Abstract
Beyond the influence of lifestyle-related risk factors for myocardial infarction (MI), the mechanisms of genetic predispositions for MI remain unclear. We sought to identify and characterize differentially expressed genes in early-onset MI in a translational approach. In an observational case–control study, transcriptomes from 112 early-onset MI individuals showed upregulated G protein-coupled receptor 15 (GPR15) expression in peripheral blood mononuclear cells compared to controls (fold change = 1.4, p = 1.87 × 10−7). GPR15 expression correlated with intima-media thickness (β = 0.8498, p = 0.111), C-reactive protein (β = 0.2238, p = 0.0052), ejection fraction (β = −0.9991, p = 0.0281) and smoking (β = 0.7259, p = 2.79 × 10−10). The relation between smoking and MI was diminished after the inclusion of GPR15 expression as mediator in mediation analysis (from 1.27 (p = 1.9 × 10−5) to 0.46 (p = 0.21)). The DNA methylation of two GPR15 sites was 1%/5% lower in early-onset MI individuals versus controls (p = 2.37 × 10−6/p = 0.0123), with site CpG3.98251219 significantly predicting risk for incident MI (hazard ratio = 0.992, p = 0.0177). The nucleotide polymorphism rs2230344 (C/T) within GPR15 was associated with early-onset MI (odds ratio = 3.61, p = 0.044). Experimental validation showed 6.3-fold increased Gpr15 expression in an ischemic mouse model (p < 0.05) and 4-fold increased Gpr15 expression in cardiomyocytes under ischemic stress (p < 0.001). After the induction of MI, Gpr15gfp/gfp mice showed lower survival (p = 0.042) and deregulated gene expression for response to hypoxia and signaling pathways. Using a translational approach, our data provide evidence that GPR15 is linked to cardiovascular diseases, mediating the adverse effects of smoking.
1. Introduction
Cardiovascular diseases (CVD) are a significant global burden and a main cause of death worldwide [1]. The leading condition of the global CVD burden is MI, which is most commonly induced by atherosclerosis [2]. As a chronic inflammatory disease of the arteries, atherosclerosis progresses slowly throughout an individual’s lifetime, and lifestyle factors, such as smoking, diet and physical inactivity, promote its progression [3,4,5]. Therefore, MI in young adults is a rare phenomenon compared to patients older than 50 years [6]. Traditional cardiovascular risk factors, such as diabetes, hypertension and hypercholesterolemia as well as a family history of ischemic heart disease and obesity, are more prevalent in early-onset MI subjects [7,8] and a specific genetic background is likely in those patients [9].
In recent years, the application of genome-wide association studies (GWAS) has led to the discovery of more than hundred genetic loci contributing to the risk of CVD [10]. However, a substantial proportion of the CVD heritability is still unexplained and the functional role of most GWAS loci is unknown [10]. To date, only a few variants have been found that specifically explain the risk of premature CVD [11] and explanations about the molecular mechanisms of the genetic predisposition remain elusive [12].
G protein-coupled receptors (GPCRs) are important cell surface receptors, mediating signals between cells and environment. More than 200 GPCRs are expressed in the heart, where they are of prime importance for cardiovascular homeostasis, regulating essential functions, such as contractility, heart rate and vascular tone [13,14,15]. Despite many GPCRs being involved in CVD, only about 4% of all GPCRs are targeted by clinical therapeutics [13]. Amongst them are important cardiovascular drugs, such as β-blockers, angiotensin II receptor blockers and angiotensin-converting enzyme inhibitors [15]. Several GPCRs have already been linked to cardiovascular risk factors, such as the GPR15 gene (encoding the G protein-coupled receptor 15), whose DNA methylation sites showed strong association with smoking [16,17], indicating a regulation of GPR15 gene expression by smoking. The further identification and characterization of GPCRs will likely promote the discovery of novel regulatory mechanisms and bears the potential for new (cardiovascular) drug targets and biomarkers. To date, no GPCR is known as a potential drug target or biomarker for early-onset MI. To our knowledge, no large-scale transcriptome profiling has been performed in early-onset MI individuals so far.
The prespecified aims of this study were to (i) identify target transcripts for MI (discovery), (ii) to independently validate the target (replication), (iii) to experimentally functionally characterize the role of the target in MI and (iv) to evaluate the potential for MI risk prediction of the target. Hence, the present study combined transcriptomic analyses in early-onset MI individuals (aged < 50 years at index event) and healthy controls with experimental analyses of mouse models, cell culture models and functional molecular analyses on the level of DNA methylation in a translational approach (Figure 1).
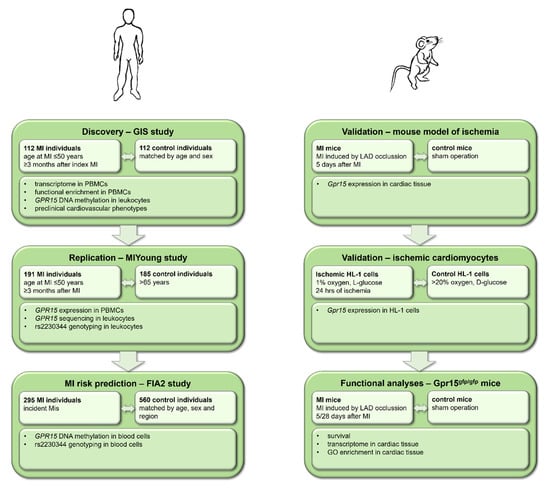
Figure 1.
Study workflow including data from human studies and mouse experiments. In a translational approach, human data from the GIS and MIYoung studies were used for the discovery and replication of results, whereas data from the FIA2 study were analyzed for myocardial infarction (MI) risk prediction. Experimental validation and functional analyses were performed in mouse models of ischemia, ischemic cardiomyocytes and Gpr15gfp/gfp mice. PBMCs = peripheral blood mononuclear cells.
