Functional Characterization of the Cystine-Rich-Receptor-like Kinases (CRKs) and Their Expression Response to Sclerotinia sclerotiorum and Abiotic Stresses in Brassica napus
Abstract
1. Introduction
2. Result
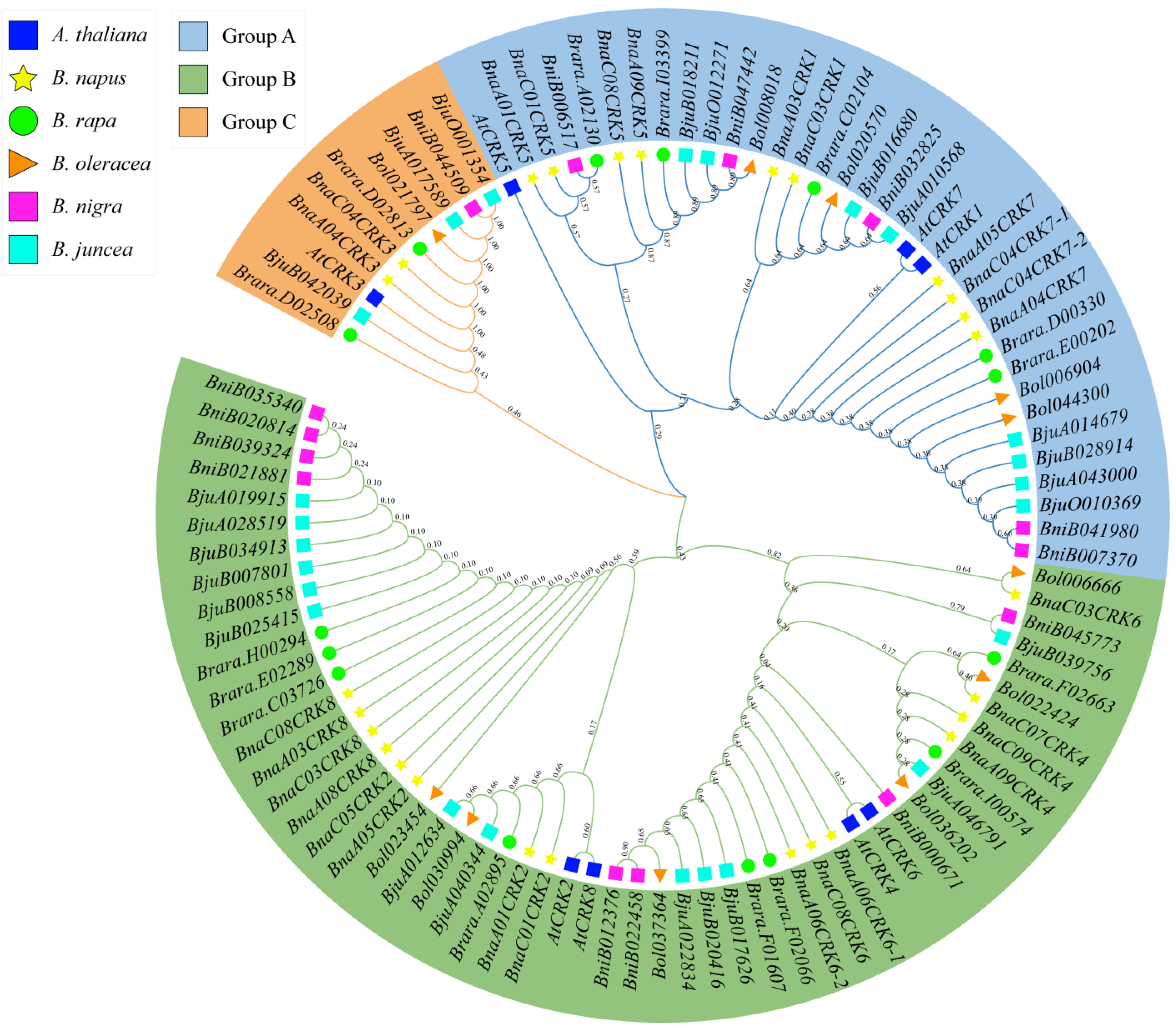
2.1. Identification, Phylogenetic Analysis, and Structural Characterization of B. napus CRK Family
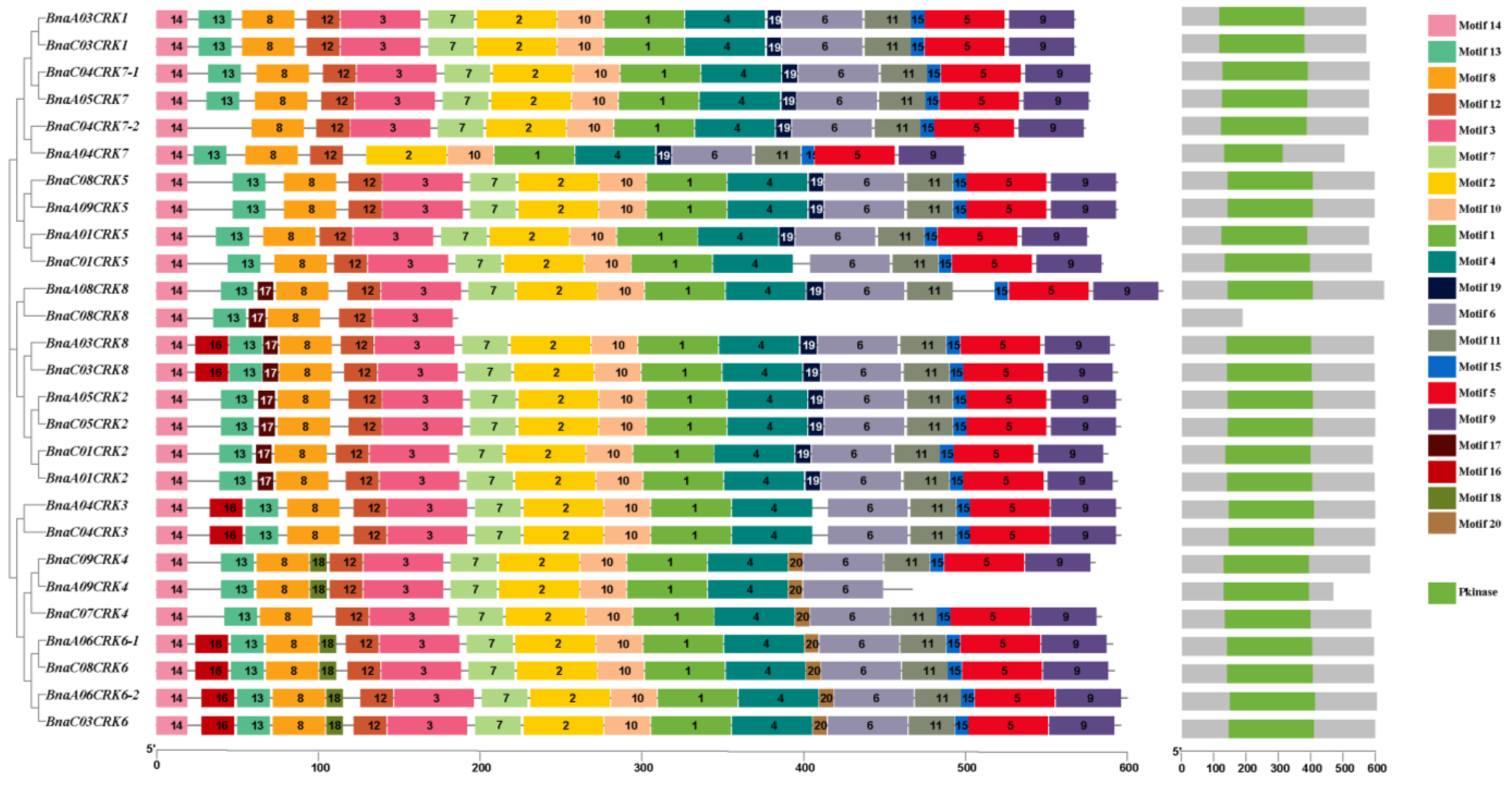
2.2. Motif Prediction and Distribution Analysis of BnaCRKs
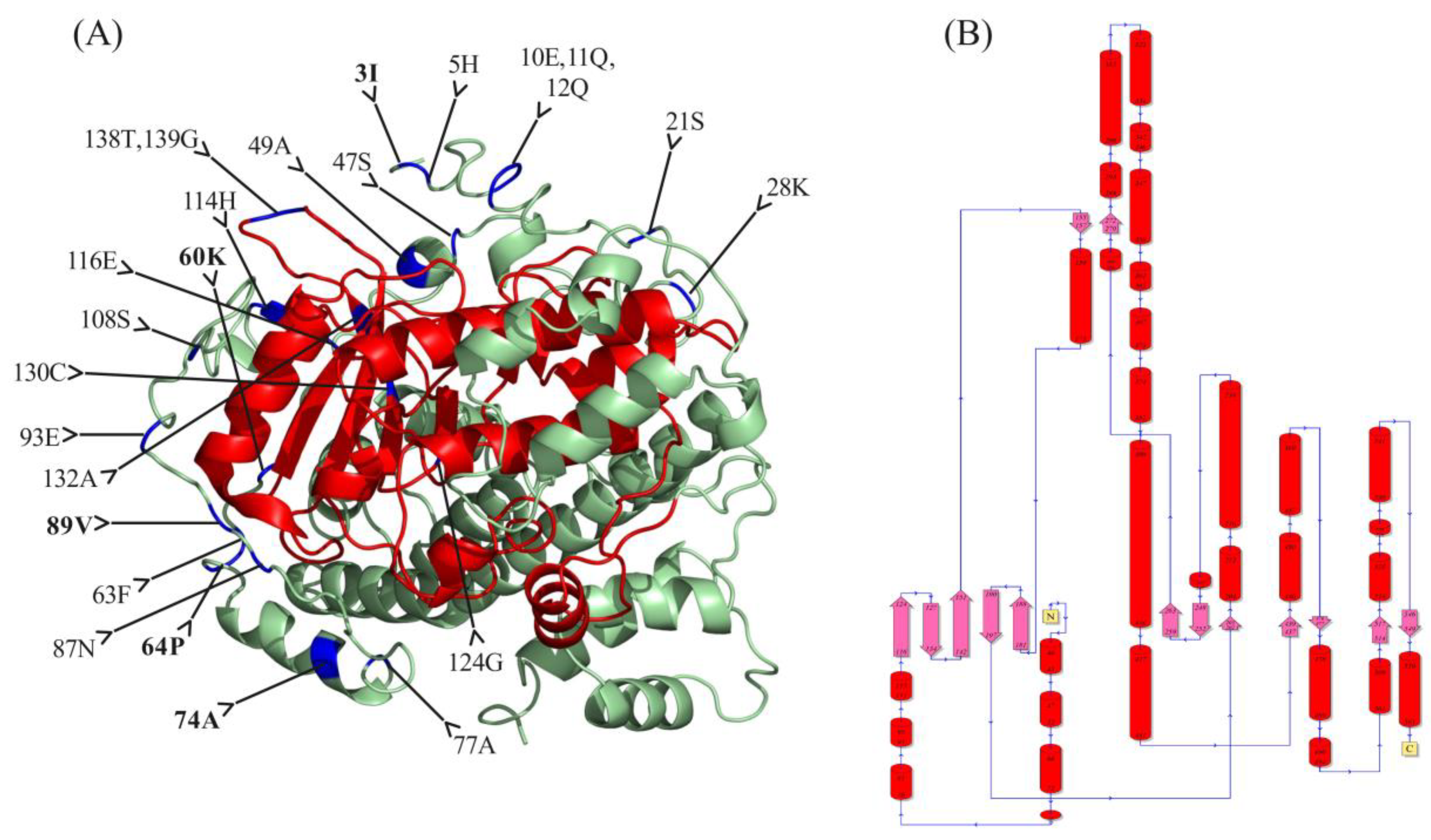
2.3. Functional Divergence Analysis and Protein 3D Structure Prediction
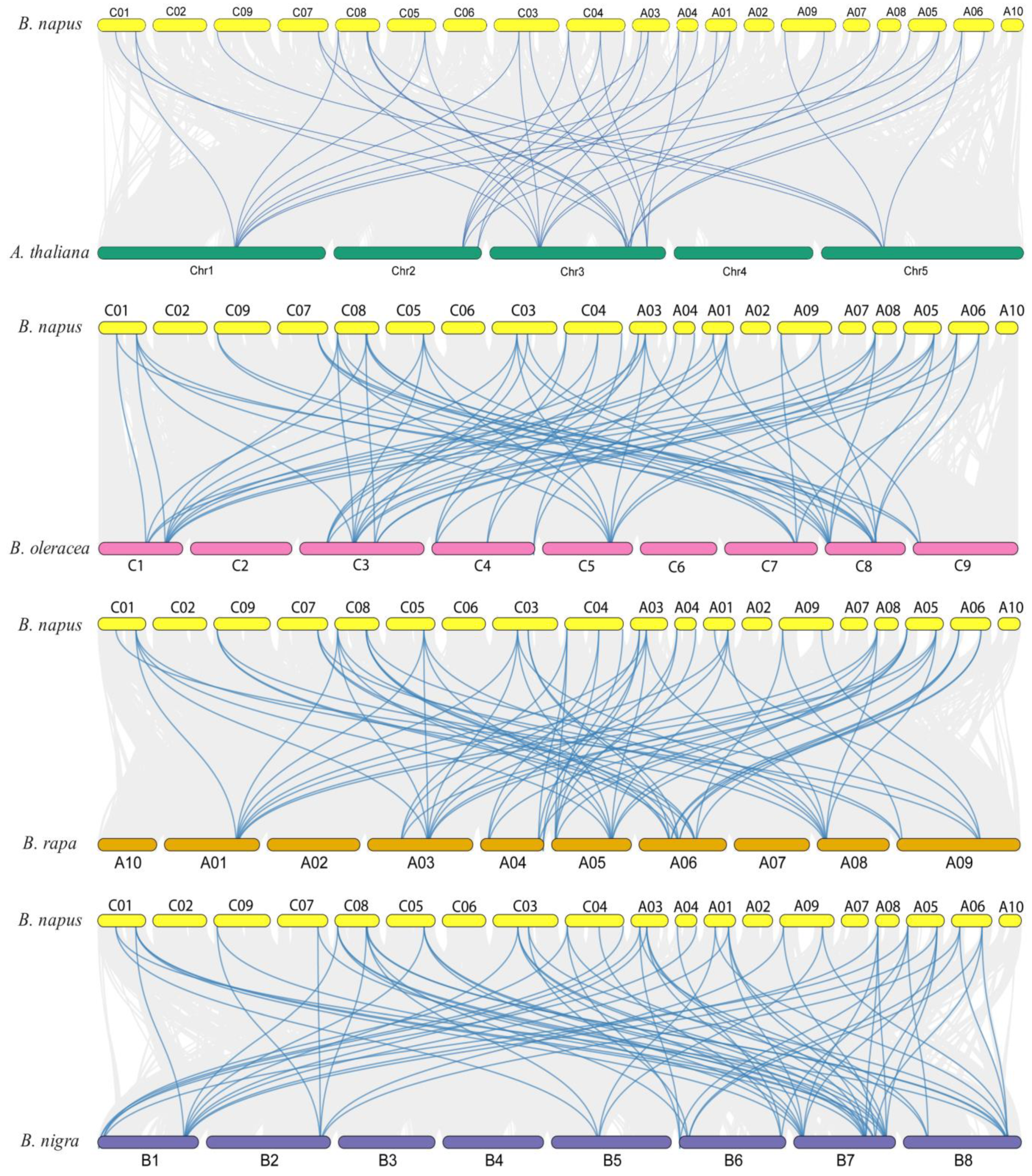
2.4. Synteny and Duplication Prediction of BnaCRK Gene Family in B. napus
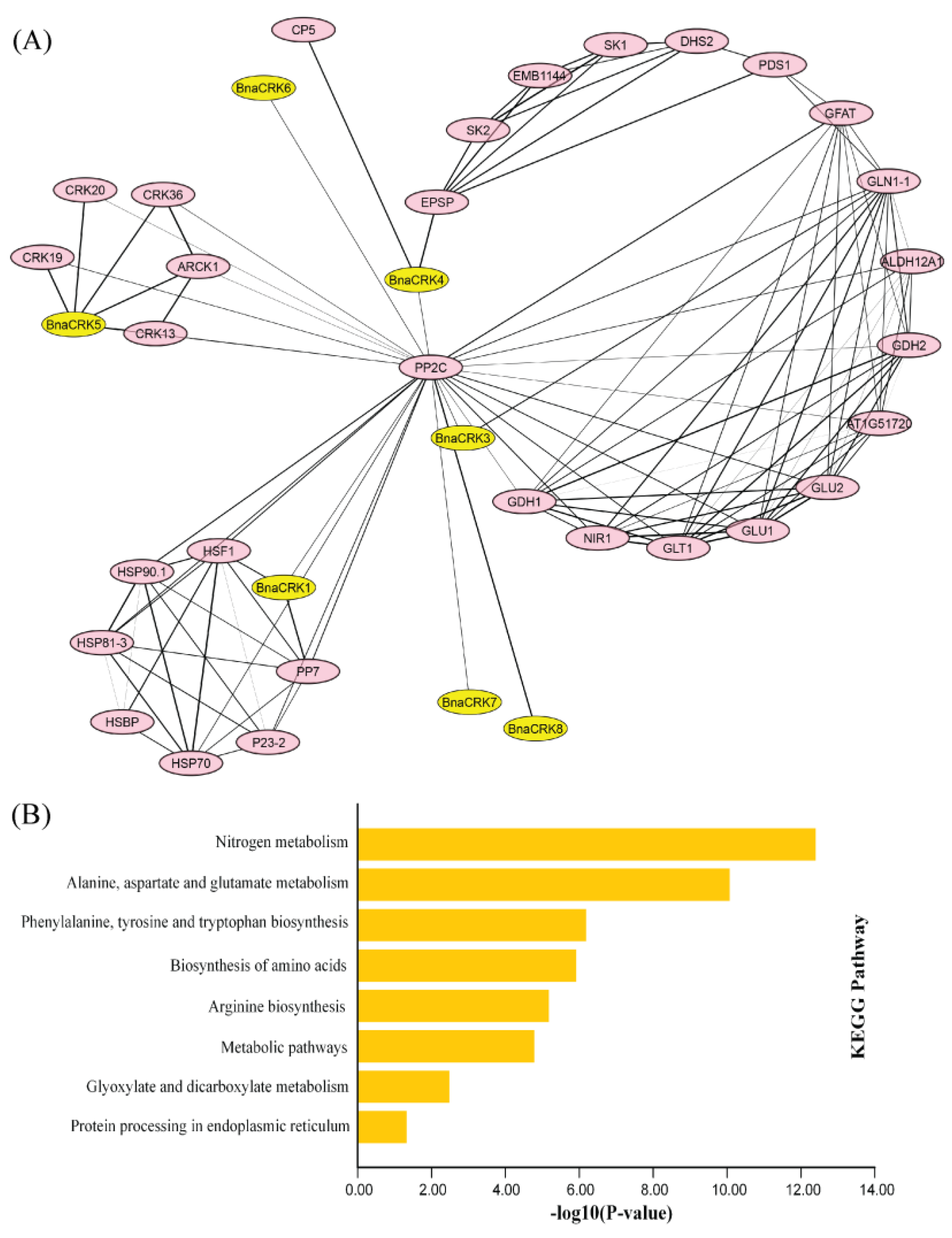
2.5. BnaCRK Protein-Protein Interaction Network Prediction
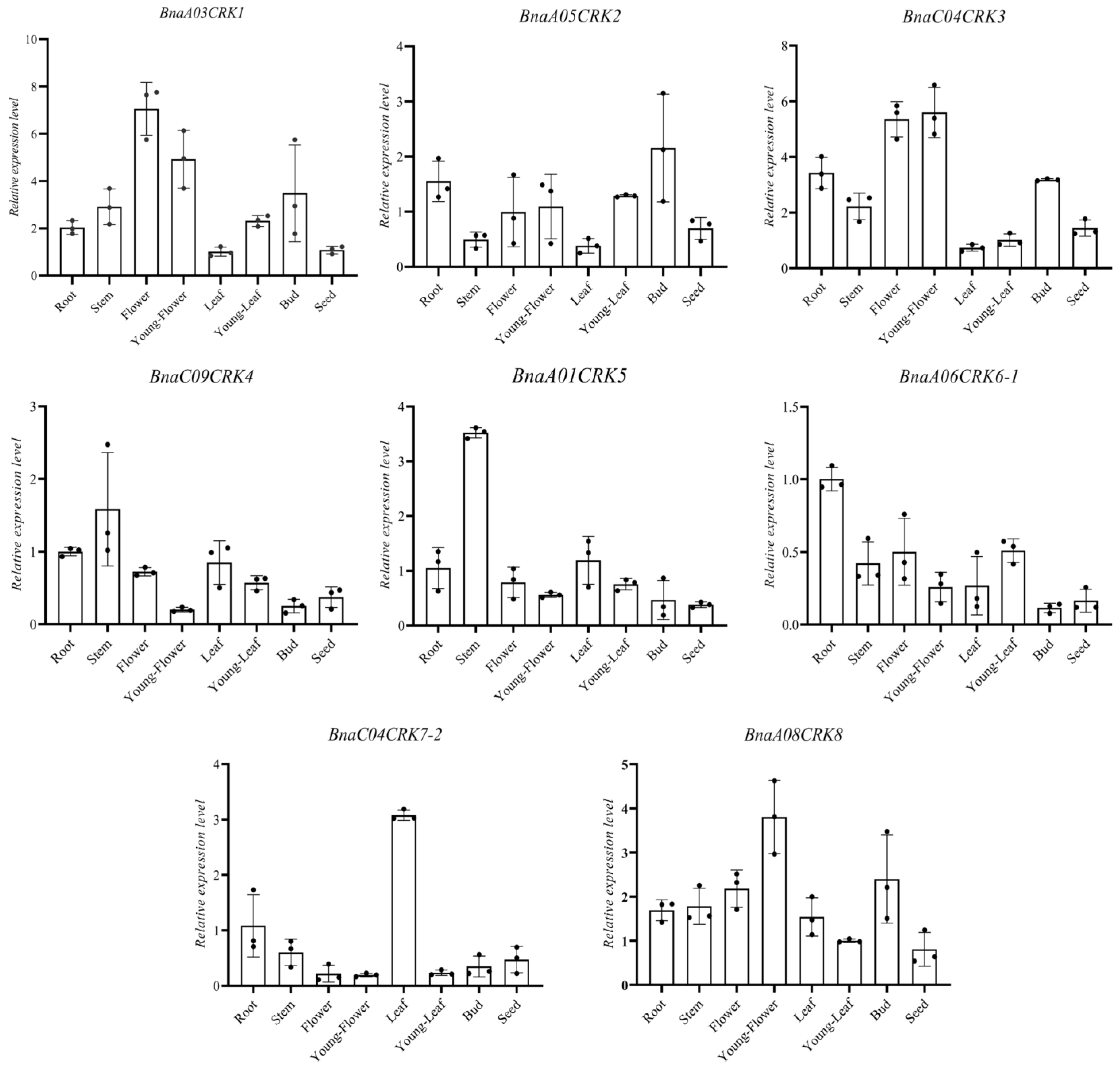
2.6. Expression Profile of the BnaCRKs in Different Organs of B. napus
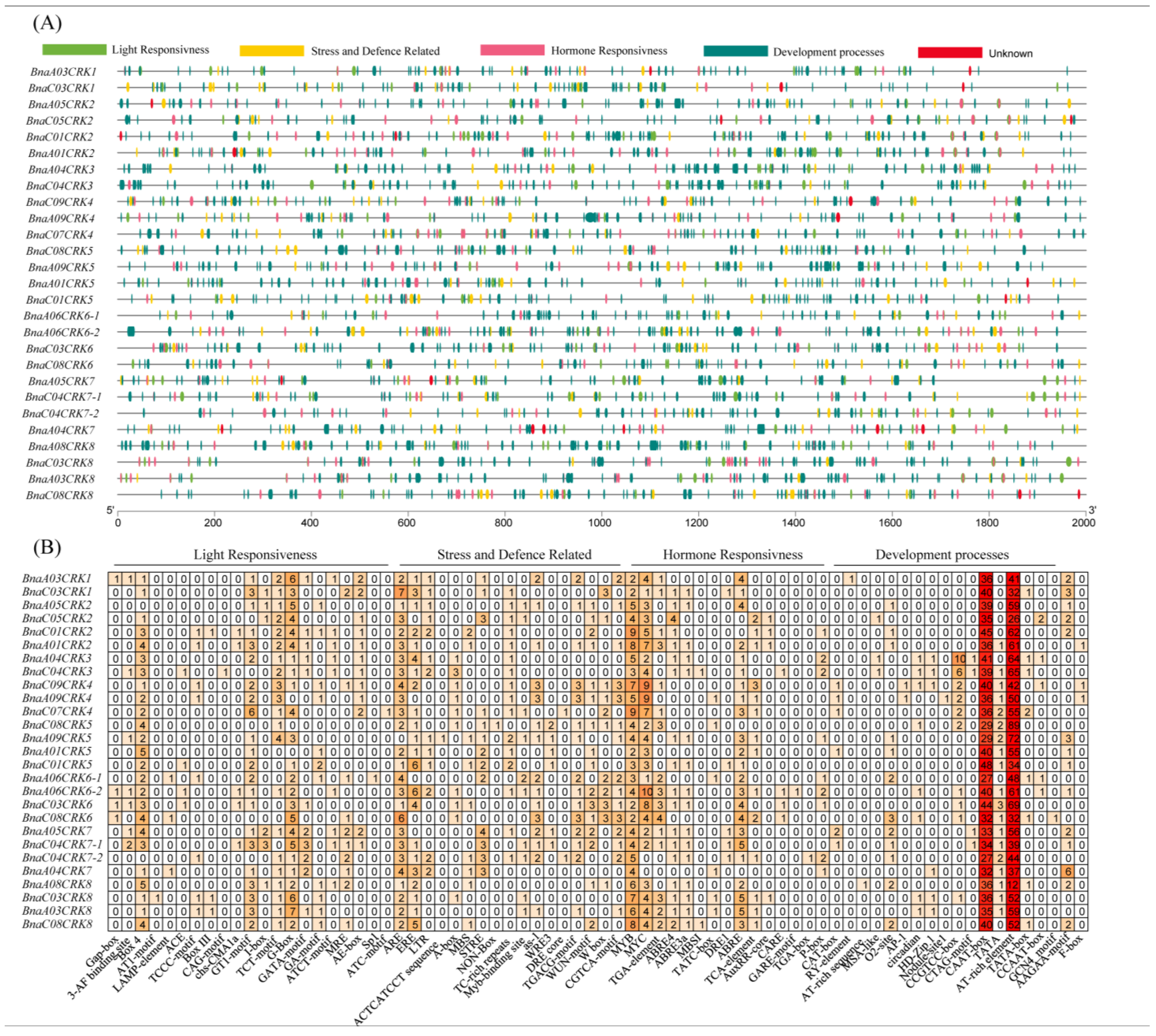
2.7. Prediction of Cis-Acting Elements in the Promoter Region of BnaCRKs
2.8. Expression Profile of the BnaCRKs under Abiotic Stresses and S. sclerotiorum Infection
2.9. Prediction of Potential MicroRNAs Targeting BnaCRKs Gene Family in B. napus
2.10. SNP Polymorphism
3. Discussion
4. Materials and Methods
4.1. Identification, Phylogenetic Analysis, and Structural Characterization of B. napus CRK Family
4.2. Motif Composition, Genomic Distribution, Site-Specific Assessment, and Synteny Analysis of CRK Gene Family in B. napus
4.3. Functional Divergence and Three-Dimensional Architecture of CRK in B. napus
4.4. BnaCRKs-Target Interaction, Cis-Acting Elements, and Micro-RNA Target Site Prediction
4.5. B. napus Growth Condition and Expression Analysis of Selected BnaCRKs under Different Stresses
4.6. RNA-Seq and SNP Distribution Analysis of BnaCRKs
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cárdenas-Aguiar, E.; Suárez, G.; Paz-Ferreiro, J.; Askeland, M.; Méndez, A.; Gascó, G. Remediation of mining soils by combining Brassica napus growth and amendment with chars from manure waste. Chemosphere 2020, 261, 127798. [Google Scholar] [CrossRef] [PubMed]
- Bolton, M.D.; Thomma, B.P.; Nelson, B.D. Sclerotinia sclerotiorum (Lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Mol. Plant Pathol. 2006, 7, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Al-lami, H.F.; You, M.P.; Mohammed, A.E.; Barbetti, M.J. Virulence variability across the Alternaria spp. population determines incidence and severity of alternaria leaf spot on rapeseed. Plant Pathol. 2020, 69, 506–517. [Google Scholar] [CrossRef]
- Chen, W.; Provart, N.J.; Glazebrook, J.; Katagiri, F.; Chang, H.-S.; Eulgem, T.; Mauch, F.; Luan, S.; Zou, G.; Whitham, S.A. Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 2002, 14, 559–574. [Google Scholar] [CrossRef]
- Mysore, K.S.; Crasta, O.R.; Tuori, R.P.; Folkerts, O.; Swirsky, P.B.; Martin, G.B. Comprehensive transcript profiling of Pto-and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002, 32, 299–315. [Google Scholar] [CrossRef]
- Cheng, S.-H.; Willmann, M.R.; Chen, H.-C.; Sheen, J. Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 2002, 129, 469–485. [Google Scholar] [CrossRef]
- Tang, D.; Wang, G.; Zhou, J.-M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637. [Google Scholar] [CrossRef]
- Shiu, S.H.; Bleecker, A.B. Expansion of the receptor-like kinase/Pelle gene family and receptor-like proteins in Arabidopsis. Plant Physiol 2003, 132, 530–543. [Google Scholar] [CrossRef]
- Shiu, S.-H.; Karlowski, W.M.; Pan, R.; Tzeng, Y.-H.; Mayer, K.F.; Li, W.-H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell 2004, 16, 1220–1234. [Google Scholar] [CrossRef]
