Shedding Light on Osteosarcoma Cell Differentiation: Impact on Biomineralization and Mitochondria Morphology
Abstract
1. Introduction
2. Results
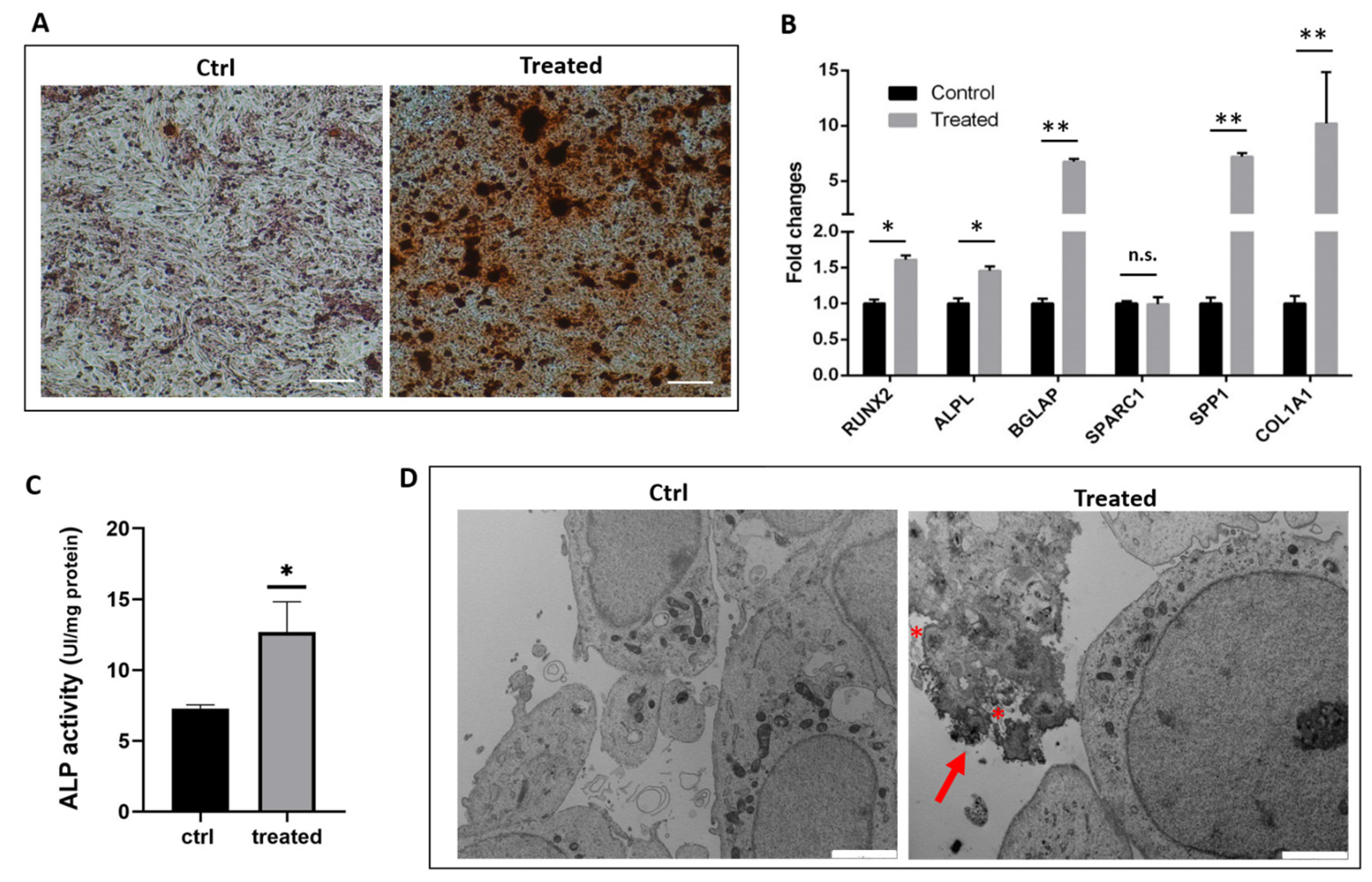
2.1. Gene Expression Analysis and Functional Characterization of SaOS-2 Cells after Osteogenic Induction
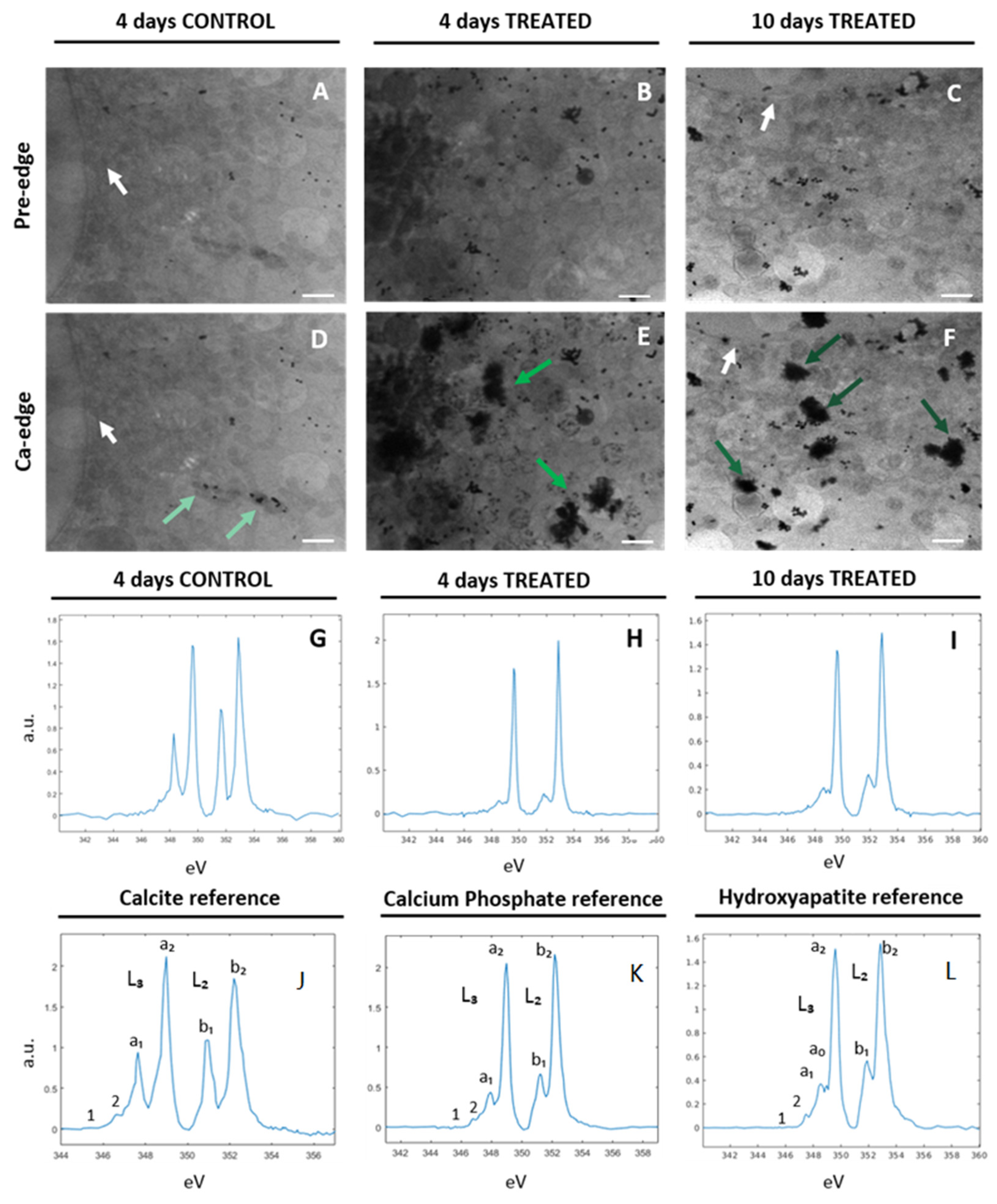
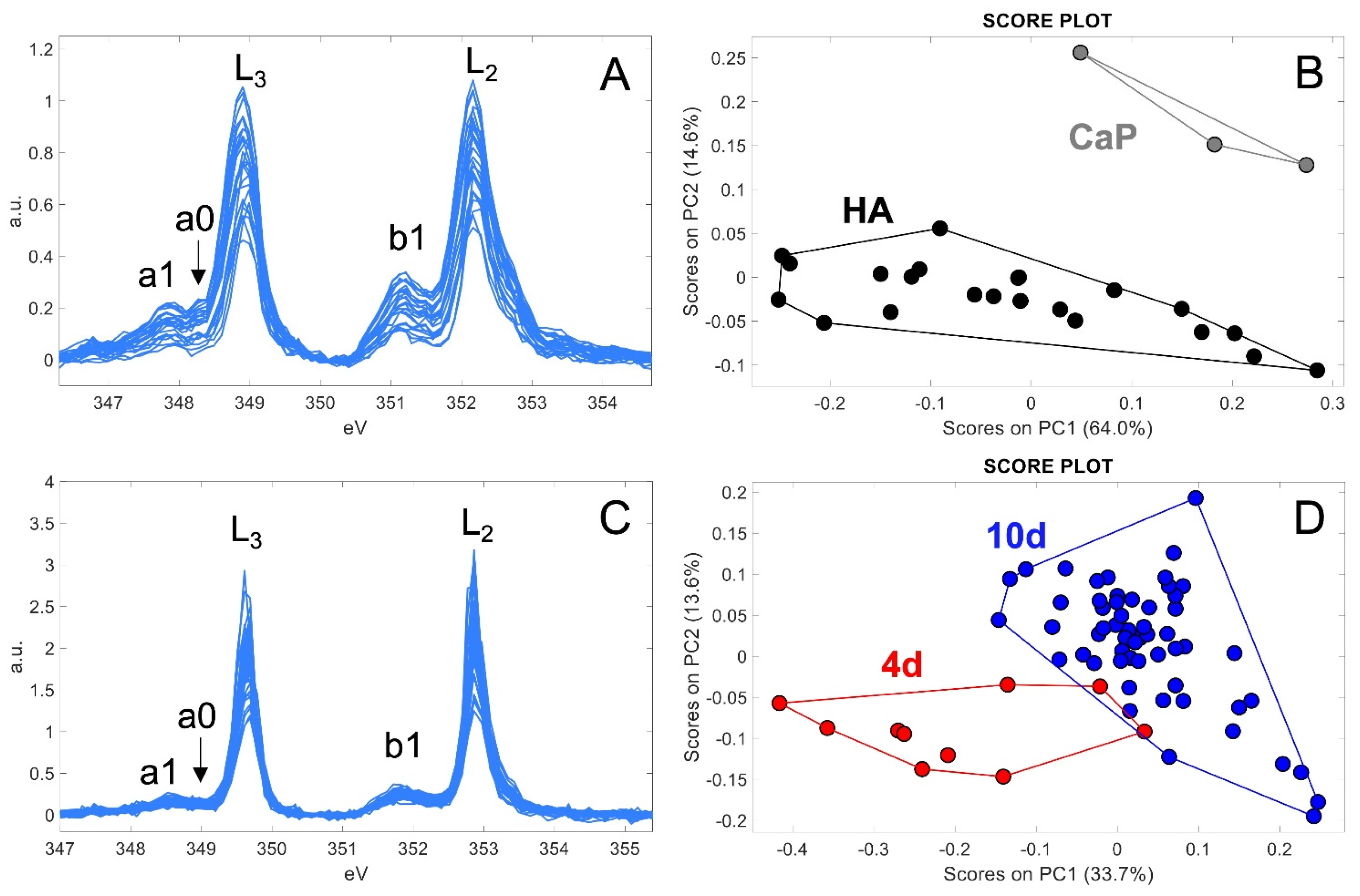
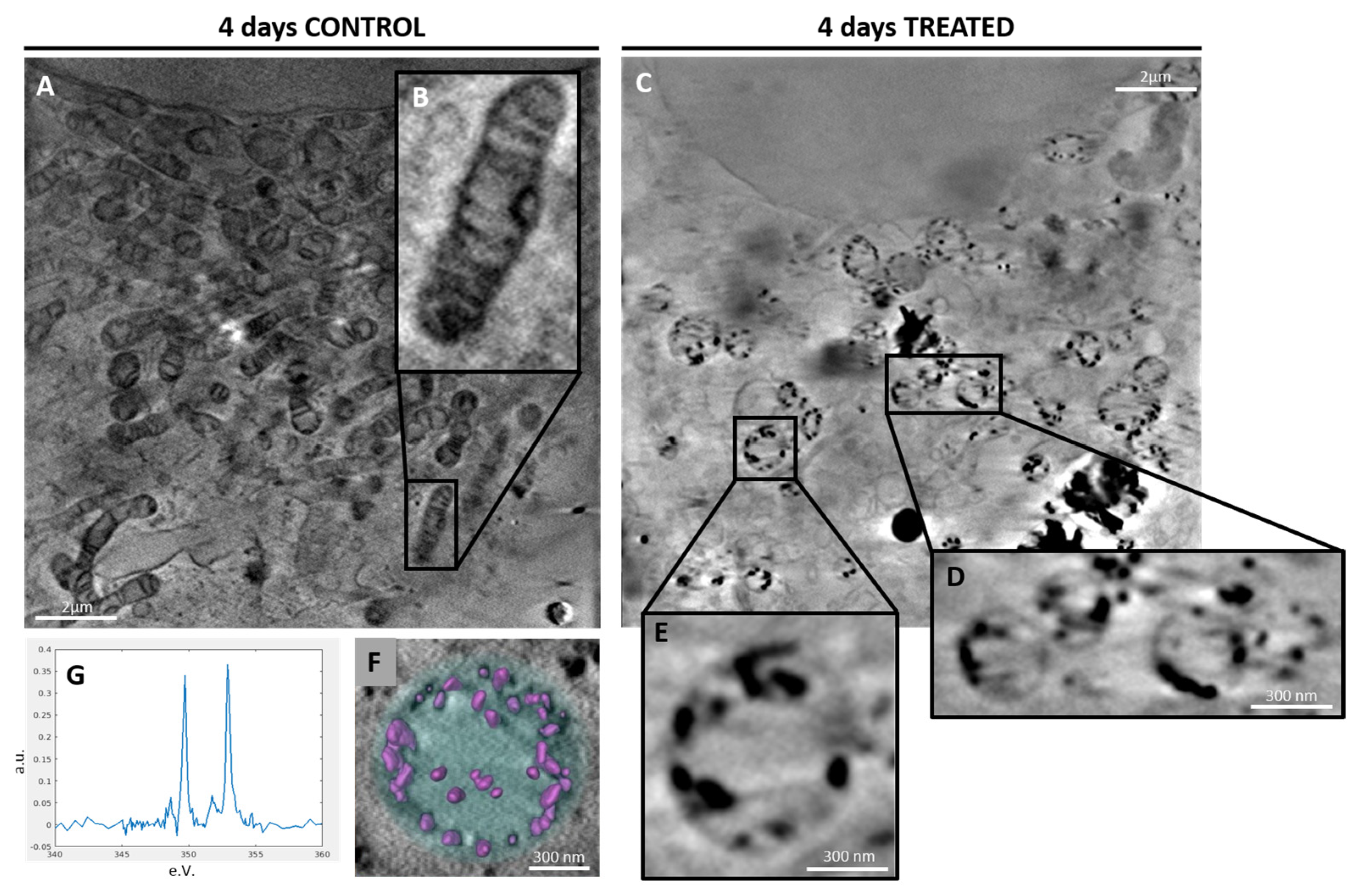
2.2. Characterization of Ca-Depositions by Cryo-XANES and Cryo-SXT
2.2.1. Ca Distribution and Chemical Fingerprint of Ca Depositions
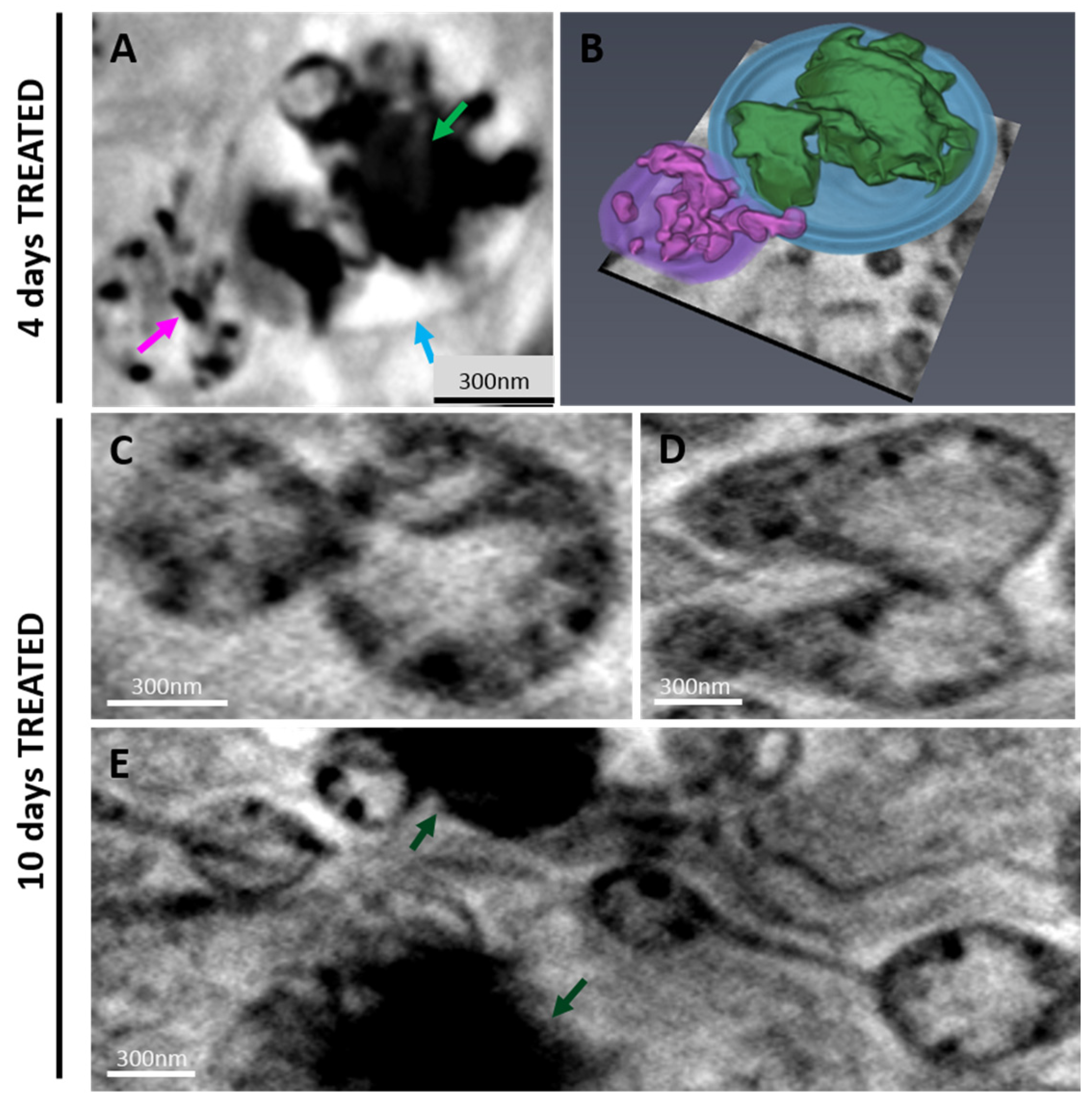
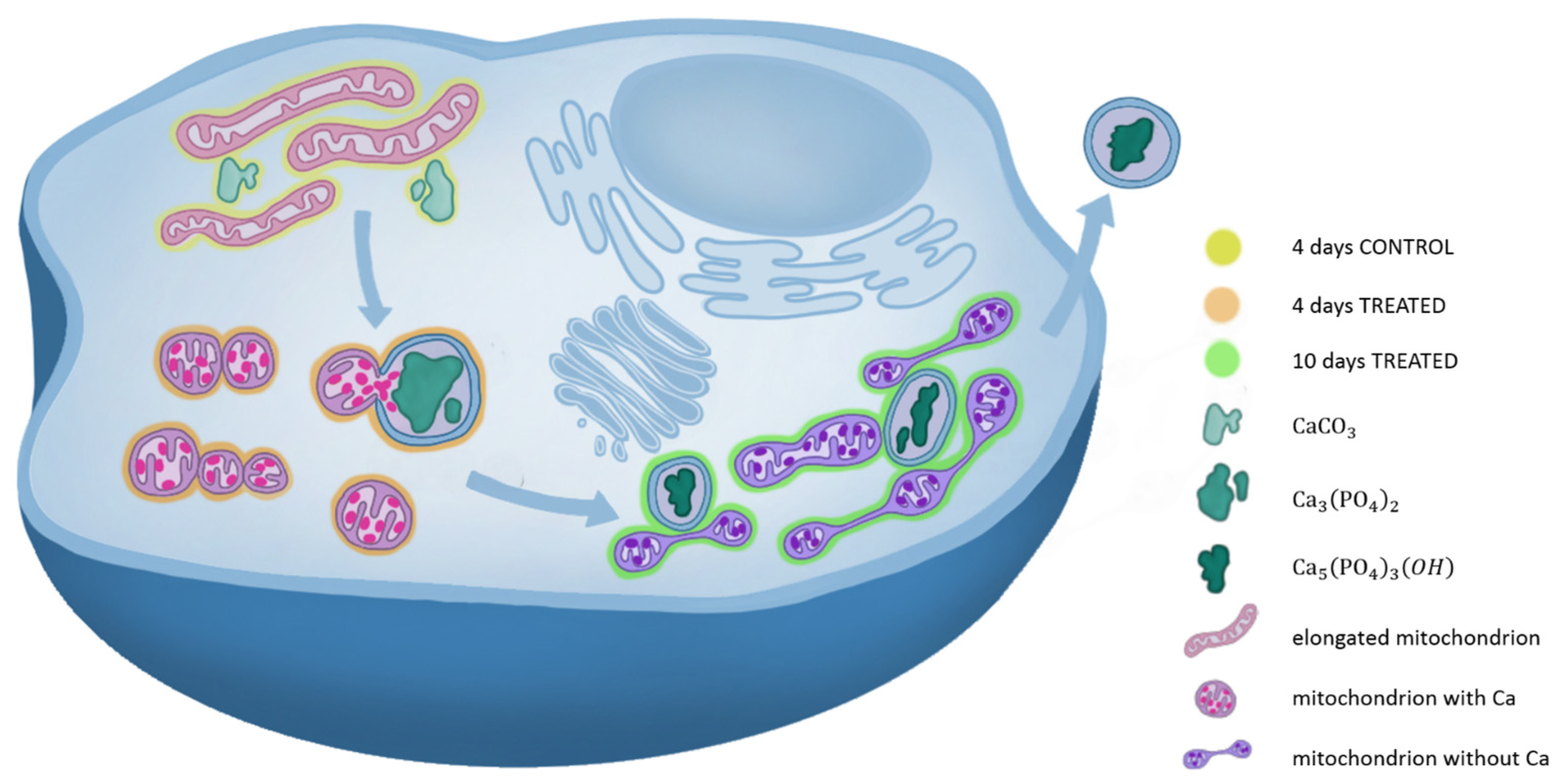
2.2.2. 3D Cell Ultrastructure Analysis
3. Discussion
4. Materials and Methods
4.1. SaOS-2 Cell Culture and Osteogenic Induction
4.2. Alizarin Red Staining
4.3. Alkaline Phosphatase Activity Assay
4.4. Gene Expression Analysis
4.5. Transmission Electron Microscopy (TEM)
4.6. X-Ray Absorption Near-Edge Spectro-Microscopy (XANES-Imaging) and Cryo-Soft-X-Ray Tomography (Cryo-SXT)
4.7. Principal Components Analysis (PCA)
