Development of a Duplex LAMP Assay with Probe-Based Readout for Simultaneous Real-Time Detection of Schistosoma mansoni and Strongyloides spp. -A Laboratory Approach to Point-Of-Care
Abstract
1. Introduction
2. Results
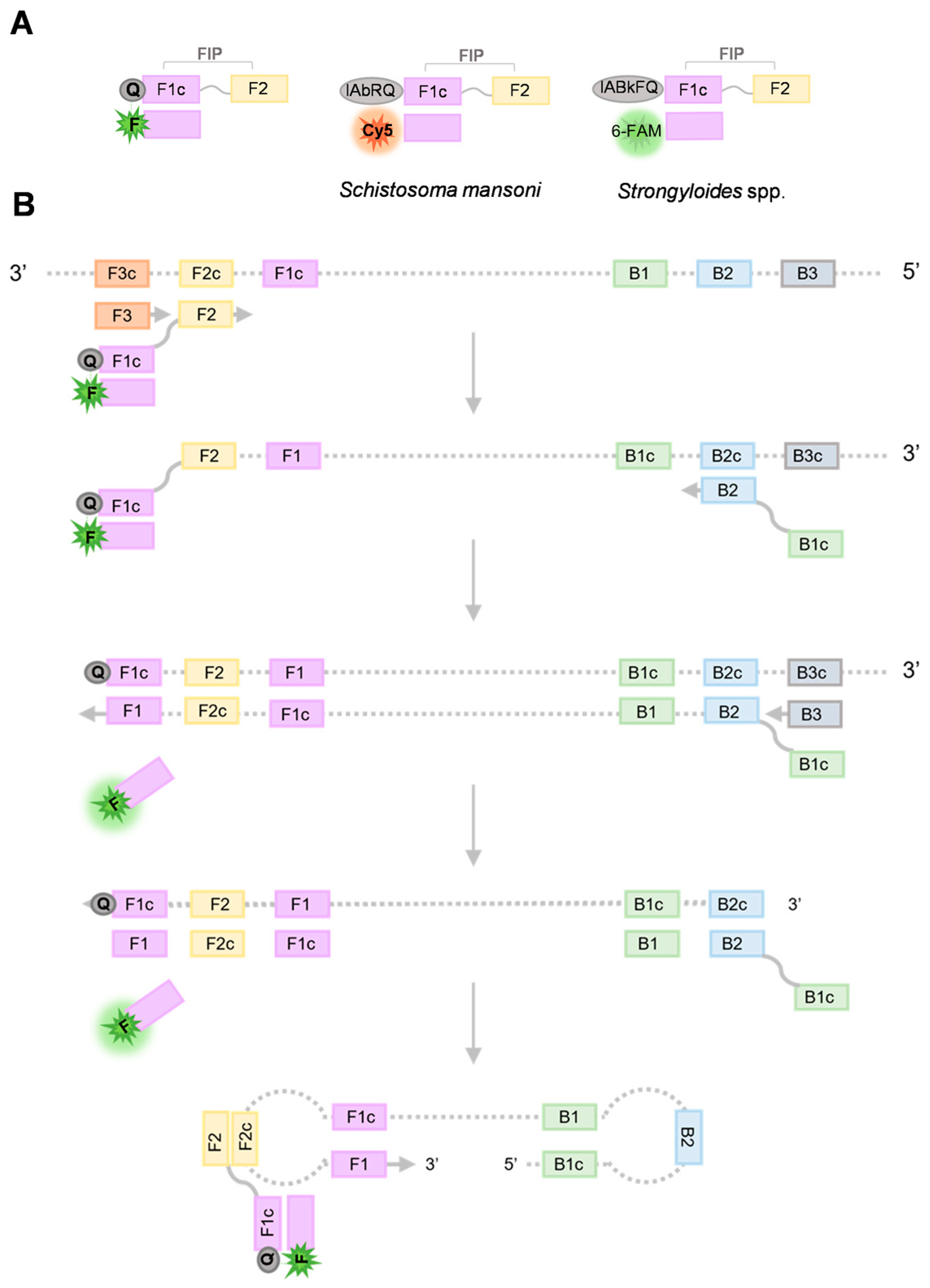
2.1. Setting up and Operation of DARQ-LAMP
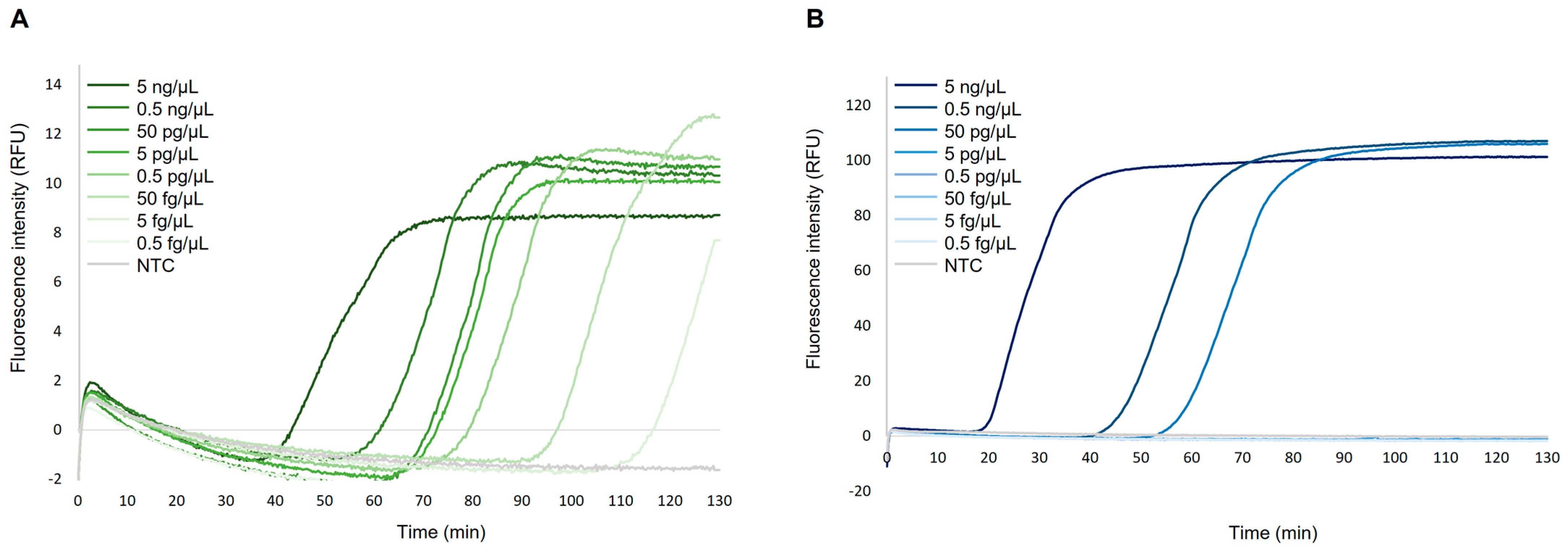
2.2. Sensitivity Assessment of Simplex DARQ-LAMP Assays
2.3. Operation of Duplex DARQ-LAMP Assay
2.4. Sensitivity Assessment of Duplex DARQ-LAMP Assay
2.5. Stability and Functionality over Time of Duplex Dry-DARQ-LAMP
3. Discussion
4. Materials and Methods
4.1. Parasites DNA Standard Samples
4.2. DARQ LAMP Primer Design
4.3. DARQ-LAMP Reactions
4.4. Sensitivity of Simplex and Duplex DARQ-LAMP Reactions
4.5. Stabilization of DARQ-LAMP Reaction Mixes for Room-Temperature Storage: Dry-DARQ-LAMP
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Njiru, Z.K. Loop-mediated isothermal amplification technology: Towards point of care diagnostics. PLoS Negl. Trop. Dis. 2012, 6, e1572. [Google Scholar] [CrossRef]
- Gong, P.; Zhang, T.; Chen, F.; Wang, L.; Jin, S.; Bai, X. Advances in loop-mediated isothermal amplification: Integrated with several point-of-care diagnostic methods. Anal. Methods 2014, 6, 7585–7589. [Google Scholar] [CrossRef]
- Glökler, J.; Lim, T.S.; Ida, J.; Frohme, M. Isothermal amplifications–a comprehensive review on current methods. Crit. Rev. Biochem. Mol. Biol. 2021, 56, 543–586. [Google Scholar] [CrossRef]
- Walker, G.T.; Little, M.C.; Nadeau, J.G.; Shank, D.D. Isothermal in vitro amplification of DNA by a restriction enzyme/DNA polymerase system. Proc. Natl. Acad. Sci. USA 1992, 89, 392–396. [Google Scholar] [CrossRef] [PubMed]
- Vincent, M.; Xu, Y.; Kong, H. Helicase-dependent isothermal DNA amplification. EMBO Rep. 2004, 5, 795–800. [Google Scholar] [CrossRef]
- Ali, M.M.; Li, F.; Zhang, Z.; Zhang, K.; Kang, D.K.; Ankrum, J.A.; Le, X.C.; Zhao, W. Rolling circle amplification: A versatile tool for chemical biology, materials science and medicine. Chem. Soc. Rev. 2014, 43, 3324–3341. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Macdonald, J.; Von Stetten, F. Review: A comprehensive summary of a decade development of the recombinase polymerase amplification. Analyst 2019, 144, 31–67. [Google Scholar] [CrossRef] [PubMed]
- Compton, J. Nucleic acid sequence-based amplificacion. Nature 1991, 350, 91–92. [Google Scholar] [CrossRef]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef]