2. Results
2.1. Expression of GRP15 Is Increased in Early-Onset Myocardial Infarction
To identify transcripts associated with early-onset myocardial infarction on the level of mRNA, a microarray transcriptomics analysis was conducted in a discovery cohort (GIS) and validated by TaqMan qPCR in an independent cohort (MIYoung). In both the discovery and validation cohort, mRNA isolated from PBMCs was used to perform gene expression analyses.
In the discovery cohort, gene expression data of 112 early-onset MI individuals and 112 age and sex-matched healthy controls were compared using a case–control approach. The characteristics of the GIS cohort are provided in Table S1. Median age at the index event was 43 years for subjects suffering early-onset MI. Controls had a lower prevalence of classical cardiovascular risk factors, such as diabetes, obesity, hypertension and a family history of MI and higher LDL-cholesterol levels.
Out of 27,068 transcripts, 183 transcripts were differentially expressed between early-onset MI individuals and controls, with 82 transcripts being upregulated and 101 transcripts being downregulated in the early-onset MI group (Figure 2A, Table S2). Among all genes analyzed, GPR15 (encoding the G protein-coupled receptor 15) showed the highest differential expression with a fold change of 1.43 (p = 1.87 × 10−7, Figure 2B).
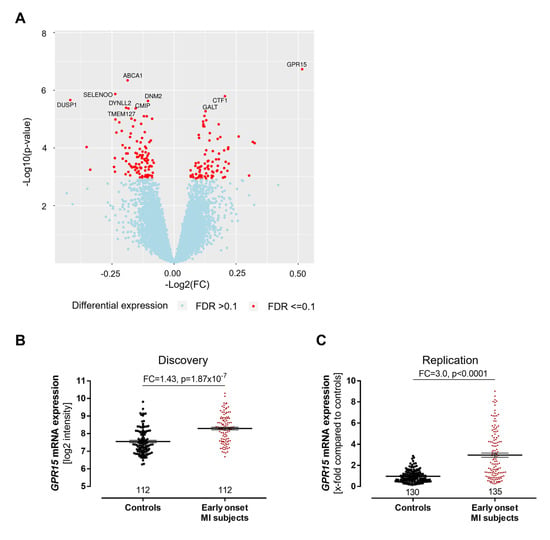
Figure 2.
Increased GPR15 expression after myocardial infarction. (A) In a transcriptome analysis of peripheral blood mononuclear cells (PBMCs) from 112 early-onset myocardial infarction (MI) patients and 112 age- and gender-matched controls from the GIS discovery cohort, GPR15, ABCA1, SELO, CTF1 and DUSP1 were the top differentially expressed genes with GPR15 that displayed the highest fold change and lowest p-value. Linear mixed model adjusted for hypertension, smoking status, body mass index, diabetes and low-density lipoprotein/high-density lipoprotein ratio. FDR = false discovery rate using the q-value method. (B) In the GIS discovery cohort, GPR15 mRNA expression in PBMCs was increased 1.4-fold in early-onset MI individuals (<50 years) compared to sex- and age-matched controls as determined by microarray. Linear mixed regression model adjusted for hypertension, smoking status, body mass index, diabetes and low-density lipoprotein/high-density lipoprotein ratio and technical covariates. (C) Increased GPR15 expression was validated by TaqMan qPCR in the MIYoung replication cohort in PBMCs from early-onset MI individuals compared to healthy controls over 65 years. Mann–Whitney U test. Data are shown as mean ± standard error of the mean.
Functional enrichment analysis was performed to investigate the involvement of differentially expressed genes in biological pathways. Consistently, pathway analysis revealed key cardiovascular and inflammatory signaling processes, driven by G protein signaling, to be affected in individuals with early-onset MI. The top pathways were (i) the P2Y purigenic receptor signaling pathway (8 out of 125 pathway genes), (ii) the GNRH signaling pathway (8 out of 135 pathway genes) and (iii) the renin–angiotensin signaling pathway (7 out of 113 pathway genes), indicating the important role of G protein signaling in the pathophysiology of MI.
The validation of these findings was performed by TaqMan qPCR in an independent cohort study of early-onset MI subjects, the MIYoung study, including 191 early-onset MI subjects and 185 healthy controls. The study characteristics of the MIYoung cohort are provided in Table S3. Similar to the results from our discovery cohort, GPR15 expression was higher in early-onset MI individuals compared to the controls (3.0-fold, p < 0.0001, Figure 2C).
2.2. Relation of GPR15 Expression with DNA Methylation, Myocardial Infarction and Smoking
As previously described, GPR15 mRNA expression and GPR15 DNA methylation significantly associate with smoking and with each other [17,18]. The GPR15 DNA methylation sites CpG1, CpG2 and CpG3 are located within the coding sequence of the GPR15 gene (Figure S1). To analyze whether the changes of GPR15 expression in early-onset MI subjects are affected by smoking, the expression of GPR15 in PBMCs was compared between early-onset MI subjects and controls—stratified by smoking status in the GIS study. GPR15 showed higher expression in the MI individuals (p = 0.004 for never smokers and p = 0.024 for current smokers, Figure 3A). In MI individuals, not only was GPR15 mRNA increased but leukocytic GPR15 DNA methylation was also significantly lower compared to controls (p = 0.0021 for CpG1 and p = 0.0504 for CpG3, Figure 3B). Even after adjusting for smoking, the DNA methylation of CpG1 and CpG3 was significantly different between MI individuals and controls (p = 2.4 × 10−6 and p = 0.0123, respectively). Furthermore, in early-onset MI individuals, GPR15 was not only associated to MI but also to preclinical cardiovascular phenotypes, including ejection fraction, intima-media thickness and CRP as a surrogate marker for inflammation. The correlation to intima-media-thickness remained significant after adjustment for smoking (β = 0.8021, p = 0.0086, Figure 3C). To assess whether the observed association between smoking and MI was mediated by GPR15, causal mediation analysis was performed in the GIS-GHS study (Figure 3D). The effect of smoking on MI was fully mediated for current smokers and partially mediated for ex-smokers by GPR15 mRNA expression (coefficients changed from 1.27 (p = 1.9 × 10−5) to 0.46 (p = 0.21) and from 1.64 (p = 1 × 10−8) to 1.27 (p = 4.3 × 10−5), respectively, when including GPR15 mRNA expression to the linear model). The regression of smoking to GPR15 mRNA expression (coefficient for current smoking 1.31, p < 2 × 10−16; coefficient for ex-smokers 0.74, p = 2.7 × 10−10) and GPR15 mRNA expression to MI (coefficient 0.64, p = 2.8 × 10−4) were significant. The total effect of smoking mediated by GPR15 mRNA expression was 0.43 (p < 2 × 10−16) and the unstandardized indirect effect was 0.11 (95% CI: 0.057–0.18, p < 2 × 10−16), i.e., 26.2% of the total (95% CI 13–51%).