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Brown, H.K.; Schiavone, K.; Gouin, F.; Heymann, M.F.; Heymann, D. Biology of Bone Sarcomas and New Therapeutic Developments. Calcif. Tissue Int. 2018, 102, 174–195. [Google Scholar] [CrossRef]
- Lamplot, J.D.; Denduluri, S.; Qin, J.; Li, R.; Liu, X.; Zhang, H.; Chen, X.; Wang, N.; Pratt, A.; Shui, W.; et al. The Current and Future Therapies for Human Osteosar-coma. Curr. Cancer Ther. Rev. 2013, 9, 55–77. [Google Scholar] [PubMed]
- Yan, G.-N.; Lv, Y.-F.; Guo, Q.-N.; Yan, G.-N.; Lv, Y.-F.; Guo, Q.-N. Advances in osteosarcoma stem cell research and opportunities for novel therapeutic targets. Cancer Lett. 2016, 370, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Cao, J.; Zhang, N.; Yang, B.; He, Q.; Shao, X.; Ying, M. Advances in differentiation therapy for osteosarcoma. Drug. Discov. Today 2020, 25, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Isakoff, M.S.; Bielack, S.S.; Meltzer, P.; Gorlick, R. Osteosarcoma: Current treatment and a collaborative pathway to success. J. Clin. Oncol. 2015, 33, 3029–3035. [Google Scholar] [CrossRef]
- Kansara, M.; Teng, M.W.; Smyth, M.J.; Thomas, D.M. Translational biology of osteosarcoma. Nat. Rev. Cancer 2014, 14, 722–735. [Google Scholar] [CrossRef]