- Becherer, L.; Borst, N.; Bakheit, M.; Frischmann, S.; Zengerle, R.; von Stetten, F. Loop-mediated isothermal amplification (LAMP)—Review and classification of methods for sequence-specific detection. Anal. Methods 2020, 12, 717–746. [Google Scholar] [CrossRef]
- Parida, M.; Posadas, G.; Inoue, S.; Hasebe, F.; Morita, K. Real-Time Reverse Transcription Loop-Mediated Isothermal Amplification for Rapid Detection of West Nile Virus. J. Clin. Microbiol. 2004, 42, 257–263. [Google Scholar] [CrossRef] [PubMed]
- Rako, L.; Agarwal, A.; Semeraro, L.; Broadley, A.; Rodoni, B.C.; Blacket, M.J. A LAMP (loop-mediated isothermal amplification) test for rapid identification of Khapra beetle (Trogoderma granarium). Pest Manag. Sci. 2021, 77, 5509–5521. [Google Scholar] [CrossRef]
- Lee, S.H.; Ahn, G.; Kim, M.S.; Jeong, O.C.; Lee, J.H.; Kwon, H.G.; Kim, Y.H.; Ahn, J.Y. Poly-adenine-Coupled LAMP Barcoding to Detect Apple Scar Skin Viroid. ACS Comb. Sci. 2018, 20, 472–481. [Google Scholar] [CrossRef]
- García-Bernalt, J.; Fernández-Soto, P.; Domínguez-Gil, M.; Belhassen-García, M.; Muñoz, J.L.; Muro, A. A Simple, Affordable, Rapid, Stabilized, Colorimetric, Versatile RT-LAMP Assay to Detect SARS-CoV-2. Diagnostics 2021, 11, 438. [Google Scholar] [CrossRef] [PubMed]
- Kreft, R.; William Costerton, J.; Ehrlich, G.D. PCR is changing clinical diagnostics. Microbe 2013, 8, 15–20. [Google Scholar] [CrossRef]
- Zhang, H.; Xu, Y.; Fohlerova, Z.; Chang, H.; Iliescu, C.; Neuzil, P. LAMP-on-a-chip: Revising microfluidic platforms for loop-mediated DNA amplification. Trends Anal. Chem. 2019, 113, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Augustine, R.; Hasan, A.; Das, S.; Ahmed, R.; Mori, Y.; Notomi, T.; Kevadiya, B.D.; Thakor, A.S. Loop-mediated isothermal amplification (Lamp): A rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology 2020, 9, 182. [Google Scholar] [CrossRef] [PubMed]
- Wong, Y.P.; Othman, S.; Lau, Y.L.; Radu, S.; Chee, H.Y. Loop-mediated isothermal amplification (LAMP): A versatile technique for detection of micro-organisms. J. Appl. Microbiol. 2018, 124, 626–643. [Google Scholar] [CrossRef]
- Goto, M.; Honda, E.; Ogura, A.; Nomoto, A.; Hanaki, K.I. Colorimetric detection of loop-mediated isothermal amplification reaction by using hydroxy naphthol blue. Biotechniques 2009, 46, 167–172. [Google Scholar] [CrossRef]
- Mori, Y.; Kitao, M.; Tomita, N.; Notomi, T. Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 2004, 59, 145–157. [Google Scholar] [CrossRef]
