Transcriptional Regulation of Siglec-15 by ETS-1 and ETS-2 in Hepatocellular Carcinoma Cells
Abstract
1. Introduction
2. Results
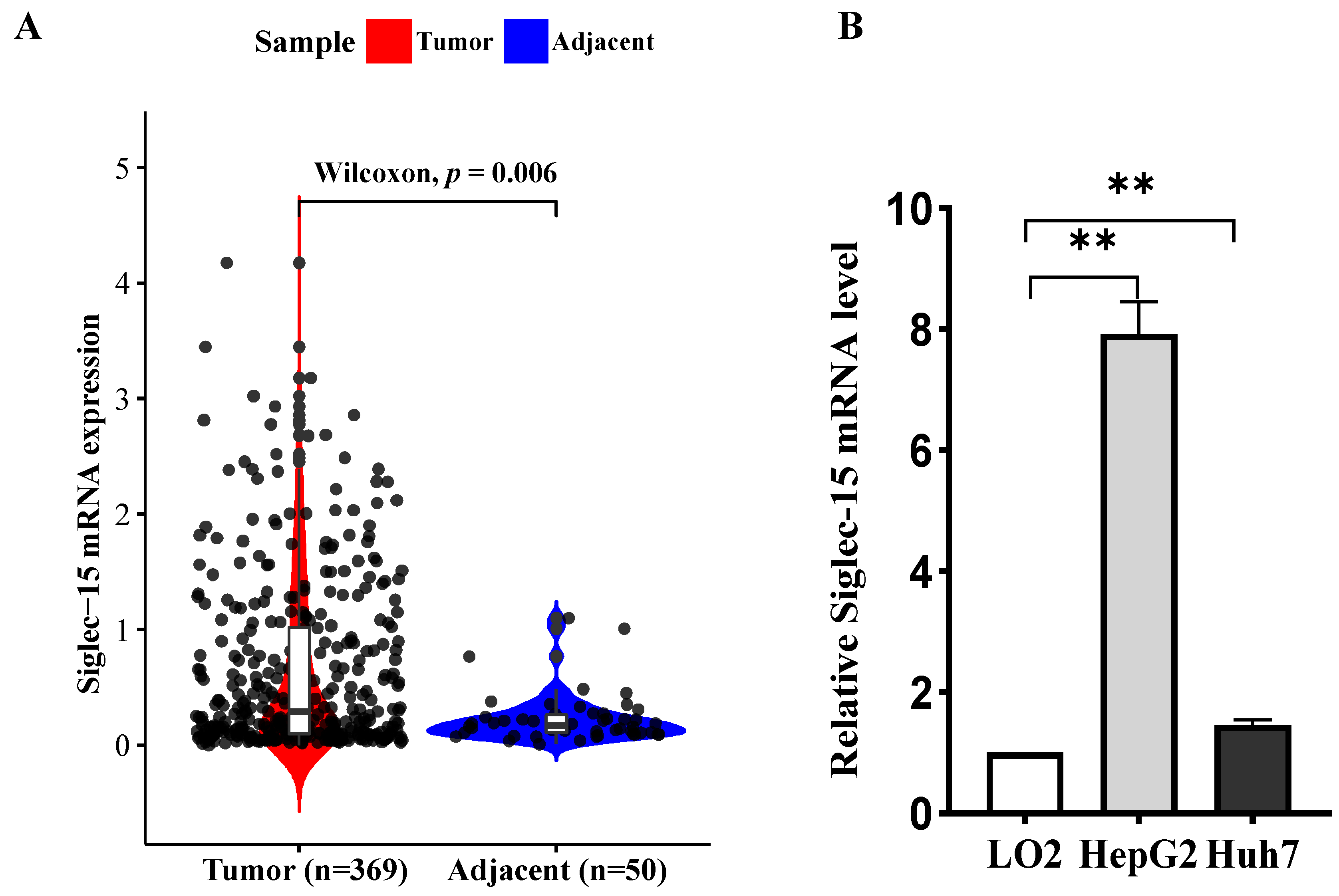
2.1. Siglec-15 Is Highly Expressed in the HCC Tissues and Cells
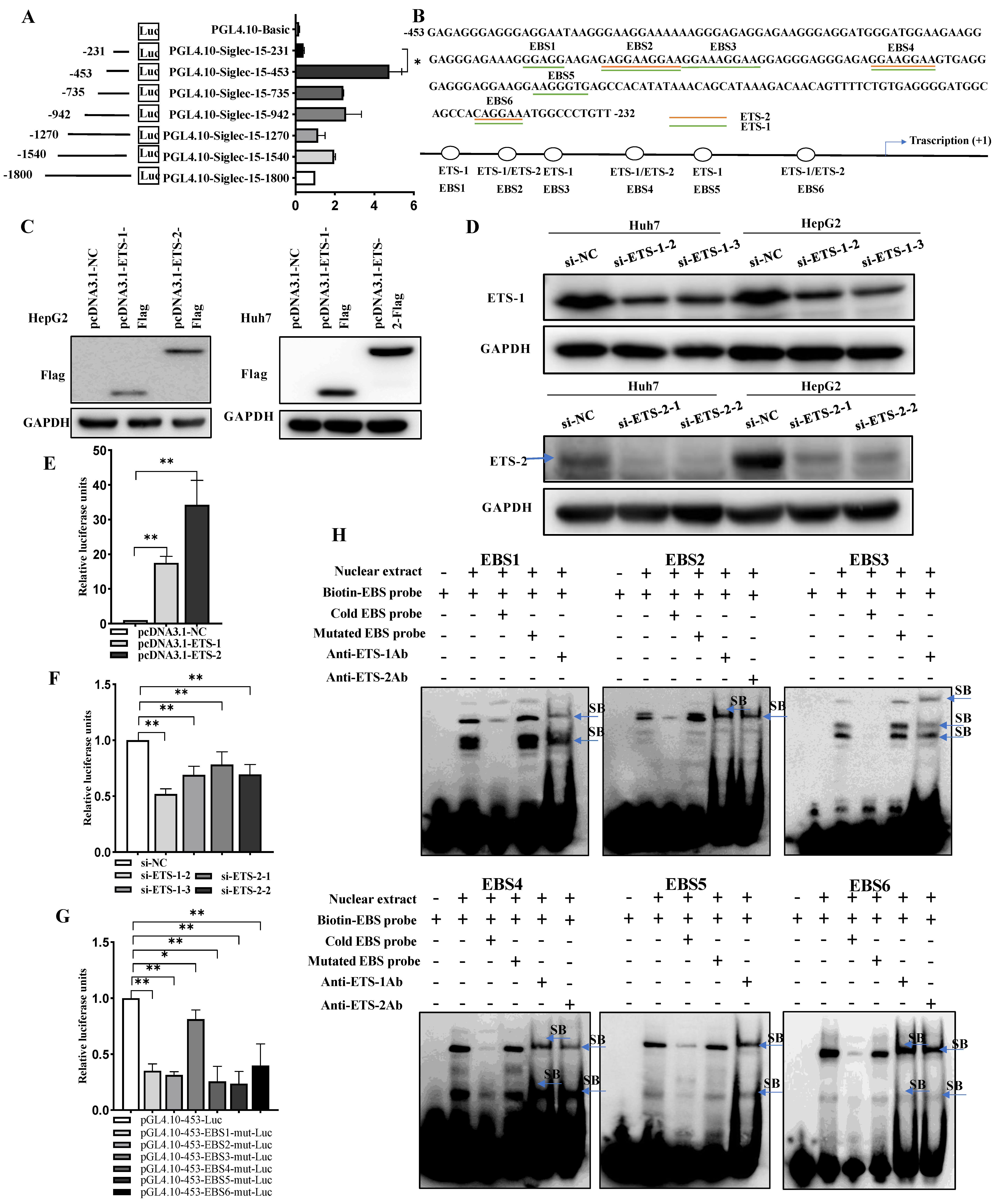
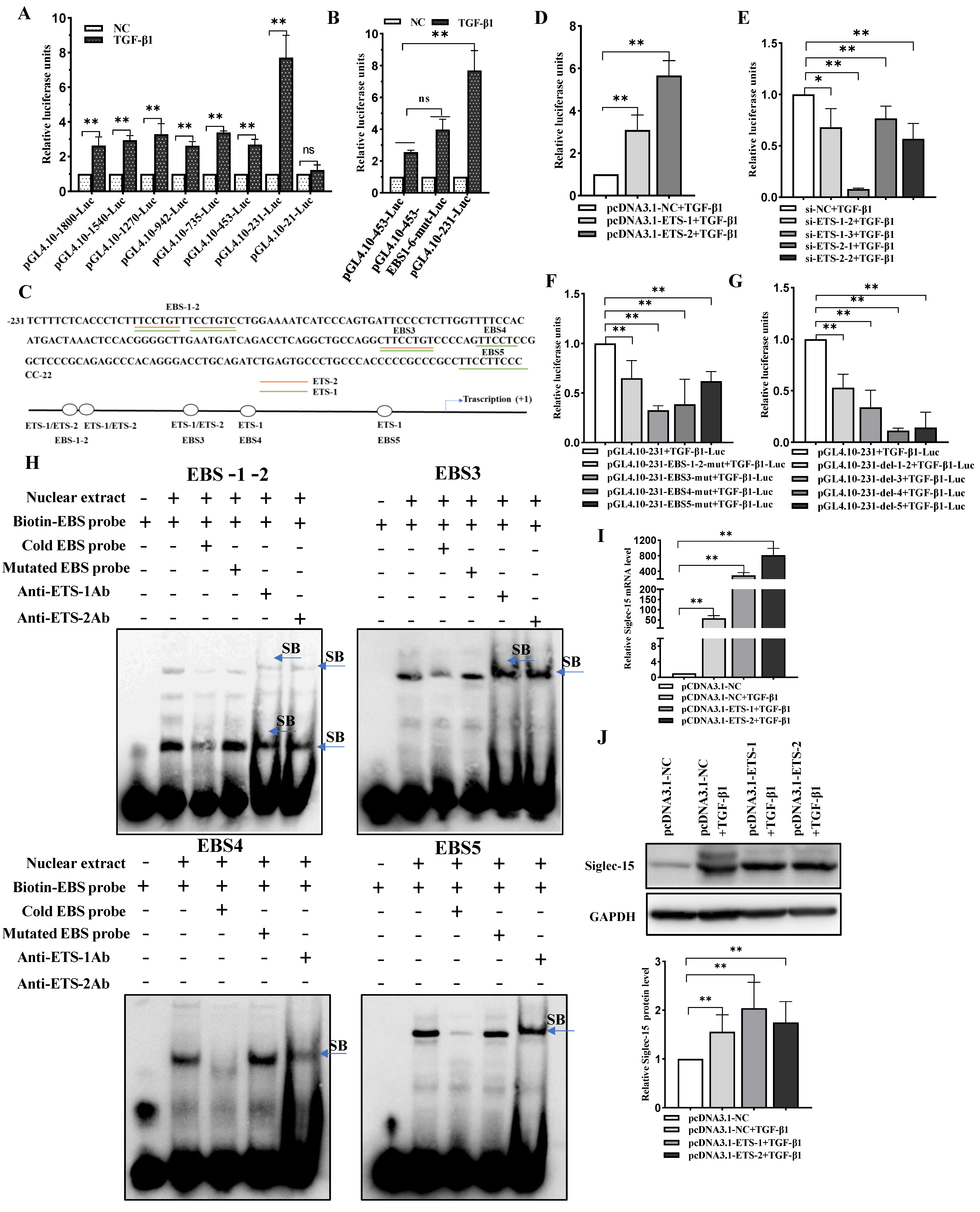
2.2. Siglec-15 Gene Promoter Activity in HCC Cells and Putative Transcription Factor Binding Sites
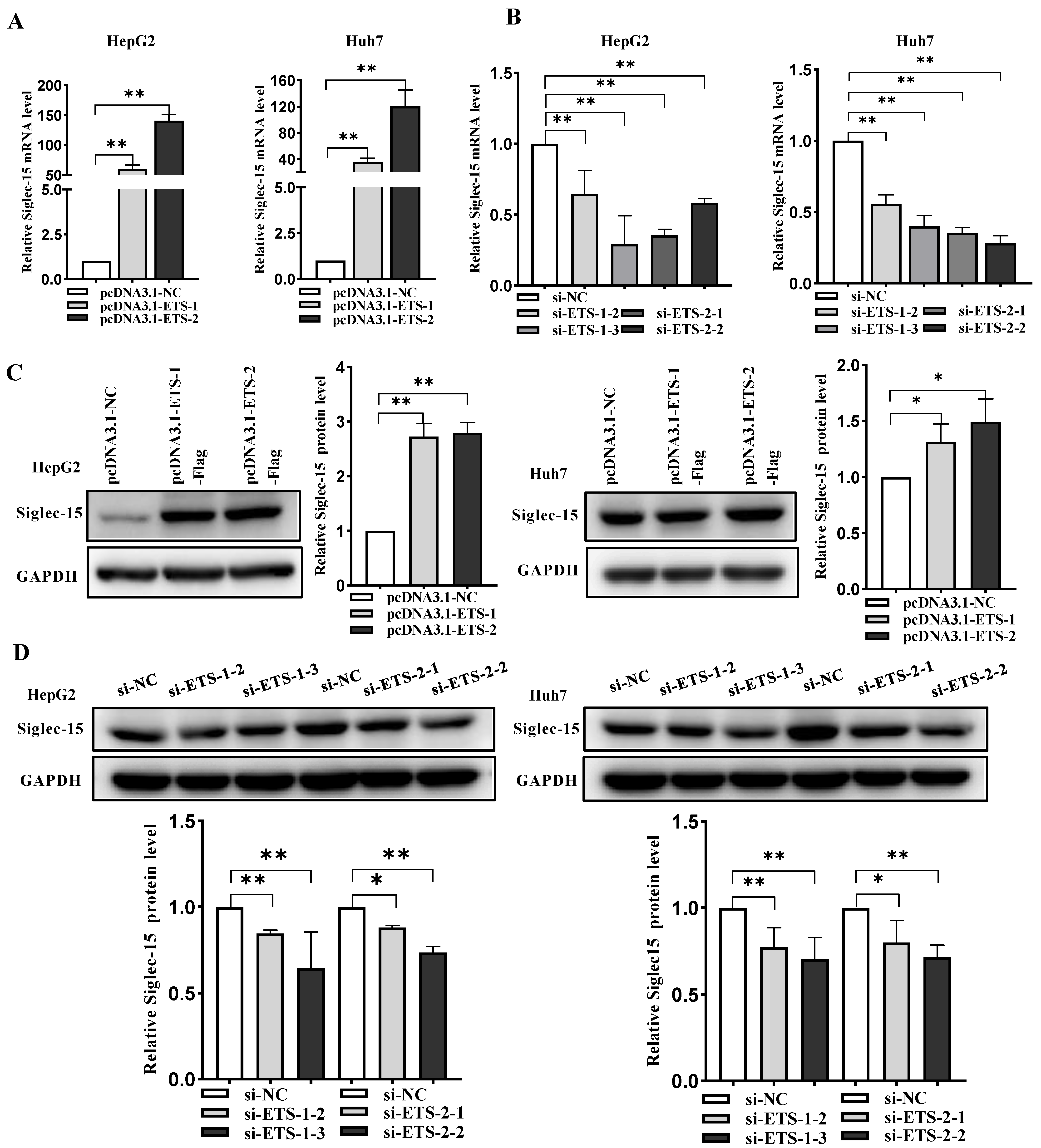
2.3. ETS-1 and ETS-2 Enhance Transcription of Siglec-15 in HCC Cells
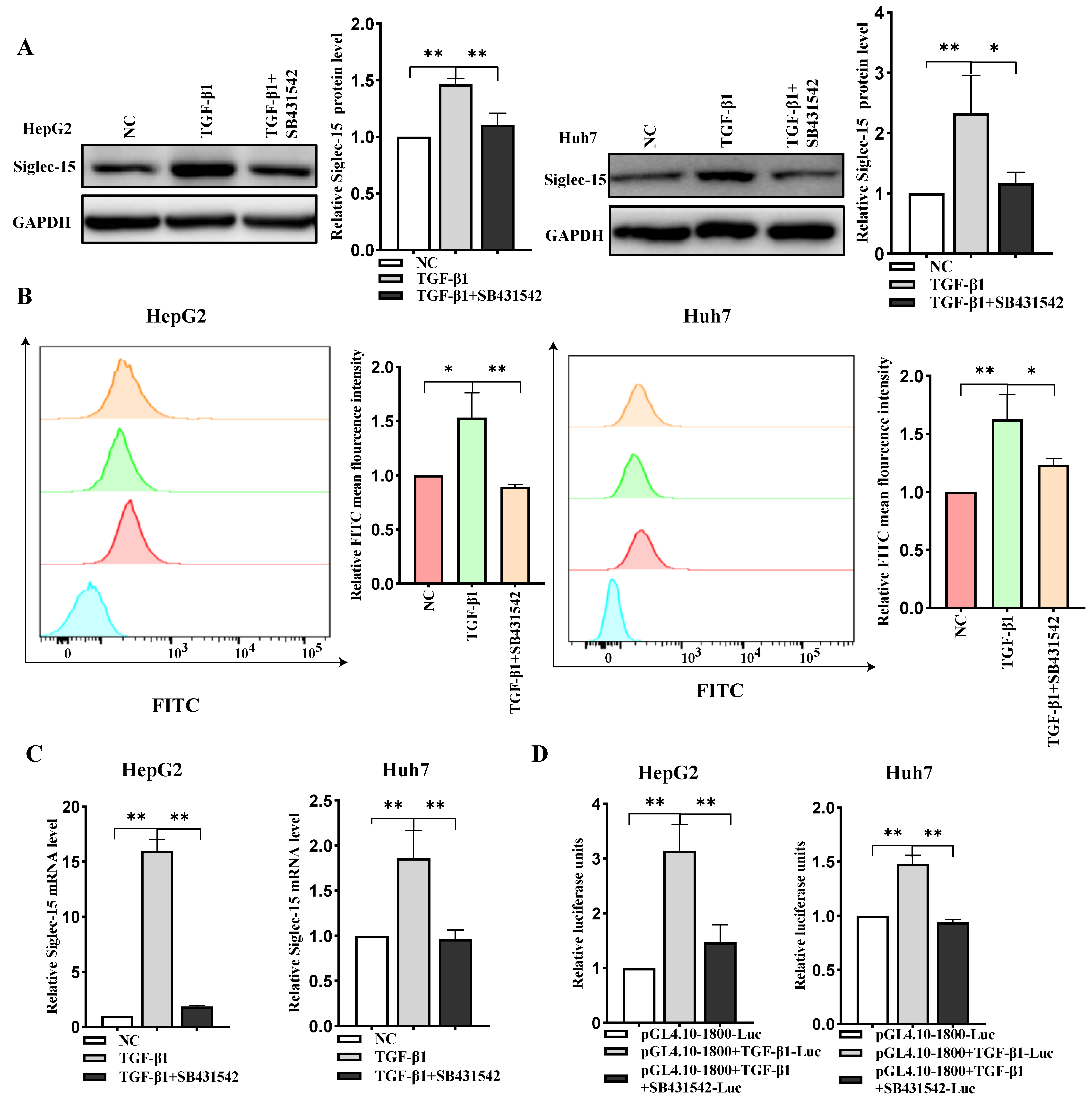
2.4. TGF-β1 Stimulates Siglec-15 Expression in HCC Cells
2.5. ETS-1/ETS-2 Is Indispensable for TGF-β1-Induced Transcriptional Activation of Siglec-15
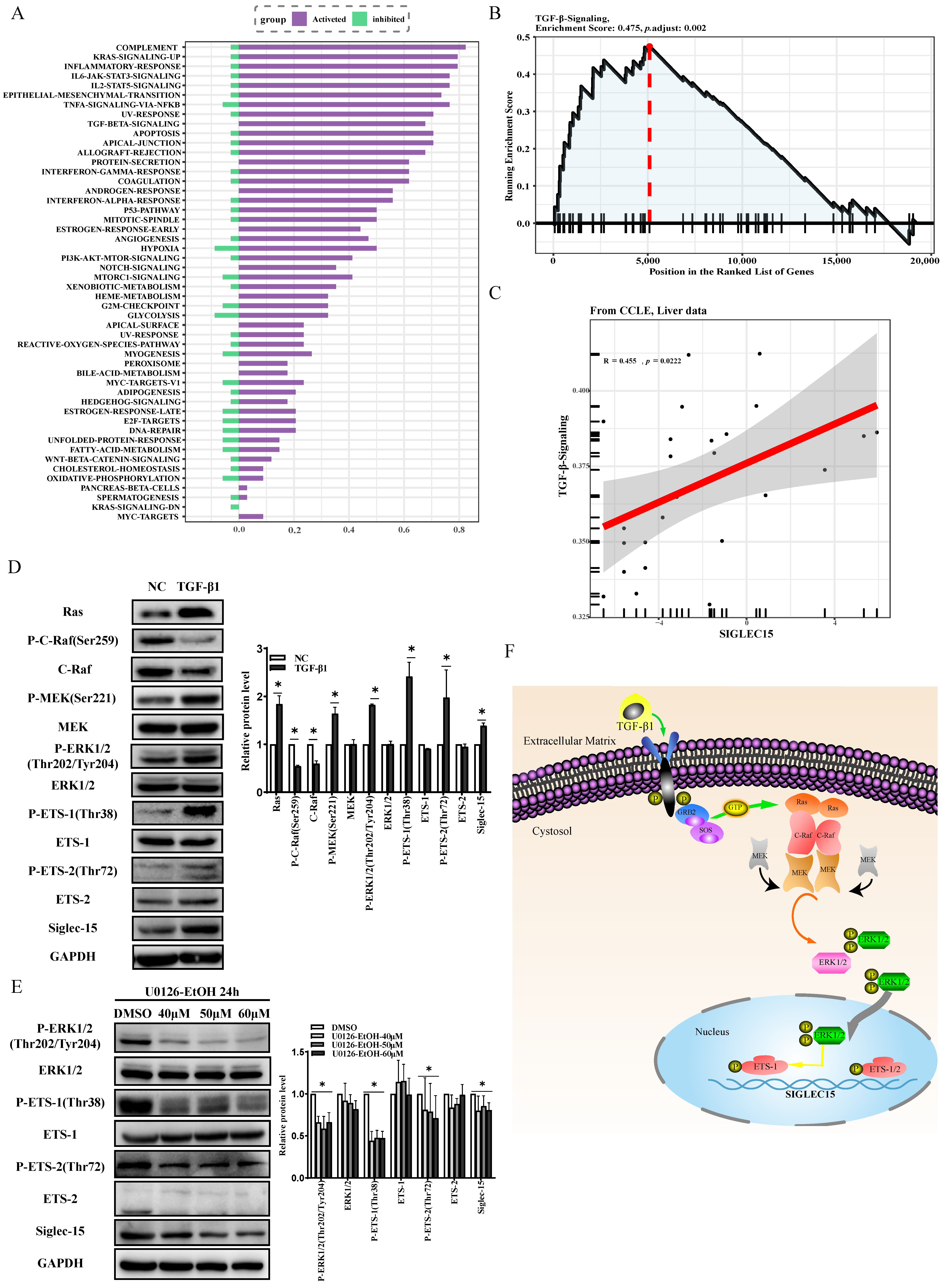
2.6. TGF-β1 Induces Transcriptional Activation of Siglec-15 via MAPK Signaling Pathway
3. Discussion
4. Materials and Methods