- Sun, X.; Wang, G.-L. Genome-wide identification, characterization and phylogenetic analysis of the rice LRR-kinases. PloS One 2011, 6, e16079. [Google Scholar] [CrossRef]
- Liu, T.; Liu, Z.; Song, C.; Hu, Y.; Han, Z.; She, J.; Fan, F.; Wang, J.; Jin, C.; Chang, J. Chitin-induced dimerization activates a plant immune receptor. science 2012, 336, 1160–1164. [Google Scholar] [CrossRef] [PubMed]
- Chinchilla, D.; Bauer, Z.; Regenass, M.; Boller, T.; Felix, G. The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 2006, 18, 465–476. [Google Scholar] [CrossRef]
- Ederli, L.; Madeo, L.; Calderini, O.; Gehring, C.; Moretti, C.; Buonaurio, R.; Paolocci, F.; Pasqualini, S. The Arabidopsis thaliana cysteine-rich receptor-like kinase CRK20 modulates host responses to Pseudomonas syringae pv. tomato DC3000 infection. J. Plant Physiol. 2011, 168, 1784–1794. [Google Scholar] [CrossRef] [PubMed]
- Baba, A.I.; Rigó, G.; Ayaydin, F.; Rehman, A.U.; Andrási, N.; Zsigmond, L.; Valkai, I.; Urbancsok, J.; Vass, I.; Pasternak, T. Functional Analysis of the Arabidopsis thaliana CDPK-Related Kinase Family: At CRK1 Regulates Responses to Continuous Light. Int. J. Mol. Sci. 2018, 19, 1282. [Google Scholar] [CrossRef] [PubMed]
- Yadeta, K.A.; Elmore, J.M.; Creer, A.Y.; Feng, B.; Franco, J.Y.; Rufian, J.S.; He, P.; Phinney, B.; Coaker, G. A Cysteine-Rich Protein Kinase Associates with a Membrane Immune Complex and the Cysteine Residues Are Required for Cell Death. Plant Physiol 2017, 173, 771–787. [Google Scholar] [CrossRef]
- Vaattovaara, A.; Brandt, B.; Rajaraman, S.; Safronov, O.; Veidenberg, A.; Luklová, M.; Kangasjärvi, J.; Löytynoja, A.; Hothorn, M.; Salojärvi, J. Mechanistic insights into the evolution of DUF26-containing proteins in land plants. Commun. Biol. 2019, 2, 1–18. [Google Scholar] [CrossRef]
- Leclercq, J.; Ranty, B.; Sanchez-Ballesta, M.-T.; Li, Z.; Jones, B.; Jauneau, A.; Pech, J.-C.; Latché, A.; Ranjeva, R.; Bouzayen, M. Molecular and biochemical characterization of LeCRK1, a ripening-associated tomato CDPK-related kinase. J. Exp. Bot. 2005, 56, 25–35. [Google Scholar] [CrossRef]
- Renna, L.; Stefano, G.; Majeran, W.; Micalella, C.; Meinnel, T.; Giglione, C.; Brandizzi, F. Golgi traffic and integrity depend on N-myristoyl transferase-1 in Arabidopsis. Plant Cell 2013, 25, 1756–1773. [Google Scholar] [CrossRef]
- Li, T.-G.; Zhang, D.-D.; Zhou, L.; Kong, Z.-Q.; Hussaini, A.S.; Wang, D.; Li, J.-J.; Short, D.P.; Dhar, N.; Klosterman, S.J. Genome-wide identification and functional analyses of the CRK gene family in cotton reveals GbCRK18 confers verticillium wilt resistance in Gossypium barbadense. Front. Plant Sci. 2018, 9, 1266. [Google Scholar] [CrossRef]
- Wang, J.-P.; Xu, Y.-P.; Munyampundu, J.-P.; Liu, T.-Y.; Cai, X.-Z. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: Genome-wide identification and functional analyses in disease resistance. Mol. Genet. Genom. 2016, 291, 661–676. [Google Scholar] [CrossRef]
- Tyagi, S.; Sharma, A.; Singh, K.; Upadhyay, S.K. Genomic dissection and transcriptional profiling of Cysteine-rich receptor-like kinases in five cereals and functional characterization of TaCRK68-A. Int. J. Biol. Macromol. 2019, 134, 316–329. [Google Scholar]
- Zuo, C.; Liu, H.; Lv, Q.; Chen, Z.; Tian, Y.; Mao, J.; Chu, M.; Ma, Z.; An, Z.; Chen, B. Genome-wide analysis of the apple (Malus domestica) cysteine-rich receptor-like kinase (CRK) family: Annotation, genomic organization, and expression profiles in response to fungal infection. Plant Mol. Biol. Report. 2020, 38, 14–24. [Google Scholar] [CrossRef]