- Wagner, E.R.; He, B.C.; Chen, L.; Zuo, G.W.; Zhang, W.; Shi, Q.; Luo, Q.; Luo, X.; Liu, B.; Luo, J.; et al. Therapeutic implications of PPARγ in human osteo-sarcoma. PPAR Res. 2010, 2010, 11–13. [Google Scholar] [CrossRef]
- Wang, J.; Liu, S.; Shi, J.; Li, J.; Wang, S.; Liu, H.; Zhao, S.; Duan, K.; Pan, X.; Yi, Z. The Role of miRNA in the Diagnosis, Prognosis, and Treatment of Osteosarcoma. Cancer Biother. Radiopharm. 2019, 34, 605–613. [Google Scholar] [CrossRef]
- Beird, H.C.; Bielack, S.S.; Flanagan, A.M.; Gill, J.; Heymann, D.; Janeway, K.A.; Livingston, J.A.; Roberts, R.D.; Strauss, S.J.; Gorlick, R. Osteosarcoma. Nat. Rev. Dis. Prim. 2022, 8, 77. Available online: https://www.nature.com/articles/s41572-022-00409-y (accessed on 20 April 2023). [CrossRef]
- Seo, B.J.; Yoon, S.H.; Do, J.T. Mitochondrial dynamics in stem cells and differentiation. Int. J. Mol. Sci. 2018, 19, 3893. [Google Scholar] [CrossRef]
- Chang, C.R.; Blackstone, C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann. N. Y. Acad. Sci. 2010, 1201, 34–39. [Google Scholar] [CrossRef] [PubMed]