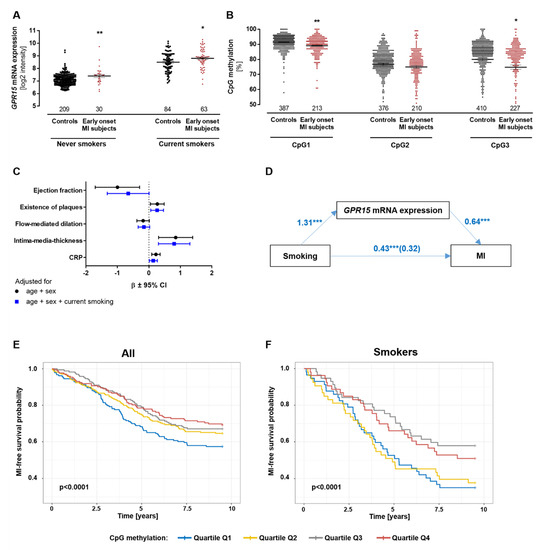
Figure 3.
Risk for myocardial infarction predicted by GPR15 DNA methylation independent of smoking status. (A) In the GIS discovery cohort, early-onset MI subjects showed significantly higher GPR15 mRNA expression levels in BPMCs compared to controls, independent of smoking status. Mann–Whitney U test. (B) DNA methylation in leukocytes was 2–5% lower in MI cases compared to controls (Cpg1 p = 0.0021/p = 2.37 × 10−6, CpG3 p = 0.0201/p = 0.123, both adjusted for smoking). Linear mixed models adjusted for age/sex; Bonferroni correction, * = p < 0.05, ** = p < 0.01 compared to respective controls; mean ± standard error of the mean. (C) Correlation between GPR15 expression in PBMCs and preclinical cardiovascular phenotypes. Intima-media-thickness was significantly associated with GPR15 expression after adjusting for current smoking status (p = 0.0086). n = 234, linear mixed model, significance after Bonferroni correction, CRP = C-reactive protein (log scale), β = standardized beta coefficient, CI = confidence interval. (D) The effect of current smoking on MI was fully mediated by GPR15 expression, as the coefficient changed from 1.27 (significant p = 1.9 × 10−5) to 0.46 (non-significant p = 0.21) when including GPR15 expression. The total effect of current smoking mediated by GPR15 expression was 0.43 (p < 2 × 10−16; average direct effects = 0.32, p < 2 × 10−16) and the unstandardized indirect effect was 0.11 (95% CI: 0.057–0.18, p < 2 × 10−16); 1000 simulations with bootstrapping).*** = p < 0.001 (E,F) In the FIA2 study with incident MI cases and matched controls, MI-free survival probability was significantly higher in subjects with high versus low methylation of the GPR15 DNA methylation site CpG3 in all samples (E) and current smokers only (F) as visualized in quartiles and log-rank tests.
2.3. GPR15 DNA Methylation Can Predict Risk of Myocardial Infarction
Based on the finding that the GPR15 DNA methylation of sites CpG1 and CpG3 associates with early-onset MI, MI risk prediction properties were calculated for GPR15 DNA methylation levels and incident MI individuals. Associations between the percentages of GPR15 DNA methylation and incident MI were analyzed in the prospective nested case–control FIA2 study (comprising MI individuals and matched controls; FIA2 study characteristics are given in Table S4). The GPR15 DNA methylation of CpG1 and CpG3 was significantly lower in current smokers compared to never smokers and ex-smokers (Figure S2), validating previous results [17]. CpG3 methylation significantly predicted the risk for incident MI after adjustment for smoking (HR = 0.992, p = 0.0177, Table S5, Figure S3). As shown in Kaplan–Meier curves in Figure 3E/F, MI risk was lower in individuals with higher CpG3 DNA methylation levels compared to individuals with lower CpG3 DNA methylation levels. This was shown in the overall cohort (Figure 3E) as well as in smokers only (Figure 3F; p < 0.0001). Hence, the probability of MI-free survival was higher in individuals with higher CpG3 DNA methylation levels compared to individuals with lower CpG3 DNA methylation levels.
2.4. GPR15 SNP rs2230344 Associates with Early-Onset Myocardial Infarction
To identify genetic variants linked to GPR15 expression and myocardial infarction, the entire GPR15 gene was sequenced in a subgroup of early-onset MI individuals and healthy controls from the MIYoung study. The subgroup consisted of n = 32 individuals having had an MI at an age below 40 years. The characteristics of this subgroup of early-onset MI individuals and controls (n = 32) are given in Table S6. The single nucleotide polymorphism (SNP) rs2230344 (C/T) showed a minor allele frequency (T) in early-onset MI individuals of 9% compared to 22% in healthy controls (OR = 3.74, p = 0.048 for number of C alleles; adjusted for BMI). Validation by qPCR showed consistent results, with a rs2230344-T minor allele frequency of 13% for early-onset MI individuals and 23% for healthy controls (OR = 3.61, p = 0.044 for number of C alleles; adjusted for BMI).
rs2230344 is located in close proximity to the GPR15 DNA methylation sites (Figure S1, USCS Genome Browser [19]) and thus might influence GPR15 mRNA expression or DNA methylation. However, no significant association was shown between rs2230344 and GPR15 mRNA expression or DNA methylation in the MIYoung study (Figure S4, Table S7) nor did the rs2230344 minor T allele predict the MI risk in the FIA2 study (Table S8).