4.1. Siglec-15 Promoter Luciferase Reporter Constructs
4.2. Overexpression Constructs
4.3. RNA Interference
4.4. Dual-Luciferase Reporter Assay
4.5. Cell Culture and Reagents
4.6. Western Blot Analysis
4.7. RNA Isolation and Real-Time Quantitative Polymerase Chain Reaction
4.8. Electrophoretic Mobility Shift Assay (EMSA)
4.9. Flow Cytometry
4.10. Analysis of Database
4.11. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chen, L.; Han, X. Anti-PD-1/PD-L1 therapy of human cancer: Past, present, and future. J. Clin. Investig. 2015, 125, 3384–3391. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yuan, R.; Song, W.; Sun, J.; Liu, D.; Li, Z. PD-1, PD-L1 (B7-H1) and Tumor-Site Immune Modulation Therapy: The Historical Perspective. J. Hematol. Oncol. 2017, 10, 34. [Google Scholar] [CrossRef] [PubMed]
- Macauley, M.S.; Crocker, P.R.; Paulson, J.C. Siglec-mediated regulation of immune cell function in disease. Nat. Rev. Immunol. 2014, 14, 653–666. [Google Scholar] [CrossRef] [PubMed]
- Van de Wall, S.; Santegoets, K.C.M.; van Houtum, E.J.H.; Büll, C.; Adema, G.J. Sialoglycans and Siglecs Can Shape the Tumor Immune Microenvironment. Trends Immunol. 2020, 41, 274–285. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Liu, L.N.; Flies, D.B.; Nie, X.; Toki, M.; Zhang, J.; Song, C.; Zarr, M.; Zhou, X.; et al. Siglec-15 as an immune suppressor and potential target for normalization cancer immunotherapy. Nat. Med. 2019, 25, 656–666. [Google Scholar] [CrossRef]
- Ren, X. Immunosuppressive checkpoint Siglec-15: A vital new piece of the cancer immunotherapy jigsaw puzzle. Cancer Biol. Med. 2019, 16, 205–210. [Google Scholar] [CrossRef]
- Cao, G.; Xiao, Z.; Yin, Z. Normalization cancer immunotherapy: Blocking Siglec-15! Signal Transduct. Target. Ther. 2019, 4, 10. [Google Scholar] [CrossRef] [PubMed]