- Wrzaczek, M.; Brosché, M.; Salojärvi, J.; Kangasjärvi, S.; Idänheimo, N.; Mersmann, S.; Robatzek, S.; Karpiński, S.; Karpińska, B.; Kangasjärvi, J. Transcriptional regulation of the CRK/DUF26 group of receptor-like protein kinases by ozone and plant hormones in Arabidopsis. BMC Plant Biol. 2010, 10, 95. [Google Scholar] [CrossRef]
- Chen, K.; Du, L.; Chen, Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol. Biol. 2003, 53, 61–74. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Fan, B.; Du, L.; Chen, Z. Activation of hypersensitive cell death by pathogen-induced receptor-like protein kinases from Arabidopsis. Plant Mol. Biol. 2004, 56, 271–283. [Google Scholar] [CrossRef] [PubMed]
- Yeh, Y.-H.; Chang, Y.-H.; Huang, P.-Y.; Huang, J.-B.; Zimmerli, L. Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front. Plant Sci. 2015, 6, 322. [Google Scholar] [CrossRef]
- Liu, H.T.; Gao, F.; Li, G.L.; Han, J.L.; Liu, D.L.; Sun, D.Y.; Zhou, R.G. The calmodulin-binding protein kinase 3 is part of heat-shock signal transduction in Arabidopsis thaliana. Plant J. 2008, 55, 760–773. [Google Scholar] [CrossRef]
- Li, R.-J.; Hua, W.; Lu, Y.-T. Arabidopsis cytosolic glutamine synthetase AtGLN1; 1 is a potential substrate of AtCRK3 involved in leaf senescence. Biochem. Biophys. Res. Commun. 2006, 342, 119–126. [Google Scholar] [CrossRef]
- Tanaka, H.; Osakabe, Y.; Katsura, S.; Mizuno, S.; Maruyama, K.; Kusakabe, K.; Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Abiotic stress-inducible receptor-like kinases negatively control ABA signaling in Arabidopsis. Plant J. 2012, 70, 599–613. [Google Scholar] [CrossRef]
- Zhang, X.; Yang, G.; Shi, R.; Han, X.; Qi, L.; Wang, R.; Xiong, L.; Li, G. Arabidopsis cysteine-rich receptor-like kinase 45 functions in the responses to abscisic acid and abiotic stresses. Plant Physiol. Biochem. 2013, 67, 189–198. [Google Scholar] [CrossRef]
- Lu, K.; Liang, S.; Wu, Z.; Bi, C.; Yu, Y.-T.; Wang, X.-F.; Zhang, D.-P. Overexpression of an Arabidopsis cysteine-rich receptor-like protein kinase, CRK5, enhances abscisic acid sensitivity and confers drought tolerance. J. Exp. Bot. 2016, 67, 5009–5027. [Google Scholar] [CrossRef] [PubMed]
- Bourdais, G.; Burdiak, P.; Gauthier, A.; Nitsch, L.; Salojärvi, J.; Rayapuram, C.; Idänheimo, N.; Hunter, K.; Kimura, S.; Merilo, E. Large-scale phenomics identifies primary and fine-tuning roles for CRKs in responses related to oxidative stress. PLoS Genet. 2015, 11, e1005373. [Google Scholar] [CrossRef] [PubMed]
- Idänheimo, N.; Gauthier, A.; Salojärvi, J.; Siligato, R.; Brosché, M.; Kollist, H.; Mähönen, A.P.; Kangasjärvi, J.; Wrzaczek, M. The Arabidopsis thaliana cysteine-rich receptor-like kinases CRK6 and CRK7 protect against apoplastic oxidative stress. Biochem. Biophys. Res. Commun. 2014, 445, 457–462. [Google Scholar] [CrossRef] [PubMed]
- Rayapuram, C.; Jensen, M.K.; Maiser, F.; Shanir, J.V.; Hornshøj, H.; Rung, J.H.; Gregersen, P.L.; Schweizer, P.; Collinge, D.B.; Lyngkjaer, M.F. Regulation of basal resistance by a powdery mildew-induced cysteine-rich receptor-like protein kinase in barley. Mol. Plant Pathol. 2012, 13, 135–147. [Google Scholar] [CrossRef]
- Yang, K.; Rong, W.; Qi, L.; Li, J.; Wei, X.; Zhang, Z. Isolation and characterization of a novel wheat cysteine-rich receptor-like kinase gene induced by Rhizoctonia cerealis. Sci. Rep. 2013, 3, 1–10. [Google Scholar] [CrossRef]
- Hrabak, E.M.; Chan, C.W.; Gribskov, M.; Harper, J.F.; Choi, J.H.; Halford, N.; Kudla, J.; Luan, S.; Nimmo, H.G.; Sussman, M.R. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003, 132, 666–680. [Google Scholar] [CrossRef]
- Xiao, X.H.; Yang, M.; Sui, J.L.; Qi, J.Y.; Fang, Y.J.; Hu, S.N.; Tang, C.R. The calcium-dependent protein kinase (CDPK) and CDPK-related kinase gene families in Hevea brasiliensis—Comparison with five other plant species in structure, evolution, and expression. FEBS Open Biol. 2017, 7, 4–24. [Google Scholar] [CrossRef]