2.5. Experimental Validation of Increased Gpr15 Expression after Ischemia
To assess our findings of an increased GPR15 expression after MI in experimental models, Gpr15 mRNA expression was measured in a mouse model of infarction-related ischemia as well as in an ischemic cell culture model. Cardiac Gpr15 expression significantly increased five days after MI in the scared infarct zones (IZ) (6.32-fold, p < 0.05, Figure 4A). On cellular level, Gpr15 mRNA expression was determined in a model of ischemic HL-1 cardiomyocytes. Here, the 24 h induction of ischemic stress significantly increased Gpr15 mRNA expression 4-fold in a cell culture model of ischemic cardiomyocytes compared to controls (p < 0.001, Figure 4B), affirming the increased cardiac Gpr15 expression in infarcted mouse hearts.
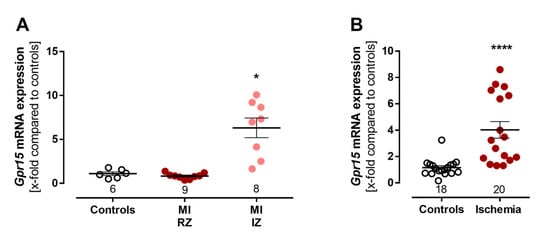
Figure 4.
Experimental validation of increased Gpr15 expression after induced myocardial infarction. (A) Validation of increased Gpr15 expression in a mouse model of myocardial infarction. Cardiac Gpr15 expression was increased 6.3-fold in the infarct zone (IZ), but not in the remote zone (RZ), five days after MI in male C57BL/6J (B6) mice. Kruskal–Wallis test with Dunn’s multiple comparisons test. (B) Validation of increased cardiac Gpr15 expression in a cell culture model of ischemic cardiomyocytes. HL-1 cells were incubated with 20% oxygen/D-glucose (control) and 1% oxygen/L-glucose (ischemia) for 24 h. Gpr15 expression was increased 4-fold in ischemic cardiomyocytes (n = 4 experiments). Mann–Whitney U test. Expression levels were normalized to the reference 18S rRNA and to the expression levels of the control group and plotted as x-fold expression using the formula 2−ΔΔCt. * = p < 0.05, **** = p < 0.0001 compared to the respective control group. Data are shown as mean ± standard error of the mean.
2.6. Gpr15 Knockout Affects Survival and Cardiac Remodeling in a Murine Model of Myocardial Infarction
In humans, we showed increased GPR15 expression after myocardial infarction, combined with altered G protein signaling pathways. To characterize the role of Gpr15 in MI and to identify molecular pathways through which Gpr15 acts in the heart, Gpr15gfp/gfp mice were subjected to MI induced by LAD ligation (Figure S5). Gpr15gfp/gfp mice did not show significant hemodynamic differences compared to wildtype (WT) mice. However, Kaplan–Meier survival curves revealed a significantly lower survival 28 days post MI in Gpr15gfp/gfp compared to WT mice (p = 0.0424, Figure 5A).
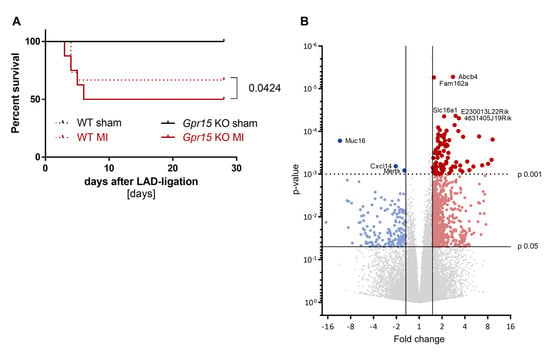
Figure 5.
Reduced survival and differential gene expression in Gpr15gfp/gfp mice after induced myocardial infarction. (A) Survival was slightly lower in Gpr15gfp/gfp mice compared to wildtype mice (p = 0.0424, n = 5/6 sham and 15/16 MI). log-rank test (B) Gene expression measured by 3′ mRNA sequencing of cardiac tissue from the IZ from nine Bl6 wildtype (WT) and eight Gpr15gfp/gfp mice 5 days after MI surgery. Downregulated genes in cardiac tissue from Gpr15gfp/gfp compared to WT mice are depicted in blue, while red highlights upregulated genes. Fold change and p-value are displayed in the volcano plot to identify significant differences in gene expression. Further restricted by p-value ≤ 0.001, 84 top differentially expressed genes were identified (81 upregulated and three downregulated genes). Gene symbols for five upregulated genes with the lowest p-value and for the downregulated genes are displayed. Red and blue colors highlight significantly regulated genes with p-value < 0.05 (small dots) or p-value < 0.001 (large dots).
To identify molecular pathways affected by Gpr15, RNA from cardiac tissue from the IZ from WT and Gpr15gfp/gfp mice 5 days after MI was analyzed by 3’ mRNA sequencing. In the IZ, 822 genes were identified as significant differentially expressed (p < 0.05) between Gpr15gfp/gfp and WT mice (Figure 5B, Table S9). Out of these 822 genes, 656 were significantly upregulated (red) in Gpr15gfp/gfp mice and 166 genes were significantly downregulated (blue). The top differentially expressed genes were Myl7, Myl4 (upregulation) and Epo and Ccrl2 (downregulation). The resulted GO term analyses for biological processes that were differentially regulated in the IZ are plotted in Figure 6, the GO term analyses for molecular functions and for cellular components are plotted in Figures S6 and S7, respectively. The identified GO terms affected by Gpr15 knockout include desensitization of GPCR, response to hypoxia, intracellular signal transduction, adaptation of signaling pathway and multicellular organismal signaling (Figure 6).
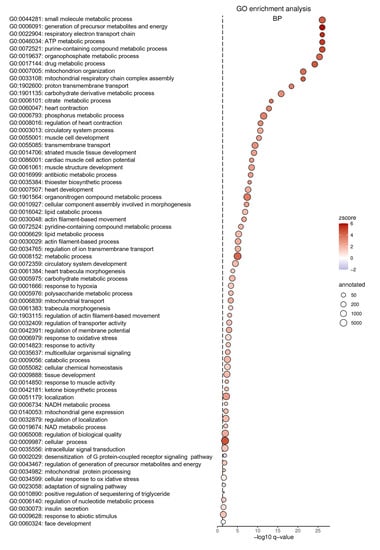
Figure 6.