- Oscorbin, I.P.; Belousova, E.A.; Zakabunin, A.I.; Boyarskikh, U.A.; Filipenko, M.L. Comparison of fluorescent intercalating dyes for quantitative loop-mediated isothermal amplification (qLAMP). Biotechniques 2016, 61, 20–25. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Warmt, C.; Henkel, J.; Schrick, L.; Nitsche, A.; Bier, F.F. Lateral flow–based nucleic acid detection of SARS-CoV-2 using enzymatic incorporation of biotin-labeled dUTP for POCT use. Anal. Bioanal. Chem. 2022, 414, 3177–3186. [Google Scholar] [CrossRef] [PubMed]
- Selvam, K.; Najid, M.A.; Khalid, M.; Mohamad, S.; Palaz, F.; Ozsoz, M.; Aziah, I. RT-LAMP CRISPR-Cas12/13-Based SARS-CoV-2 Detection Methods. Diagnostics 2021, 11, 1646. [Google Scholar] [CrossRef] [PubMed]
- Garg, N.; Ahmad, F.J.; Kar, S. Recent advances in loop-mediated isothermal amplification (LAMP) for rapid and efficient detection of pathogens. Curr. Res. Microb. Sci. 2022, 3, 100120. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; Zhang, Y.; Evans, T.C. Simultaneous multiple target detection in real-time loop-mediated isothermal amplification. Biotechniques 2012, 53, 81–89. [Google Scholar] [CrossRef]
- Petney, T.N.; Andrews, R.H. Multiparasite communities in animals and humans: Frequency, structure and pathogenic significance. Int. J. Parasitol. 1998, 28, 377–393. [Google Scholar] [CrossRef]
- Vaumourin, E.; Vourc’h, G.; Gasqui, P.; Vayssier-Taussat, M. The importance of multiparasitism: Examining the consequences of co-infections for human and animal health. Parasites Vectors 2015, 8, 545. [Google Scholar] [CrossRef]
- Cox, F.E.G. Concomitant infections, parasites and immune responses. Parasitology 2001, 122, S23–S38. [Google Scholar] [CrossRef]
- Gazzinelli-Guimaraes, P.H.; Nutman, T.B. Helminth parasites and immune regulation [version 1; peer review: 2 approved]. F1000Research 2018, 7, 1685. [Google Scholar] [CrossRef]
- Gryseels, B. Schistosomiasis. Infect. Dis. Clin. N. Am. 2012, 26, 383–397. [Google Scholar] [CrossRef]
- Colley, D.G.; Bustinduy, A.L.; Secor, W.E.; King, C.H. Human schistosomiasis. Lancet 2014, 383, 2253–2264. [Google Scholar] [CrossRef] [PubMed]
- Schär, F.; Trostdorf, U.; Giardina, F.; Khieu, V.; Muth, S.; Marti, H.; Vounatsou, P.; Odermatt, P. Strongyloides stercoralis: Global Distribution and Risk Factors. PLoS Negl. Trop. Dis. 2013, 7, e2288. [Google Scholar] [CrossRef] [PubMed]
- Buonfrate, D.; Bisanzio, D.; Giorli, G.; Odermatt, P.; Fürst, T.; Greenaway, C.; French, M.; Reithinger, R.; Gobbi, F.; Montresor, A.; et al. The global prevalence of Strongyloides stercoralis infection. Pathogens 2020, 9, 468. [Google Scholar] [CrossRef]