- Zhang, H.; Wei, C.; Yang, X.; Chen, H.; Yang, Y.; Mo, Y.; Li, H.; Zhang, Y.; Ma, J.; Yang, J. Genome-wide identification and expression analysis of calcium-dependent protein kinase and its related kinase gene families in melon (Cucumis melo L.). PLoS ONE 2017, 12, e0176352. [Google Scholar] [CrossRef]
- Delgado-Cerrone, L.; Alvarez, A.; Mena, E.; Ponce de León, I.; Montesano, M. Genome-wide analysis of the soybean CRK-family and transcriptional regulation by biotic stress signals triggering plant immunity. PloS One 2018, 13, e0207438. [Google Scholar] [CrossRef]
- Zhao, P.; Liu, Y.; Kong, W.; Ji, J.; Cai, T.; Guo, Z. Genome-wide identification and characterization of calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in Medicago truncatula. Int. J. Mol. Sci. 2021, 22, 1044. [Google Scholar] [CrossRef]
- Rout, S.S.; Rout, P.; Uzair, M.; Kumar, G.; Nanda, S. Genome-wide identification and expression analysis of CRK gene family in chili pepper (Capsicum annuum L.) in response to Colletotrichum truncatum infection. J. Hortic. Sci. Biotechnol. 2022, 1–13. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B. Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef] [PubMed]
- Song, J.-M.; Guan, Z.; Hu, J.; Guo, C.; Yang, Z.; Wang, S.; Liu, D.; Wang, B.; Lu, S.; Zhou, R. Eight high-quality genomes reveal pan-genome architecture and ecotype differentiation of Brassica napus. Nat. Plants 2020, 6, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Song, J.M.; Liu, D.X.; Xie, W.Z.; Yang, Z.; Guo, L.; Liu, K.; Yang, Q.Y.; Chen, L.L. BnPIR: Brassica napus pan-genome information resource for 1689 accessions. Plant Biotechnol. J. 2021, 19, 412. [Google Scholar] [CrossRef] [PubMed]
- Kudla, J.; Batistič, O.; Hashimoto, K. Calcium signals: The lead currency of plant information processing. Plant Cell 2010, 22, 541–563. [Google Scholar] [CrossRef] [PubMed]
- Linder, M.E.; Deschenes, R.J. Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 2007, 8, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Moriya, K.; Nagatoshi, K.; Noriyasu, Y.; Okamura, T.; Takamitsu, E.; Suzuki, T.; Utsumi, T. Protein N-myristoylation plays a critical role in the endoplasmic reticulum morphological change induced by overexpression of protein Lunapark, an integral membrane protein of the endoplasmic reticulum. PLoS ONE 2013, 8, e78235. [Google Scholar] [CrossRef]
- Gu, X.; Zou, Y.; Su, Z.; Huang, W.; Zhou, Z.; Arendsee, Z.; Zeng, Y. An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 2013, 30, 1713–1719. [Google Scholar] [CrossRef]
- Moore, R.C.; Purugganan, M.D. The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. USA 2003, 100, 15682–15687. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef]
- Nekrutenko, A.; Makova, K.D.; Li, W.-H. The KA/KS ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Res. 2002, 12, 198–202. [Google Scholar] [CrossRef]
- Nielsen, R.; Yang, Z. Likelihood models for detecting positively selected amino acid sites and applications to the HIV-1 envelope gene. Genetics 1998, 148, 929–936. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z. User guide PAML: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol 2009, 3, dob0515. [Google Scholar] [CrossRef]
- Gao, F.; Chen, C.; Arab, D.A.; Du, Z.; He, Y.; Ho, S.Y. EasyCodeML: A visual tool for analysis of selection using CodeML. Ecol. Evol. 2019, 9, 3891–3898. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.-C.; Lu, Y.-T. Loss of AtCRK1 gene function in Arabidopsis thaliana decreases tolerance to salt. J. Plant Biol. 2013, 56, 306–314. [Google Scholar] [CrossRef]
