is controlled by a dose-sensing autoregulatory loop. Sci. Signal. 2019, 12, eaau3568. [Google Scholar] [CrossRef]
- Rushworth, L.K.; Hindley, A.D.; Neill, E.O.; Kolch, W. Regulation and role of Raf-1/B-Raf heterodimerization. Mol. Cell. Biol. 2006, 26, 2262–2272. [Google Scholar] [CrossRef]
- Brooks, A.J.; Dai, W.; O’mara, M.L.; Abankwa, D.; Chhabra, Y.; Pelekanos, R.A.; Gardon, O.; Tunny, K.A.; Blucher, K.M.; Morton, C.J.; et al. Mechanism of Activation of Protein Kinase JAK2 by the Growth Hormone Receptor. Science 2014, 344, 1249783. [Google Scholar] [CrossRef] [PubMed]
- Oliver, M.R.; Horne, C.R.; Shrestha, S.; Keown, J.R.; Liang, L.-Y.; Young, S.N.; Sandow, J.J.; Webb, A.I.; Goldstone, D.C.; Lucet, I.S.; et al. Granulovirus PK-1 kinase activity relies on a side-to-side dimerization mode centered on the regulatory αC helix. Nat. Commun. 2021, 12, 1002. [Google Scholar] [CrossRef] [PubMed]
- Mace, P.D.; Murphy, J.M. There’s more to death than life: Noncatalytic functions in kinase and pseudokinase signaling. J. Biol. Chem. 2021, 296, 100705. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, H.; Nolan, A.; Naveed, B.; Hoshino, Y.; Segal, L.N.; Fujita, Y.; Rom, W.N.; Weiden, M.D. Neutrophils activate alveolar macrophages by producing caspase-6−mediated cleavage of IL-1 receptor-associated kinase-M. J. Immunol. 2010, 186, 403–410. [Google Scholar] [CrossRef] [PubMed]
- Su, J.; Xie, Q.; Wilson, I.; Li, L. Differential regulation and role of interleukin-1 receptor associated kinase-M in innate immunity signaling. Cell. Signal. 2007, 19, 1596–1601. [Google Scholar] [CrossRef] [PubMed]
- Lyn-Kew, K.; Rich, E.; Zeng, X.; Wen, H.; Kunkel, S.L.; Newstead, M.W.; Bhan, U.; Standiford, T.J. IRAK-M regulates chromatin remodeling in lung macrophages during experimental sepsis. PLoS ONE 2010, 5, e11145. [Google Scholar] [CrossRef] [PubMed]
- Lyroni, K.; Patsalos, A.; Daskalaki, M.G.; Doxaki, C.; Soennichsen, B.; Helms, M.; Liapis, I.; Zacharioudaki, V.; Kampranis, S.C.; Tsatsanis, C. Epigenetic and transcriptional regulation of IRAK-M expression in macrophages. J. Immunol. 2017, 198, 1297–1307. [Google Scholar] [CrossRef]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acid. Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Berman, H.M.; Westbrook, J.; Feng, Z.; Gilliland, G.; Bhat, T.N.; Weissig, H.; Shindyalov, I.N.; Bourne, P.E. The Protein Data Bank. Nucleic Acid. Res. 2000, 28, 235–242. [Google Scholar] [CrossRef]
- Desta, I.; Porter, K.A.; Xia, B.; Kozakov, D.; Vajda, S. Performance and its limits in rigid body protein-protein docking. Structure 2020, 28, 1071–1081. [Google Scholar] [CrossRef]
- Grosdidier, A.; Zoete, V.; Michielin, O. SwissDock, a protein-small molecule docking web service based on EADock DSS. Nucleic Acid. Res. 2011, 39, 270–277. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Turek, I.; Nguyen, T.H.; Galea, C.; Abad, I.; Freihat, L.; Manallack, D.T.; Velkov, T.; Irving, H. Mutations in the Vicinity of the IRAK3 Guanylate Cyclase Center Impact Its Subcellular Localization and Ability to Modulate Inflammatory Signaling in Immortalized Cell Lines. Int. J. Mol. Sci. 2023, 24, 8572. https://doi.org/10.3390/ijms24108572
Turek I, Nguyen TH, Galea C, Abad I, Freihat L, Manallack DT, Velkov T, Irving H. Mutations in the Vicinity of the IRAK3 Guanylate Cyclase Center Impact Its Subcellular Localization and Ability to Modulate Inflammatory Signaling in Immortalized Cell Lines. International Journal of Molecular Sciences. 2023; 24(10):8572. https://doi.org/10.3390/ijms24108572
Chicago/Turabian StyleTurek, Ilona, Trang H. Nguyen, Charles Galea, Isaiah Abad, Lubna Freihat, David T. Manallack, Tony Velkov, and Helen Irving. 2023. "Mutations in the Vicinity of the IRAK3 Guanylate Cyclase Center Impact Its Subcellular Localization and Ability to Modulate Inflammatory Signaling in Immortalized Cell Lines" International Journal of Molecular Sciences 24, no. 10: 8572. https://doi.org/10.3390/ijms24108572
APA StyleTurek, I., Nguyen, T. H., Galea, C., Abad, I., Freihat, L., Manallack, D. T., Velkov, T., & Irving, H. (2023). Mutations in the Vicinity of the IRAK3 Guanylate Cyclase Center Impact Its Subcellular Localization and Ability to Modulate Inflammatory Signaling in Immortalized Cell Lines. International Journal of Molecular Sciences, 24(10), 8572. https://doi.org/10.3390/ijms24108572