Altered GO terms in Gpr15gfp/gfp mice after induced myocardial infarction. Significant gene ontology (GO) terms for biological processes (BP) clustered utilizing Revigo. Differentially expressed GO terms for Gpr15gfp/gfp mice include response to hypoxia, desensitization of GPCRs and intracellular signal transduction. The GO term ID and the description were plotted according to their p-value as dot plot. On the x-axis, the negative decadic logarithm of the p-value of the GO terms is shown. The individual GO terms are displayed on the y-axis, sorted according to their p-value. The size of the dots represents the number of all genes that are annotated in the respective GO term. The color of the dots is based on the GO term’s z-score. Whether the majority of genes in a GO term is up- or downregulated is indicated by the color gradient from red (upregulated) to blue (downregulated), respectively.
3. Discussion
Understanding the molecular mechanisms of CVD is of great importance for the identification of novel biomarkers and the development of therapeutic targets for CVD. In this study, transcriptomic analyses in early-onset MI individuals were performed to identify novel genes related to CVD. The G protein-coupled receptor 15 had previously been implicated to play a role in inflammatory diseases [20,21,22,23,24]. Even though the identification of GPR15 signaling, as well as the identification of GPR15 ligand-binding, has progressed significantly within the last years and GPR15 activation has been demonstrated in vascular endothelial cells, insights into the physiological and pathophysiological role of GPR15 remain scarce [25]. Here we show that (i) GPR15 is upregulated in early-onset MI subjects; (ii) GPR15 mRNA expression mediates the effect of smoking on MI; (iii) GPR15 DNA methylation predicts risk for MI, independently of smoking; (iv) SNP rs2230344 is associated with early-onset MI; and v) Gpr15 knockout reduces survival after MI and affects response to hypoxia and GPCR pathways in an experimental mouse model of myocardial infarction.
To identify molecular mechanisms for CVD, we focused on RNA expression in early-onset MI individuals, being under stable disease conditions for at least three months after the index event to prevent a bias by post-acute MI inflammatory and remodeling processes. The GPR15 gene expression was most significantly upregulated in early-onset MI individuals. Additionally, GPCR pathways were shown to be regulated in early-onset MI individuals, indicating an important role of G protein signaling in the pathophysiology of MI. Consistently, GPCRs are known to play important roles in cardiovascular function and CVD [14]. GPR15 itself is a GPCR as well, which supports its connection to CVD. Furthermore, GPR15 has been described to act as a T cell homing receptor and to be involved in inflammatory diseases [20,21,22,23,24]. The development of CVD involves chronic inflammatory processes and inflammation also plays an important role after MI [3,26]. In early-onset MI individuals, GPR15 expression was also associated with subclinical cardiovascular phenotypes and inflammation, suggesting a potential involvement in or the potential use of GPR15 as a surrogate marker for the development of early-onset MI.
Consistent with the transcriptome analysis, the increased GPR15 expression was replicated in a second cohort with early-onset MI individuals. The controls in the replication cohort were older than 65 years and with increasing age, the influence of genetic CVD heritability is less prominent than the influence of cardiovascular risk factors, suggesting that GPR15 might play a role in CVD on a genetic level. Furthermore, increased cardiac Gpr15 expression was validated in a mouse model of MI as well as in ischemic cardiomyocytes. These data indicate a specific response to conditions of MI regarding myocardial ischemia, which is in agreement with the human data. Consequently, our data imply that GPR15 might play a role in the pathogenesis of acute MI and in the conditions of ischemia, such as artery narrowing by plaques.
Previously, we described a substantial association between smoking, GPR15 DNA methylation and GPR15 mRNA expression [17]. DNA methylation within the gene body, as measured in the current study, has been described in relation to mRNA expression levels [27,28]. Specifically, for GPR15, tobacco smoking seems to alter blood cell composition, thereby affecting GPR15 expression levels [29]. In the current study, we show not only higher GPR15 mRNA expression but also lower GPR15 DNA methylation in early-onset MI individuals. Smoking itself poses a strong risk factor for CVD [5]. Interestingly, the associations between GPR15 mRNA expression and GPR15 DNA methylation with myocardial infarction were found to be independent of smoking status. The effect of smoking on MI, however, was mediated by GPR15 mRNA expression. Hence, our data suggest that changes in GPR15 expression or DNA methylation and association to early-onset MI might mediate smoking effects while not excluding other unknown mechanisms.
On the genetic level, our data indicated the lower allele frequencies of the minor allele of SNP rs2230344 in early-onset MI individuals. Potentially, rs2230344 might influence the development of early-onset MI via an effect on GPR15 DNA methylation, as previously described in African Americans [30] and/or by affecting GPR15 expression. Our data, however, did not reveal an association between rs2230344 and GPR15 DNA methylation. Nevertheless, rs2230344 is located within a region of the GPR15 gene coding for a transmembrane domain. The minor T allele is a missense variant, which leads to an amino acid exchange from proline to serine. The lower abundance of the T allele in early-onset MI subjects might cause changes in protein conformation and thus the integration of the GPR15 protein into the plasma membrane or even affect GPR15 protein function.
In order to characterize the role of Gpr15 in MI and to identify underlying molecular mechanisms, we experimentally assessed the role of Gpr15 in a mouse model of MI induced by LAD. After the induction of MI, Gpr15gfp/gfp mice had reduced survival compared to wildtype mice. On a molecular level, we showed that genes deregulated in Gpr15gfp/gfp mice after MI induction belong to the GO terms desensitization of GPCR, signaling pathways (such as the adaptation of signaling pathway, intracellular signal transduction) and response to hypoxia pathways. Similar to the molecular mechanisms in the Gpr15gfp/gfp mice, in the human early-onset MI transcriptome analysis, GPCR signaling pathways were affected. Taken together, these mouse data indicate that Gpr15 might be involved in signaling processes in response to hypoxia after MI, potentially explaining why Gpr15gfp/gfp mice showed reduced survival. Given the level of significance, the relevance and reproducibility of the reduced survival rate has to be elucidated in further studies.