- Schweighofer, A.; Hirt, H.; Meskiene, I. Plant PP2C phosphatases: Emerging functions in stress signaling. Trends Plant Sci. 2004, 9, 236–243. [Google Scholar] [CrossRef]
- Yadav, A.; Garg, T.; Singh, H.; Yadav, S.R. Tissue-specific expression pattern of calcium-dependent protein kinases-related kinases (CRKs) in rice. Plant Signal. Behav. 2020, 15, 1809846. [Google Scholar] [CrossRef]
- Boudsocq, M.; Willmann, M.R.; McCormack, M.; Lee, H.; Shan, L.; He, P.; Bush, J.; Cheng, S.-H.; Sheen, J. Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 2010, 464, 418–422. [Google Scholar] [CrossRef]
- Liu, H.-H.; Tian, X.; Li, Y.-J.; Wu, C.-A.; Zheng, C.-C. Microarray-based analysis of stress-regulated microRNAs in Arabidopsis thaliana. Rna 2008, 14, 836–843. [Google Scholar] [CrossRef]
- Zhou, L.; Liu, Y.; Liu, Z.; Kong, D.; Duan, M.; Luo, L. Genome-wide identification and analysis of drought-responsive microRNAs in Oryza sativa. J. Exp. Bot. 2010, 61, 4157–4168. [Google Scholar] [CrossRef]
- Jeyaraj, A.; Liu, S.; Zhang, X.; Zhang, R.; Shangguan, M.; Wei, C. Genome-wide identification of microRNAs responsive to Ectropis oblique feeding in tea plant (Camellia sinensis L.). Sci. Rep. 2017, 7, 1–16. [Google Scholar] [CrossRef]
- Chen, L.; Chen, L.; Zhang, X.; Liu, T.; Niu, S.; Wen, J.; Yi, B.; Ma, C.; Tu, J.; Fu, T. Identification of miRNAs that regulate silique development in Brassica napus. Plant Sci. 2018, 269, 106–117. [Google Scholar] [CrossRef] [PubMed]
- Jian, H.; Wang, J.; Wang, T.; Wei, L.; Li, J.; Liu, L. Identification of rapeseed microRNAs involved in early stage seed germination under salt and drought stresses. Front. Plant Sci. 2016, 7, 658. [Google Scholar] [CrossRef] [PubMed]
- Joshi, R.K.; Megha, S.; Basu, U.; Rahman, M.H.; Kav, N.N. Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS ONE 2016, 11, e0158784. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.; Koh, C.; Feurtado, J.A.; Tsang, E.W.; Cutler, A.J. MicroRNAs and their putative targets in Brassica napusseed maturation. BMC Genom. 2013, 14, 140. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Hao, M.; Wang, W.; Mei, D.; Wells, R.; Liu, J.; Wang, H.; Sang, S.; Tang, M.; Zhou, R. Integrative RNA-and miRNA-profile analysis reveals a likely role of BR and auxin signaling in branch angle regulation of B. napus. Int. J. Mol. Sci. 2017, 18, 887. [Google Scholar] [CrossRef]
- Cao, J.-Y.; Xu, Y.-P.; Zhao, L.; Li, S.-S.; Cai, X.-Z. Tight regulation of the interaction between Brassica napus and Sclerotinia sclerotiorum at the microRNA level. Plant Mol. Biol. 2016, 92, 39–55. [Google Scholar] [CrossRef]
- Pradhan, S.K.; Pandit, E.; Pawar, S.; Baksh, S.Y.; Mukherjee, A.K.; Mohanty, S.P. Development of flash-flood tolerant and durable bacterial blight resistant versions of mega rice variety ‘Swarna’ through marker-assisted backcross breeding. Sci. Rep. 2019, 9, 12810. [Google Scholar] [CrossRef]
- Khan, S.A.; Li, M.Z.; Wang, S.M.; Yin, H.J. Revisiting the Role of Plant Transcription Factors in the Battle against Abiotic Stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef]
- Nguyen, H.C.; Lin, K.H.; Ho, S.L.; Chiang, C.M.; Yang, C.M. Enhancing the abiotic stress tolerance of plants: From chemical treatment to biotechnological approaches. Physiol Plant 2018, 164, 452–466. [Google Scholar] [CrossRef]
- Li, Y.-B.; Han, L.-B.; Wang, H.-Y.; Zhang, J.; Sun, S.-T.; Feng, D.-Q.; Yang, C.-L.; Sun, Y.-D.; Zhong, N.-Q.; Xia, G.-X. The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 2016, 170, 2392–2406. [Google Scholar] [CrossRef]
- Liu, Y.; Xu, A.; Liang, F.; Yao, X.; Wang, Y.; Liu, X.; Zhang, Y.; Dalelhan, J.; Zhang, B.; Qin, M. Screening of clubroot-resistant varieties and transfer of clubroot resistance genes to Brassica napus using distant hybridization. Breed. Sci. 2018, 68, 258–267. [Google Scholar] [CrossRef] [PubMed]