- Da Silva Santos, L.; Wolff, H.; Chappuis, F.; Albajar-Viñas, P.; Vitoria, M.; Tran, N.T.; Baggio, S.; Togni, G.; Vuilleumier, N.; Girardin, F.; et al. Coinfections between persistent parasitic neglected tropical diseases and viral infections among prisoners from Sub-Saharan Africa and Latin America. J. Trop. Med. 2018, 2018, 7218534. [Google Scholar] [CrossRef] [PubMed]
- Agbata, E.N.; Morton, R.L.; Bisoffi, Z.; Bottieau, E.; Greenaway, C.; Biggs, B.A.; Montero, N.; Tran, A.; Rowbotham, N.; Arevalo-Rodriguez, I.; et al. Effectiveness of screening and treatment approaches for schistosomiasis and strongyloidiasis in newly-arrived migrants from endemic countries in the EU/EEA: A systematic review. Int. J. Environ. Res. Public Health 2019, 16, 11. [Google Scholar] [CrossRef]
- Abruzzi, A.; Fried, B. Coinfection of Schistosoma (Trematoda) with Bacteria, Protozoa and Helminths. Adv. Parasitol. 2011, 77, 1–85. [Google Scholar]
- Posey, D.L.; Blackburn, B.G.; Weinberg, M.; Flagg, E.W.; Ortega, L.; Wilson, M.; Secor, W.E.; Sanders-Lewis, K.; Won, K.; Maguire, J.H. High prevalence and presumptive treatment of schistosomiasis and strongyloidiasis among African refugees. Clin. Infect. Dis. 2007, 45, 1310–1315. [Google Scholar] [CrossRef]
- Fernández-Soto, P.; Gandasegui Arahuetes, J.; Sánchez Hernández, A.; López Abán, J.; Vicente Santiago, B.; Muro, A. A Loop-Mediated Isothermal Amplification (LAMP) Assay for Early Detection of Schistosoma mansoni in Stool Samples: A Diagnostic Approach in a Murine Model. PLoS Negl. Trop. Dis. 2014, 8, e3126. [Google Scholar] [CrossRef]
- Fernández-Soto, P.; Sánchez-Hernández, A.; Gandasegui, J.; Bajo Santos, C.; López-Abán, J.; Saugar, J.M.; Rodríguez, E.; Vicente, B.; Muro, A. Strong-LAMP: A LAMP Assay for Strongyloides spp. Detection in Stool and Urine Samples. Towards the Diagnosis of Human Strongyloidiasis Starting from a Rodent Model. PLoS Negl. Trop. Dis. 2016, 10, e0004836. [Google Scholar] [CrossRef]
- Gandasegui, J.; Fernández-Soto, P.; Muro, A.; Simões Barbosa, C.; Lopes de Melo, F.; Loyo, R.; de Souza Gomes, E.C. A field survey using LAMP assay for detection of Schistosoma mansoni in a low-transmission area of schistosomiasis in Umbuzeiro, Brazil: Assessment in human and snail samples. PLoS Negl. Trop. Dis. 2018, 12, e0006314. [Google Scholar] [CrossRef]