In our study, Gpr15 was involved in immune response mechanisms in a mouse model of MI. Consistently, our human data showed an association between GPR15 mRNA expression with CRP as a marker of inflammation. This is in line with previous human and mouse studies, which show an involvement of GPR15 in inflammatory diseases, such as inflammatory bowel disease and rheumatoid arthritis [20,21]. Whether or not, and under which conditions, GPR15 could have pro- or anti-inflammatory properties, possibly varying between conditions and species [31], is still to be determined and requires further investigations.
Our presented work comprises large-scale human data, including longitudinal data and different omics data, combined with experimental models. Unique data from early-onset prevalent and incident MI individuals showed changes in GPR15 mRNA expression as well as GPR15 DNA methylation and rs2230344 allele frequency. The DNA methylation of the GPR15 CpG3 site even predicted the MI risk. Nevertheless, limitations include sample availability, ethnicity and effect sizes. Human samples were available from Caucasian individuals. Ethnicity largely affects molecular mechanisms, e.g., as the association between the SNP rs2230344 genotype and GPR15 DNA methylation has previously been reported for African Americans but not for Caucasians [30]. As only DNA but no RNA is available in the FIA2 study, the prediction of MI risk for GPR15 mRNA levels was not possible. Changes in mRNA expression and DNA methylation might further be caused by variations in cell composition; however, no data on cell composition has been measured and is thus not available. Even though gene expression and DNA methylation changes are of small effect sizes, they are significant and persistent throughout human and mouse data. In mice, Gpr15 expression was increased in ischemic cardiac tissue. A possibility of evaluating the involvement of Gpr15 in the pathogenesis of MI, a combination of an atherosclerosis mouse model (e.g., apolipoprotein E knockout mice) and Gpr15 knockout mouse model might be suitable.
4. Materials and Methods
The present study combined transcriptomic analyses in early-onset MI individuals (aged < 50 years at index event) and healthy controls with experimental analyses of mouse models, cell culture models and functional molecular analyses on the level of DNA methylation in a translational approach. The study workflow is shown in Figure 1.
4.1. Human Cohorts
All human cohorts followed the Declaration of Helsinki. All subjects gave written informed consent. Detailed cohort descriptions are shown in Table S10.
4.1.1. Gutenberg Young Myocardial Infarction Study (GIS)
Caucasian individuals with a history of early-onset MI (≤50 years) were recruited into the Gutenberg Young Myocardial Infarction Study (GIS). Individuals were invited to participate in GIS according to the following criteria: (i) confirmed MI (positive ECG, increase in biomarkers of cardiac necrosis (troponin, creatine-kinase MB) or clinical symptoms) and proven by coronary angiography, (ii) age at myocardial infarction ≤50 years (as described previously [32]) and (iii) stable disease conditions for at least three months before study participation. The study protocols and sampling design were approved by the local ethics committee of the Medical Chamber of Rhineland-Palatinate, Germany (ethical approval code 837.211.08 (6208)). Caucasian individuals without manifest cardiovascular disease and recruited as part of the Gutenberg Health Study (GHS) [33,34] were used as controls. Out of 234 MI patients and 419 controls, 112 case-control pairs were matched by age and gender. The study protocols and sampling design were approved by the local ethics committee of the Medical Chamber of Rhineland-Palatinate, Germany (ethical approval code 837.020.07 (5555)).
The participants of GIS and GHS visited the same study center simultaneously. Self-reported smoking status was classified as follows: current smokers (including occasional smokers), ex-smokers (smoking cessation at least six weeks before study participation) and never smokers. Cumulative smoking exposure was evaluated by pack years (one pack year = smoking of 20 cigarettes per day for one year) for current smokers and ex-smokers. Peripheral blood mononuclear cells (PBMC) RNA was isolated via Trizol extraction and leukocytic DNA was isolated as described by Miller et al. from buffy-coated ethylenediamine tetra-acetic acid blood samples [35,36].
4.1.2. Young Myocardial Infarction Study (MIYoung)
The MIYoung study is a clinical cohort study from the University Heart and Vascular Center Hamburg, Medical University Hamburg-Eppendorf, including subjects with a history of early-onset MI at an age < 50 years and under stable disease conditions for at least three months (n = 191) and healthy controls over 65 years (n = 185). The study protocols and sampling design were approved by the local ethics committee of the University Medical Center Hamburg-Eppendorf, Germany (ethical approval code PV4137). Self-reported smoking status was classified as follows: current smokers, ex-smokers (smoking cessation at least six weeks before study participation) and never smokers. From all participants, PBMC RNA and leukocytic DNA was isolated.
4.1.3. First-Ever Myocardial Infarction Study 2 (FIA2)
The FIA 2 study is a prospective nested case-referent study from Northern Sweden, including incident MI cases matched with two healthy controls for age, sex, date of health examination and geographical region [37]. Subjects were recruited from the Northern Sweden MONitoring of Trends and Determinants in CArdiovascular Diseases (MONICA) project, the Västerbotten Intervention Program (VIP), and the Mammary Screening Program (MSP). MI events were identified through the screening of hospital discharge records, general practitioners’ reports and death certificates [38]. Smoking status was classified as smokers (daily smokers), ex-smokers and non-smokers. Blood samples obtained at the baseline health survey were used, and sampling was carried out 3.9 years (interquartile range 3.6 years) before the MI event. DNA was isolated by phenol-chloroform extraction or salting-out precipitation. The study was approved by the local ethics committee (Dnr 05-142M, with an additional review on 19 January 2009).