- Farmer, T.; Naslavsky, N.; Caplan, S. Tying trafficking to fusion and fission at the mighty mitochondria. Traffic 2018, 19, 569–577. [Google Scholar] [CrossRef] [PubMed]
- Murshed, M. Mechanism of Bone Mineralization. Cold Spring Harb. Perspect. Med. 2018, 8, a031229. Available online: https://pubmed.ncbi.nlm.nih.gov/29610149/ (accessed on 19 April 2023). [CrossRef] [PubMed]
- Marchegiani, F.; Cibej, E.; Vergni, P.; Tosi, G.; Fermani, S.; Falini, G. Hydroxyapatite synthesis from biogenic calcite single crystals into phosphate solutions at ambient conditions. J. Cryst. Growth 2009, 311, 4219–4225. [Google Scholar] [CrossRef]
- Sorrentino, A.; Malucelli, E.; Rossi, F.; Cappadone, C.; Farruggia, G.; Moscheni, C.; Perez-Berna, A.J.; Conesa, J.J.; Colletti, C.; Roveri, N.; et al. Calcite as a precursor of hydrox-yapatite in the early biomineralization of differentiating human bone-marrow mesenchymal stem cells. Int. J. Mol. Sci. 2021, 22, 4939. [Google Scholar] [CrossRef]
- Procopio, A.; Malucelli, E.; Pacureanu, A.; Cappadone, C.; Farruggia, G.; Sargenti, A.; Castiglioni, S.; Altamura, D.; Sorrentino, A.; Giannini, C.; et al. Chemical Fingerprint of Zn-Hydroxyapatite in the