- Fernández-Soto, P.; Gandasegui, J.; Rodríguez, C.C.; Pérez-Arellano, J.L.; Crego-Vicente, B.; Diego, J.G.B.; López-Abán, J.; Vicente, B.; Muro, A. Detection of Schistosoma mansoni-derived DNA in human urine samples by loop-mediated isothermal amplification (LAMP). PLoS ONE 2019, 14, e0214125. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Soto, P.; Celis-Giraldo, C.T.; Collar-Fernández, C.; Gorgojo, Ó.; Camargo, M.; Muñoz, J.; Salas-Coronas, J.; Patarroyo, M.A.; Muro, A. Strong -LAMP Assay Based on a Strongyloides spp.-Derived Partial Sequence in the 18S rRNA as Potential Biomarker for Strongyloidiasis Diagnosis in Human Urine Samples. Dis. Markers 2020, 2020, 5265198. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; Evans, T.C. Loop-mediated isothermal amplification for detection of nucleic acids. Curr. Protoc. Mol. Biol. 2014, 105, 1–14. [Google Scholar] [CrossRef]
- Ball, C.S.; Light, Y.K.; Koh, C.Y.; Wheeler, S.S.; Coffey, L.L.; Meagher, R.J. Quenching of Unincorporated Amplification Signal Reporters in Reverse-Transcription Loop-Mediated Isothermal Amplification Enabling Bright, Single-Step, Closed-Tube, and Multiplexed Detection of RNA Viruses. Anal. Chem. 2016, 88, 3562–3568. [Google Scholar] [CrossRef]
- Nanayakkara, I.A.; White, I.M. Demonstration of a quantitative triplex LAMP assay with an improved probe-based readout for the detection of MRSA. Analyst 2019, 144, 3878–3885. [Google Scholar] [CrossRef]
- Curtis, K.A.; Morrison, D.; Rudolph, D.L.; Shankar, A.; Bloomfield, L.S.P.; Switzer, W.M.; Owen, S.M. A multiplexed RT-LAMP assay for detection of group M HIV-1 in plasma or whole blood. J. Virol. Methods 2018, 255, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Yaren, O.; McCarter, J.; Phadke, N.; Bradley, K.M.; Overton, B.; Yang, Z.; Ranade, S.; Patil, K.; Bangale, R.; Benner, S.A. Ultra-rapid detection of SARS-CoV-2 in public workspace environments. PLoS ONE 2021, 16, e0240524. [Google Scholar] [CrossRef]
- García-Bernalt Diego, J.; Fernández-Soto, P.; Crego-Vicente, B.; Alonso-Castrillejo, S.; Febrer-Sendra, B.; Gómez-Sánchez, A.; Vicente, B.; López-Abán, J.; Muro, A. Progress in loop-mediated isothermal amplification assay for detection of Schistosoma mansoni DNA: Towards a ready-to-use test. Sci. Rep. 2019, 9, 14744. [Google Scholar] [CrossRef] [PubMed]




| Species | Target | Primers | Sequence (5 → 3) | Reference |
|---|---|---|---|---|
| Sm | MIT-Sm | F3 | TTATCGTCTATAGTACGGTAGG | Fernández-Soto et al. [38] |
| B3 | ATACTTTAACCCCCACCAA | |||
| BIP | AGAAGTGTTTAACTTGATGAAGGGGAAACAAAACCGAAACCACTA | |||
| FIP | GCCAAGTAGAGACTACAAACATCTTTGGGTAAGGTAGAAAATGTTGT | |||