- Saha, G.; Park, J.-I.; Jung, H.-J.; Ahmed, N.U.; Kayum, M.; Chung, M.-Y.; Hur, Y.; Cho, Y.-G.; Watanabe, M.; Nou, I.-S. Genome-wide identification and characterization of MADS-box family genes related to organ development and stress resistance in Brassica rapa. BMC Genom. 2015, 16, 178. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, R.; Geng, R.; Li, L.; Shan, Y.; Zhu, K.-M.; Wang, J.; Tan, X.-L. Genome-Wide Prediction, Functional Divergence and characterization of stress-responsive BZR transcription factors in B. napus. Front. Plant Sci. 2021, 12, 3077. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wan, Y.; Meng, X.; Zhang, X.; Yao, M.; Miu, W.; Zhu, D.; Yuan, D.; Lu, K.; Li, J. Genome-wide identification and analysis of MKK and MAPK gene families in Brassica species and response to stress in Brassica napus. Int. J. Mol. Sci. 2021, 22, 544. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Xie, Y.; Atif, A.; Wang, X.; Zhang, Y.; Tian, H.; Gao, Y. Identification and Expression Analysis of SLAC/SLAH Gene Family in Brassica napus L. Int. J. Mol. Sci. 2021, 22, 4671. [Google Scholar] [CrossRef]
- Acharya, B.R.; Raina, S.; Maqbool, S.B.; Jagadeeswaran, G.; Mosher, S.L.; Appel, H.M.; Schultz, J.C.; Klessig, D.F.; Raina, R. Overexpression of CRK13, an Arabidopsis cysteine-rich receptor-like kinase, results in enhanced resistance to Pseudomonas syringae. Plant J. 2007, 50, 488–499. [Google Scholar] [CrossRef]
- Zhang, X.; Han, X.; Shi, R.; Yang, G.; Qi, L.; Wang, R.; Li, G. Arabidopsis cysteine-rich receptor-like kinase 45 positively regulates disease resistance to Pseudomonas syringae. Plant Physiol. Biochem. 2013, 73, 383–391. [Google Scholar] [CrossRef]
- Lee, D.S.; Kim, Y.C.; Kwon, S.J.; Ryu, C.-M.; Park, O.K. The Arabidopsis cysteine-rich receptor-like kinase CRK36 regulates immunity through interaction with the cytoplasmic kinase BIK1. Front. Plant Sci. 2017, 8, 1856. [Google Scholar] [CrossRef]
- Du, D.; Liu, M.; Xing, Y.; Chen, X.; Zhang, Y.; Zhu, M.; Lu, X.; Zhang, Q.; Ling, Y.; Sang, X. Semi-dominant mutation in the cysteine-rich receptor-like kinase gene, ALS 1, conducts constitutive defence response in rice. Plant Biol. 2019, 21, 25–34. [Google Scholar] [CrossRef]
- Maere, S.; De Bodt, S.; Raes, J.; Casneuf, T.; Van Montagu, M.; Kuiper, M.; Van de Peer, Y. Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. USA 2005, 102, 5454–5459. [Google Scholar] [CrossRef]
- Ha, M.; Kim, E.-D.; Chen, Z.J. Duplicate genes increase expression diversity in closely related species and allopolyploids. Proc. Natl. Acad. Sci. USA 2009, 106, 2295–2300. [Google Scholar] [CrossRef] [PubMed]
- Myers, C.; Romanowsky, S.M.; Barron, Y.D.; Garg, S.; Azuse, C.L.; Curran, A.; Davis, R.M.; Hatton, J.; Harmon, A.C.; Harper, J.F. Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. Plant J. 2009, 59, 528–539. [Google Scholar] [CrossRef] [PubMed]
- Reddy, A.S.; Ali, G.S.; Celesnik, H.; Day, I.S. Coping with stresses: Roles of calcium-and calcium/calmodulin-regulated gene expression. Plant Cell 2011, 23, 2010–2032. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Huang, W. Calcium-dependent protein kinases in phytohormone signaling pathways. Int. J. Mol. Sci. 2017, 18, 2436. [Google Scholar] [CrossRef]