Early Stages of Osteogenic Differentiation. ACS Cent. Sci. 2019, 5, 1449–1460. Available online: https://pubmed.ncbi.nlm.nih.gov/31482128/ (accessed on 19 April 2023). [CrossRef]
- Boonrungsiman, S.; Gentleman, E.; Carzaniga, R.; Evans, N.D.; McComb, D.W.; Porter, A.E.; Stevens, M.M. The role of intracellular calcium phosphate in osteoblast-mediated bone apatite formation. Proc. Natl. Acad. Sci. USA 2012, 109, 14170–14175. [Google Scholar] [CrossRef]
- Tang, C.; Wei, Y.; Gu, L.; Zhang, Q.; Li, M.; Yuan, G.; He, Y.; Huang, L.; Liu, Y.; Zhang, Y. Biomineral Precursor Formation Is Initiated by Transporting Calcium and Phosphorus Clusters from the Endoplasmic Reticulum to Mitochondria. Adv. Sci. 2020, 7, 1902536. Available online: https://pubmed.ncbi.nlm.nih.gov/32328413/ (accessed on 19 April 2023). [CrossRef]
- Murray, E.; Provvedini, D.; Curran, D.; Catherwood, B.; Sussman, H.; Manolagas, S. Characterization of a human osteo-blastic osteosarcoma cell line (SAOS-2) with high bone alkaline phosphatase activity. J. Bone Miner. Res. 1987, 2, 231–238. Available online: https://pubmed.ncbi.nlm.nih.gov/2843003/ (accessed on 19 April 2023). [CrossRef]