4.2. Mouse Models
Male C57BL/6J (B6) mice at the age of 8 to 10 weeks were used for the presented animal mouse models. Maximal five mice per cage were kept in standard cages under a 12 h:12 h light:dark cycle, constant temperature and humidity and received standard food and water ad libitum. At the time of experiments, animals were six- to twelve-weeks-old. All animal investigations conform to the Guide for the Care and Use of Laboratory Animals published by the US NIH [39]. The study protocols were approved by the Hamburg Authority for Health and Consumer Protection (approval codes G060/15) and the Regional Office for Health and Social Affairs Berlin (approval code G0055/11).
Gpr15gfp/gfp mice had a GFP knock-in, resulting in a partial deletion of Gpr15 exon 1, as described previously [20]. Male wildtype and Gpr15gfp/gfp mice were housed together and were randomly allocated by housing cages to either intervention or control group. To induce MI (intervention), mice were subjected to the permanent ligation of the left anterior descending artery (LAD). Mice of the control group underwent a sham operation, followed by recovery for five (n = 6/9) or 28 days (n = 7/16). All procedures were carried out as previously described in detail [40,41]. Briefly, mice were anesthetized with isoflurane for the surgical procedure and buprenorphine was given as analgesic therapy. The heart was accessed via the third left intercostal space and the LAD was permanently ligated with a surgical suture. Sham animals underwent the same procedure except for LAD ligation. Hemodynamic measurements were performed using a microconductance catheter system positioned in the left ventricle in closed-chest animals through the right carotid artery. After the explantation of the heart, the atria and right ventricle were removed and left ventricular tissue was dissected into post-infarction zone (IZ) and unaffected surrounding LV-tissue (remote zone, RZ), immediately snap-frozen in liquid nitrogen and stored at −80 °C. Directly after explantation, hearts were inspected for a distinctly visible infarcted area. Total RNA was isolated using QIAzol lysis reagent followed by purification using the miRNeasy mini kit (Qiagen, Hilden, Germany).
4.3. Cell Culture
HL-1 cells were cultivated in Claycomb medium, including 10% FBS, 1% Penicillin-Streptomycin, 2 mM GlutaMAX and 0.1 µM noradrenalin and incubated at 37 °C in a humidified atmosphere of 5% CO2. To simulate ischemic conditions in vitro, HL-1 cells were cultivated until confluency, starved in serum-reduced medium with 0.5% FCS overnight and placed in the hypoxic chamber. Oxygen supply was reduced from 20% to 1% and controlled with an oxygen sensor and D-glucose was replaced by L-glucose to keep osmotic properties. Cells were incubated for 24 hrs. Total RNA was isolated using the RNeasy mini kit (Qiagen, Hilden, Germany).
4.4. Gene Expression Analysis
Human transcriptomes from the GIS-GHS study were analyzed using the Affymetrix GeneChip Human Exon ST1.0 Array (Applied Biosystems, Waltham, MA, USA). The measurement of mouse transcriptomes by massive analysis of cDNA Ends (MACE) 3’ mRNA sequencing was conducted by GenXPro (Frankfurt, Germany). Real-time quantitative polymerase chain reaction (qPCR) with the 7900 TaqMan system (Applied Biosystems, Waltham, MA, USA) was used to determine mRNA expression levels. RNA was reverse transcribed using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems Waltham, MA, USA), according to manufacturers’ protocols. For gene expression analyses, the GPR15 Hs00922903_s1 (human) and Gpr15 Mm03990531_s1 (mouse) expression assays were used (Applied Biosystems Waltham, MA, USA). The quantification of housekeepers GAPDH (Hs99999905_m1) or 18S rRNA (Hs99999901_s1) as internal controls was performed for each sample (Applied Biosystems Waltham, MA, USA). Data were normalized to GAPDH or 18S rRNA levels (∆Ct-values) to account for RNA input and relative gene expression levels were expressed in comparison to the corresponding untreated controls using the formula 2−∆∆Ct [42].
4.5. Analyses of DNA Methylation
Previously, the DNA methylation levels of sites CpG3.98251047, CpG3.98251179 and CpG3.98251219, located within the GPR15 gene body, were identified to differ between smokers and non-smokers [17]. The DNA methylation levels of the GPR15 DNA methylation sites CpG3.98251047, CpG3.98251179 and CpG3.98251219 (referred to as GpC1, CpG2 and CpG3, respectively) were measured using the EpiTYPER MassARRAY technology (Agena Bioscience, Hamburg, Germany) as described previously [17,43]. The GPR15 gene from chr3:98531454 to chr3:98533234 (GRCh38/hg38) was amplified by PCR and sequenced by Sanger sequencing. The genotyping of the rs2230344 SNP was carried out by qPCR using the Applied Biosystems C__22275108_10 TaqMan® SNP Genotyping Assay on a 7900 HT Real Time PCR system (Applied Biosystems, Waltham, MA, USA).
4.6. Statistical and Bioinformatical Analyses
Differential gene expression between sex and age-matched groups of the GIS-GHS study was identified by applying a linear mixed model using R-package nlme [44,45]. The model was adjusted for the classical cardiovascular risk factors hypertension, smoking status, body mass index (BMI), diabetes and LDL/high-density lipoprotein (HDL) ratio. Correction for multiple testing was performed by calculating a false discovery rate (FDR) for each gene using the q-value method the Bioconductor package q value [46,47]. The significance level for differential gene expression was set to FDR < 0.1. Gene expression differences were reported as fold changes (FC), which represent the absolute expression changes corrected for covariates used in adjustment. To explore biological functions, canonical pathways and networks, functional enrichment analyses using ingenuity pathway analysis (IPA) was performed. If genes were differentially expressed with an FDR < 0.1 (n = 183), their respective FCs and p-values were uploaded into IPA. The reference set was restricted to genes on the Affymetrix Exon ST 1.0 Array.
Differential gene expression between Gpr15gfp/gfp and wildtype mice was calculated using DESeq2 [48] and plotted using Graph Pad Prism (GraphPad Software, San Diego, CA, USA). Gene ontology (GO) annotation data are based on ENSEMBL. Subsequent GO enrichment analysis was performed using the topGO package (bioconductor). The enrichment of GO terms was calculated by the Fisher’s exact test based on transcripts with a p-value < 0.05. Due to the large number of significant GO terms, ReviGo was used for clustering [49]. Resulting GO terms were visualized by dot plots generated with ggplot2 [50]. The z-score was calculated for each GO term by subtracting the number of downregulated genes (FC < −1.5, p < 0.05) from the number of upregulated genes (FC > 1.5 and p-value < 0.05) and dividing the result by the square root of the number of annotated genes.