- Sizemore, G.M.; Pitarresi, J.R.; Balakrishnan, S.; Ostrowski, M.C. The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer 2017, 17, 337–351. [Google Scholar] [CrossRef]
- Dittmer, J. The role of the transcription factor Ets1 in carcinoma. Semin. Cancer Biol. 2015, 35, 20–38. [Google Scholar] [CrossRef]
- Du, F.; Qi, X.; Zhang, A.; Sui, F.; Wang, X.; Proud, C.G.; Lin, C.; Fan, X.; Li, J. MRTF-A-NF-κB/p65 axis-mediated PDL1 transcription and expression contributes to immune evasion of non-small-cell lung cancer via TGF-β. Exp. Mol. Med. 2021, 53, 1366–1378. [Google Scholar] [CrossRef]
- Yang, B.S.; Hauser, C.A.; Henkel, G.; Colman, M.S.; Van Beveren, C.; Stacey, K.J.; Hume, D.A.; Maki, R.A.; Ostrowski, M.C. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activities of c-Ets1 and c-Ets2. Mol. Cell. Biol. 1996, 16, 538–547. [Google Scholar] [CrossRef] [PubMed]
- Dituri, F.; Serio, G.; Filannino, D.; Mascolo, A.; Sacco, R.; Villa, E.; Giannelli, G. Circulating TGF-β1-related biomarkers in patients with hepatocellular carcinoma and their association with HCC staging scores. Cancer Lett. 2014, 353, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Takamiya, R.; Ohtsubo, K.; Takamatsu, S.; Taniguchi, N.; Angata, T. The interaction between Siglec-15 and tumor-associated sialyl-Tn antigen enhances TGF-β secretion from monocytes/macrophages through the DAP12-Syk pathway. Glycobiology 2013, 23, 178–187. [Google Scholar] [CrossRef]
- Li, B.; Zhang, B.; Wang, X.; Zeng, Z.; Huang, Z.; Zhang, L.; Wei, F.; Ren, X.; Yang, L. Expression signature, prognosis value, and immune characteristics of Siglec-15 identified by pan-cancer analysis. Oncoimmunology 2020, 9, 1807291. [Google Scholar] [CrossRef] [PubMed]
- Derynck, R.; Zhang, Y.E. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef]