- McDermott, G.; Le Gros, M.A.; Knoechel, C.G.; Uchida, M.; Larabell, C.A. Soft X-ray tomography and cryogenic light mi-croscopy: The cool combination in cellular imaging. Trends Cell Biol. 2009, 19, 587–595. Available online: https://pubmed.ncbi.nlm.nih.gov/19818625/ (accessed on 19 April 2023). [CrossRef]
- Sorrentino, A.; Nicolas, J.; Valcárcel-Fernández, R.; Chichón, F.J.; Rosanes, M.; Avila, J.; Tkachuk, A.; Irwin, J.; Ferrer, S.; Pereiro, E. MISTRAL: A transmission soft X-ray microscopy beamline for cryo nano-tomography of biological samples and magnetic domains imaging. J. Synchrotron Radiat. 2015, 22, 1112–1117. [Google Scholar] [CrossRef] [PubMed]
- De Groot, F.M.F.; Fuggle, J.C.; Thole, B.T.; Sawatzky, G.A. L2,3 X-ray-absorption edges of d0 compounds: K+, Ca2+, Sc3+, and Ti4+ in Oh (octahedral) symmetry. Phys. Rev. B 1990, 41, 928–937. Available online: https://pubmed.ncbi.nlm.nih.gov/9993787/ (accessed on 19 April 2023). [CrossRef] [PubMed]