- Rigó, G.; Ayaydin, F.; Tietz, O.; Zsigmond, L.; Kovács, H.; Páy, A.; Salchert, K.; Darula, Z.; Medzihradszky, K.F.; Szabados, L. Inactivation of plasma membrane–localized CDPK-RELATED KINASE5 decelerates PIN2 exocytosis and root gravitropic response in Arabidopsis. Plant Cell 2013, 25, 1592–1608. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, D.; Yang, B.; Liu, W.-Z.; Mu, B.; Song, H.; Chen, B.; Li, Y.; Ren, D.; Deng, H. Arabidopsis CPK6 positively regulates ABA signaling and drought tolerance through phosphorylating ABA-responsive element-binding factors. J. Exp. Bot. 2020, 71, 188–203. [Google Scholar] [CrossRef]
- Latz, A.; Mehlmer, N.; Zapf, S.; Mueller, T.D.; Wurzinger, B.; Pfister, B.; Csaszar, E.; Hedrich, R.; Teige, M.; Becker, D. Salt stress triggers phosphorylation of the Arabidopsis vacuolar K+ channel TPK1 by calcium-dependent protein kinases (CDPKs). Mol. Plant 2013, 6, 1274–1289. [Google Scholar] [CrossRef]
- Almadanim, M.C.; Alexandre, B.M.; Rosa, M.T.; Sapeta, H.; Leitão, A.E.; Ramalho, J.C.; Lam, T.T.; Negrão, S.; Abreu, I.A.; Oliveira, M.M. Rice calcium-dependent protein kinase OsCPK17 targets plasma membrane intrinsic protein and sucrose-phosphate synthase and is required for a proper cold stress response. Plant Cell Environ. 2017, 40, 1197–1213. [Google Scholar] [CrossRef]
- Lamesch, P.; Berardini, T.Z.; Li, D.; Swarbreck, D.; Wilks, C.; Sasidharan, R.; Muller, R.; Dreher, K.; Alexander, D.L.; Garcia-Hernandez, M. The Arabidopsis Information Resource (TAIR): Improved gene annotation and new tools. Nucleic Acids Res. 2012, 40, D1202–D1210. [Google Scholar] [CrossRef]
- Chen, H.; Wang, T.; He, X.; Cai, X.; Lin, R.; Liang, J.; Wu, J.; King, G.; Wang, X. BRAD V3. 0: An upgraded Brassicaceae database. Nucleic Acids Res. 2022, 50, D1432–D1441. [Google Scholar] [CrossRef] [PubMed]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef] [PubMed]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; DeWeese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2010, 39 (Suppl. 1), D225–D229. [Google Scholar] [CrossRef]
- De Castro, E.; Sigrist, C.J.; Gattiker, A.; Bulliard, V.; Langendijk-Genevaux, P.S.; Gasteiger, E.; Bairoch, A.; Hulo, N. ScanProsite: Detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006, 34 (Suppl. 2), W362–W365. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef]
- Bologna, G.; Yvon, C.; Duvaud, S.; Veuthey, A.L. N-Terminal myristoylation predictions by ensembles of neural networks. Proteomics 2004, 4, 1626–1632. [Google Scholar] [CrossRef]
- Chou, K.-C.; Shen, H.-B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular localization. PloS One 2010, 5, e11335. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 2005, 571–607. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Stern, A.; Doron-Faigenboim, A.; Erez, E.; Martz, E.; Bacharach, E.; Pupko, T. Selecton 2007: Advanced models for detecting positive and purifying selection using a Bayesian inference approach. Nucleic Acids Res. 2007, 35 (Suppl. 2), W506–W511. [Google Scholar] [CrossRef]
- Yang, J.; Yan, R.; Roy, A.; Xu, D.; Poisson, J.; Zhang, Y. The I-TASSER Suite: Protein structure and function prediction. Nat. Methods 2015, 12, 7–8. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Yi, X.; Zhang, Z.; Ling, Y.; Xu, W.; Su, Z. PNRD: A plant non-coding RNA database. Nucleic Acids Res. 2015, 43, D982–D989. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Dai, X.; Zhao, P.X. psRNATarget: A plant small RNA target analysis server. Nucleic Acids Res. 2011, 39 (Suppl. 2), W155–W159. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Sarwar, R.; Jiang, T.; Ding, P.; Gao, Y.; Tan, X.; Zhu, K. Genome-wide analysis and functional characterization of the DELLA gene family associated with stress tolerance in B. napus. BMC Plant Biol. 2021, 21, 286. [Google Scholar] [CrossRef] [PubMed]