| FIP* | IAbRQ-GCCAAGTAGAGACTACAAACATCTTTGGGTAAGGTAGAAAATGTTGT | This study | ||
| Probe | AAGATGTTTGTAGTCTCTACTTGGC—Cy5 | |||
| Str spp. | 18S rDNA | F3 | ACACGCTTTTTATACCACATT | Fernández-Soto et al. [39] |
| B3 | GTGGAGCCGTTTATCAGG | |||
| BIP | ATCAACTTTCGATGGTAGGGTATTGCCTATCCGGAGTCGAACC | |||
| FIP | ACCAGATACACATACGGTATGTTTTGGATTTGATGAAACCATTTTTTCG | |||
| FIP* | IAbFQ-ACCAGATACACATACGGTATGTTTTGGATTTGATGAAACCATTTTTTCG | This study | ||
| Probe | AAAACATACCGTATGTGTATCTGGT—6-FAM |
| Device | Channel | λex | λem | Fluorophore | Quencher |
|---|---|---|---|---|---|
| Genie® III | 1 | 470 | 510–560 | 6-FAM (λex 495–λem 520) | IAbFQ (420–620) |
| 2 | 590 | >620 | Cy5 (λex 648–λem 668) | IAbRQ (500–800) | |
| PCR max Eco48 | 1 | 452–486 | 505–545 | 6-FAM (λex 495–λem 520) | IAbFQ (420–620) |
| 2 | 452–486 | 604–644 | |||
| 3 | 542–586 | 562–596 | |||
| 4 | 542–586 | 665–105 | Cy5 (λex 648–λem 668) | IAbRQ (500–800) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Crego-Vicente, B.; Fernández-Soto, P.; García-Bernalt Diego, J.; Febrer-Sendra, B.; Muro, A. Development of a Duplex LAMP Assay with Probe-Based Readout for Simultaneous Real-Time Detection of Schistosoma mansoni and Strongyloides spp. -A Laboratory Approach to Point-Of-Care. Int. J. Mol. Sci. 2023, 24, 893. https://doi.org/10.3390/ijms24010893
Crego-Vicente B, Fernández-Soto P, García-Bernalt Diego J, Febrer-Sendra B, Muro A. Development of a Duplex LAMP Assay with Probe-Based Readout for Simultaneous Real-Time Detection of Schistosoma mansoni and Strongyloides spp. -A Laboratory Approach to Point-Of-Care. International Journal of Molecular Sciences. 2023; 24(1):893. https://doi.org/10.3390/ijms24010893
Chicago/Turabian StyleCrego-Vicente, Beatriz, Pedro Fernández-Soto, Juan García-Bernalt Diego, Begoña Febrer-Sendra, and Antonio Muro. 2023. "Development of a Duplex LAMP Assay with Probe-Based Readout for Simultaneous Real-Time Detection of Schistosoma mansoni and Strongyloides spp. -A Laboratory Approach to Point-Of-Care" International Journal of Molecular Sciences 24, no. 1: 893. https://doi.org/10.3390/ijms24010893
APA StyleCrego-Vicente, B., Fernández-Soto, P., García-Bernalt Diego, J., Febrer-Sendra, B., & Muro, A. (2023). Development of a Duplex LAMP Assay with Probe-Based Readout for Simultaneous Real-Time Detection of Schistosoma mansoni and Strongyloides spp. -A Laboratory Approach to Point-Of-Care. International Journal of Molecular Sciences, 24(1), 893. https://doi.org/10.3390/ijms24010893