- Cosmidis, J.; Benzerara, K.; Nassif, N.; Tyliszczak, T.; Bourdelle, F. Characterization of Ca-phosphate biological materi-als by scanning transmission X-ray microscopy (STXM) at the Ca L2,3−, P L2,3− and C K-edges. Acta Biomater. 2015, 12, 260–269. Available online: https://pubmed.ncbi.nlm.nih.gov/25305511/ (accessed on 19 April 2023). [CrossRef] [PubMed]
- Bravo Sagua, R.; Parra, V.; López-Crisosto, C.; Quest, A.F.G.; Díaz, P.; Lavandero, S. Calcium Transport and Signaling in Mitochondria. Compr. Physiol. 2017, 7, 623–634. Available online: https://www.researchgate.net/publication/315364457 (accessed on 20 April 2023). [PubMed]
- Gong, Y.U.T.; Killian, C.E.; Olson, I.C.; Appathurai, N.P.; Amasino, A.L.; Martin, M.C.; Holt, L.J.; Wilt, F.H.; Gilbert, P.U.P.A. Phase transitions in biogenic amorphous calcium carbonate. Proc. Natl. Acad. Sci. USA 2012, 109, 6088–6093. [Google Scholar] [CrossRef]
- Rodan, S.B.; Imai, Y.; Thiede, M.A.; Wesolowski, G.; Thompson, D.; Bar-Shavit, Z.; Shull, S.; Mann, K.; Rodan, G.A. Characterization of a Human Osteosarcoma Cell Line (Saos-2) with Osteoblastic Properties. Am. Assoc. Cancer Res. 1987, 47, 4961–4966. Available online: https://aacrjournals.org/cancerres/article/47/18/4961/491558/Characterization-of-a-Human-Osteosarcoma-Cell-Line (accessed on 20 April 2023).