- He, H.; Du, F.; He, Y.; Wei, Z.; Meng, C.; Xu, Y.; Zhou, H.; Wang, N.; Luo, X.G.; Ma, W.; et al. The Wnt-β-catenin signaling regulated MRTF-A transcription to activate migration-related genes in human breast cancer cells. Oncotarget 2018, 9, 15239–15251. [Google Scholar] [CrossRef]
- Miranda, M.Z.; Bialik, J.F.; Speight, P.; Dan, Q.; Yeung, T.; Szászi, K.; Pedersen, S.F.; Kapus, A. TGF-β1 regulates the expression and transcriptional activity of TAZ protein via a Smad3-independent, myocardin-related transcription factor-mediated mechanism. J. Biol. Chem. 2017, 292, 14902–14920. [Google Scholar] [CrossRef] [PubMed]
- Plotnik, J.P.; Budka, J.A.; Ferris, M.W.; Hollenhorst, P.C. ETS1 is a genome-wide effector of RAS/ERK signaling in epithelial cells. Nucleic Acids Res. 2014, 42, 11928–11940. [Google Scholar] [CrossRef] [PubMed]
- Charlot, C.; Dubois-Pot, H.; Serchov, T.; Tourrette, Y.; Wasylyk, B. A review of post-translational modifications and subcellular localization of Ets transcription factors: Possible connection with cancer and involvement in the hypoxic response. Methods Mol. Biol. Clifton NJ 2010, 647, 3–30. [Google Scholar] [CrossRef]
- Hollenhorst, P.C.; McIntosh, L.P.; Graves, B.J. Genomic and biochemical insights into the specificity of ETS transcription factors. Annu. Rev. Biochem. 2011, 80, 437–471. [Google Scholar] [CrossRef]
- Sia, D.; Jiao, Y.; Martinez-Quetglas, I.; Kuchuk, O.; Villacorta-Martin, C.; Castro de Moura, M.; Putra, J.; Camprecios, G.; Bassaganyas, L.; Akers, N.; et al. Identification of an Immune-specific Class of Hepatocellular Carcinoma, Based on Molecular Features. Gastroenterology 2017, 153, 812–826. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheng, K.; Wu, Y.; Lin, H.; Fang, M.; Xue, C.; Lin, X.; Lin, X. Transcriptional Regulation of Siglec-15 by ETS-1 and ETS-2 in Hepatocellular Carcinoma Cells. Int. J. Mol. Sci. 2023, 24, 792. https://doi.org/10.3390/ijms24010792
Sheng K, Wu Y, Lin H, Fang M, Xue C, Lin X, Lin X. Transcriptional Regulation of Siglec-15 by ETS-1 and ETS-2 in Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences. 2023; 24(1):792. https://doi.org/10.3390/ijms24010792
Chicago/Turabian StyleSheng, Kaiqin, Yuecheng Wu, Hanbin Lin, Menghan Fang, Chaorong Xue, Xu Lin, and Xinjian Lin. 2023. "Transcriptional Regulation of Siglec-15 by ETS-1 and ETS-2 in Hepatocellular Carcinoma Cells" International Journal of Molecular Sciences 24, no. 1: 792. https://doi.org/10.3390/ijms24010792
APA StyleSheng, K., Wu, Y., Lin, H., Fang, M., Xue, C., Lin, X., & Lin, X. (2023). Transcriptional Regulation of Siglec-15 by ETS-1 and ETS-2 in Hepatocellular Carcinoma Cells. International Journal of Molecular Sciences, 24(1), 792. https://doi.org/10.3390/ijms24010792