- Wittig, P.R.; Ambros, S.; Müller, J.T.; Bammer, B.; Álvarez-Cansino, L.; Konnerup, D.; Pedersen, O.; Mustroph, A. Two Brassica napus cultivars differ in gene expression, but not in their response to submergence. Physiol. Plant. 2021, 171, 400–415. [Google Scholar] [CrossRef] [PubMed]
- Xin, H.; Xianchao, N.; Pan, X.; Wei, L.; Min, Y.; Yu, K.; Lunwen, Q.; Wei, H. Comparative transcriptome analyses revealed conserved and novel responses to cold and freezing stress in Brassica napus L. G3: Genes Genomes Genet. 2019, 9, 2723–2737. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]



| Plant Name | Common Name | Number of CRKs | Chromosome Number | Genome Size in Mb | References |
|---|---|---|---|---|---|
| Gossypium barbadense | Cotton | 30 | 26 | 2500 | [19] |
| Arabidopsis thaliana | Thale cress | 8 | 5 | 135 | [36] |
| Solanum lycopersicum L. | Tomato | 6 | 12 | 950 | [20] |
| Malus domestica | Apple | 36 | 17 | 750 | [22] |
| Oryza sativa | Rice | 36 | 12 | 430 | [21] |
| Hevea brasiliensis | Rubber plant | 9 | 36 | 991 | [37] |
| Cucumis melo L. | Melon | 7 | 24 | 375 | [38] |
| Glycine max | Soybean | 91 | 10 | 1150 | [39] |
| Medicago truncatula | Legume | 6 | 16 | 430 | [40] |
| Sorghum bicolor | Great millet | 38 | 10 | 730 | [21] |
| Capsicum annuum L. | Bell pepper | 22 | 12 | 3058 | [41] |
| Cluster I | Cluster II | ϴI (Type I) | ϴII (Type II) | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Coefficient ϴI | S.E.ϴI | LRT | Qk > 0.7 | Critical Amino Acids | Coefficient ϴII | S.E.ϴII | Qk > 0.7 | Critical Amino Acids | ||
| Group A | Group B | 0.126034 | 0.106769 | 0.917987 | 4 | 60K, 64P, 74A, 115Y | 0.077776 | 0.119825 | 29 | 3I, 5H, 10E, 11Q, 12Q, 15Q, 16S, 19V, 21S, 23Q, 28K, 40L, 46P, 47S, 49A, 58I, 60K, 62P, 64P, 74A, 77A, 108S, 114H, 116E, 124G, 130C, 132A, 138T, 139G |
| Group A | Group C | −0.228125 | 0.027807 | nan | 2 | 89V, 117E | −0.147697 | 0.14611 | 21 | 5H, 36A, 37K, 38S, 41F, 44Y, 47S, 49A, 63F, 68S, 69S, 74A, 77A, 87N, 89V, 93E, 108S, 132A, 138T, 139L |
| Group B | Group C | 0.207912 | 0.141844 | 1.274511 | 5 | 3I,20S,39S,89V,117E, | 0.082203 | 0.129724 | 42 | 3I, 5H, 9I, 10E, 11Q, 12Q, 21S, 22Q, 24S, 26V, 28K, 29D, 36A, 37K, 38S, 41F, 44Y, 47S, 63F, 64P, 65A, 67A, 68S, 69S, 72L, 74A, 76K, 77A, 78P, 80P, 87N, 89V, 90A, 93E, 108S, 114H, 116E, 124G, 130C, 132A, 138T, 139L |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarwar, R.; Li, L.; Yu, J.; Zhang, Y.; Geng, R.; Meng, Q.; Zhu, K.; Tan, X.-L. Functional Characterization of the Cystine-Rich-Receptor-like Kinases (CRKs) and Their Expression Response to Sclerotinia sclerotiorum and Abiotic Stresses in Brassica napus. Int. J. Mol. Sci. 2023, 24, 511. https://doi.org/10.3390/ijms24010511
Sarwar R, Li L, Yu J, Zhang Y, Geng R, Meng Q, Zhu K, Tan X-L. Functional Characterization of the Cystine-Rich-Receptor-like Kinases (CRKs) and Their Expression Response to Sclerotinia sclerotiorum and Abiotic Stresses in Brassica napus. International Journal of Molecular Sciences. 2023; 24(1):511. https://doi.org/10.3390/ijms24010511
Chicago/Turabian StyleSarwar, Rehman, Lei Li, Jiang Yu, Yijie Zhang, Rui Geng, Qingfeng Meng, Keming Zhu, and Xiao-Li Tan. 2023. "Functional Characterization of the Cystine-Rich-Receptor-like Kinases (CRKs) and Their Expression Response to Sclerotinia sclerotiorum and Abiotic Stresses in Brassica napus" International Journal of Molecular Sciences 24, no. 1: 511. https://doi.org/10.3390/ijms24010511
APA StyleSarwar, R., Li, L., Yu, J., Zhang, Y., Geng, R., Meng, Q., Zhu, K., & Tan, X.-L. (2023). Functional Characterization of the Cystine-Rich-Receptor-like Kinases (CRKs) and Their Expression Response to Sclerotinia sclerotiorum and Abiotic Stresses in Brassica napus. International Journal of Molecular Sciences, 24(1), 511. https://doi.org/10.3390/ijms24010511