- Zhang, H.; Wang, T.; Gong, H.; Jiang, R.; Zhou, W.; Sun, H.; Huang, R.; Wang, Y.; Wu, Z.; Xu, W.; et al. A novel molecular classification method for osteosar-coma based on tumor cell differentiation trajectories. Bone Res. 2023, 11, 1. Available online: https://www.nature.com/articles/s41413-022-00233-w (accessed on 21 April 2023). [CrossRef]
- Li, Q.; Gao, Z.; Chen, Y.; Guan, M.X. The role of mitochondria in osteogenic, adipogenic and chondrogenic differentiation of mesenchymal stem cells. Protein Cell 2017, 8, 439–445. [Google Scholar] [CrossRef]
- Fernández-Barral, A.; Bustamante-Madrid, P.; Ferrer-Mayorga, G.; Barbáchano, A.; Larriba, M.J.; Muñoz, A. Vitamin D effects on cell differentiation and stemness in cancer. Cancers 2020, 12, 2413. [Google Scholar] [CrossRef]
- Sherman, M.H.; Ruth, T.Y.; Engle, D.D.; Ding, N.; Atkins, A.R.; Tiriac, H.; Collisson, E.A.; Connor, F.; Van Dyke, T.; Kozlov, S.; et al. Vitamin D Receptor-Mediated Stromal Repro-gramming Suppresses Pancreatitis and Enhances Pancreatic Cancer Therapy. Cell 2014, 159, 80–93. [Google Scholar] [CrossRef]
- Rowley, D.R. Reprogramming the tumor stroma: A new paradigm. Cancer Cell 2014, 26, 451–452. Available online: https://pubmed.ncbi.nlm.nih.gov/25314074/ (accessed on 21 April 2023). [CrossRef] [PubMed]
- Pasini, A.; Lovecchio, J.; Ferretti, G.; Giordano, E. Medium perfusion flow improves osteogenic commitment of human stromal cells. Stem Cells Int. 2019, 2019, 1304194. [Google Scholar] [CrossRef] [PubMed]
- Penner-Hahn, J.E. X-ray Absorption Spectroscopy. Compr. Coord. Chem. II 2003, 2, 159–186. [Google Scholar]
- Sorrentino, A.; Simonelli, L.; Kazzazi, A.; Laszczynski, N.; Birrozzi, A.; Mullaliu, A.; Pereiro, E.; Passerini, S.; Giorgetti, M.; Tonti, D. Soft X-ray Transmission Micros-copy on Lithium-Rich Layered-Oxide Cathode Materials. Appl. Sci. 2021, 11, 2791. Available online: https://www.mdpi.com/2076-3417/11/6/2791/htm (accessed on 19 April 2023). [CrossRef]
- MessaoudiI, C.; Boudier, T.; Sorzano, C.O.S.; Marco, S. TomoJ: Tomography software for three-dimensional reconstruc-tion in transmission electron microscopy. BMC Bioinform. 2007, 8, 288. [Google Scholar] [CrossRef]
- Sciutto, G.; Oliveri, P.; Prati, S.; Quaranta, M.; Bersani, S.; Mazzeo, R. An advanced multivariate approach for processing X-ray fluorescence spectral and hyperspectral data from non-invasive in situ analyses on painted surfaces. Anal. Chim. Acta 2012, 752, 30–38. Available online: https://pubmed.ncbi.nlm.nih.gov/23101649/ (accessed on 19 April 2023). [CrossRef]
| Gene | Sequence 5′-3′ | |
|---|---|---|
| GAPDH | F | ACAGTTGCCATGTAGACC |
| R | TTGAGCACAGGGTACTTTA | |
| HPRT1 | F | ATAAGCCAGACTTTGTTGG |
| R | ATAGGACTCCAGATGTTTCC | |
| RUNX2 | F | AAGCTTGATGACTCTAAACC |
| R | TCTGTAATCTGACTCTGTCC | |
| ALPL | F | TCTTCACATTTGGTGGATAG |
| R | ATGGAGATATTCTCTCGTTC | |
| SPARC | F | AGTATGTGTAACAGGAGGAC |
| R | AATGTTGCTAGTGTGATTGG | |
| COL1A1 | F | CCAGCAAATGTTCCTTTTTG |
| R | AAAATTCACAAGTCCCCATC | |
| BGLAP | F | TTCTTTCCTCTTCCCCTTG |
| R | CCTCTTCTTGAGTTTATTTGG | |
| SPP1 | F | GACCAAGGAAAACTCACTAC |
| R | CTGTTTAACTGGTATGGCAC | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rossi, F.; Picone, G.; Cappadone, C.; Sorrentino, A.; Columbaro, M.; Farruggia, G.; Catelli, E.; Sciutto, G.; Prati, S.; Oliete, R.; et al. Shedding Light on Osteosarcoma Cell Differentiation: Impact on Biomineralization and Mitochondria Morphology. Int. J. Mol. Sci. 2023, 24, 8559. https://doi.org/10.3390/ijms24108559
Rossi F, Picone G, Cappadone C, Sorrentino A, Columbaro M, Farruggia G, Catelli E, Sciutto G, Prati S, Oliete R, et al. Shedding Light on Osteosarcoma Cell Differentiation: Impact on Biomineralization and Mitochondria Morphology. International Journal of Molecular Sciences. 2023; 24(10):8559. https://doi.org/10.3390/ijms24108559
Chicago/Turabian StyleRossi, Francesca, Giovanna Picone, Concettina Cappadone, Andrea Sorrentino, Marta Columbaro, Giovanna Farruggia, Emilio Catelli, Giorgia Sciutto, Silvia Prati, Robert Oliete, and et al. 2023. "Shedding Light on Osteosarcoma Cell Differentiation: Impact on Biomineralization and Mitochondria Morphology" International Journal of Molecular Sciences 24, no. 10: 8559. https://doi.org/10.3390/ijms24108559
APA StyleRossi, F., Picone, G., Cappadone, C., Sorrentino, A., Columbaro, M., Farruggia, G., Catelli, E., Sciutto, G., Prati, S., Oliete, R., Pasini, A., Pereiro, E., Iotti, S., & Malucelli, E. (2023). Shedding Light on Osteosarcoma Cell Differentiation: Impact on Biomineralization and Mitochondria Morphology. International Journal of Molecular Sciences, 24(10), 8559. https://doi.org/10.3390/ijms24108559