Nonparametric Mann–Whitney U test and Kruskal–Wallis test with Dunn’s multiple comparisons test were used to compare two or more than two groups, respectively. Linear mixed regression models were adjusted for batch, using the random variable, and age and sex as covariates, except for the MIYoung study, where no adjustment for age was performed. Values for C-reactive protein were log-transformed. For rs2230344 allele association with early-onset MI in the MIYoung study, logistic regressions with MI as the dependent variable and rs2230344 as the independent variable of interest were performed and adjusted for BMI. To assess whether the percentage of CpG methylation levels or rs2230344 minor T allele were significantly related to increased risk of MI, mixed effect Cox models were applied using the R coxme package. For significant DNA methylation sites, Kaplan–Meier curves were generated and tested by log-rank tests using the R survival package. The survival of mice after LAD-ligation was plotted as a Kaplan–Meier curve and analyzed by log-rank test using GraphPad Prism. Causal mediation analysis in the GIS-GHS study—with smoking as the independent variable, MI as the dependent variable and GPR15 mRNA as the potential mediator—was conducted using the R package mediation [51]. Unstandardized indirect effects were computed by bootstrapping samples (1000 simulations), and the 95% confidence interval (CI) as indirect effects at the 2.5th and 97.5th percentiles. Linear models for mediators and independent variables vs. MI were computed adjusting for age, sex, diabetes status, hypertension status, HDL and LDL.
Statistical analyses were performed and figures were prepared using R version 3.4.3 and GraphPad prism version 6.05 for Windows (GraphPad Software, San Diego, CA, USA) [44]. The threshold for statistical significance was set at p ≤ 0.05 (two-sided testing) after Bonferroni correction to adjust for multiple testing when applicable.
5. Conclusions
In summary, for the first time this study showed an involvement of GPR15 in myocardial infarction, adding an important piece of the puzzle to understanding the physiological and pathophysiological role of GPR15. A proposed model linking of GPR15 and MI is depicted in Figure 7. Future analyses are needed to show the exact molecular pathways linking GPR15 in CVD pathogenesis and whether GPR15 might be causal for MI development. With this knowledge, GPR15 bears potential as a valuable therapeutic target for CVD prevention or treatment, in particular as protein-binding GPCRs, such as GPR15, bind only a limited number of ligands (51). As GPR15 DNA methylation predicted MI risk and GPR15 expression was associated with preclinical cardiovascular phenotypes, the potential of GPR15 as a potential biomarker for CVD risk prediction merits further evaluation.
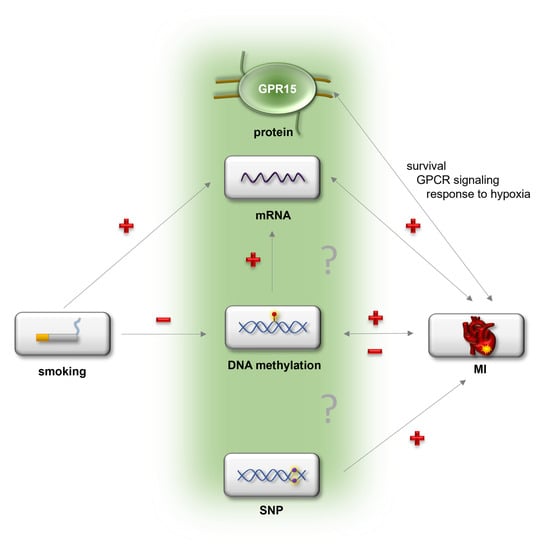
Figure 7.
Proposed model of the potential connection between GPR15 and myocardial infarction. Possibly, smoking as a major risk factor for myocardial infarction (MI) might change blood cell composition and thereby GPR15 mRNA expression as well as GPR15 DNA methylation. Myocardial infarction is associated with elevated GPR15 mRNA expression as well as decreased GPR15 DNA methylation. The effect of smoking on MI is mediated by GPR15 mRNA expression. Gpr15 affects survival as well as signaling and gene expression related to signaling and response to hypoxia after MI. The SNP rs2230344 within the GPR15 gene is associated with early-onset MI. “+” = upregulation, “−” = downregulation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms24010180/s1.
Author Contributions
Conceptualization of work: T.H., C.M., D.L. and T.Z.; Supervision: T.H. and T.Z.; Data collection: T.H., C.M., S.V., P.K., P.S.W., J.A., S.S. and T.Z.; Formal analysis: T.H., C.M. and B.S.; Methodology: S.V.K.; Data analysis and data interpretation: T.H., C.M., B.S., M.W., S.V., F.J.K., M.K., P.S.W., K.J.L., J.A., S.S., D.L. and T.Z.; Visualization: T.H., C.M., B.S. and S.V.; Resources: T.H., M.W., F.J.K., S.V.K., P.S.W., K.J.L., J.A., S.S., D.L. and T.Z.; Writing—original draft: T.H., D.L. and T.Z.; Writing—critical review and editing: T.H., C.M., B.S., P.K., M.W., F.J.K., M.K., S.V., S.V.K., P.S.W., K.J.L., J.A., S.S., D.L. and T.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the German Centre for Cardiovascular Research (DZHK e.V.), grant 81x2710163_81604/151 (T.H.); German Centre for Cardiovascular Research (DZHK e.V.), grant 81Z1710101 (T.Z.); German Centre for Cardiovascular Research (DZHK e.V.), grant 81Z0710102 (T.Z.); German Centre for Cardiovascular Research (DZHK e.V.), grant 81Z2710107, 81Z0710108 (D.L.); University Medical Center Hamburg Eppendorf (UKE), grant Research Promotion Fund of the Faculty of Medicine (FFM) (T.H.); Else Kröner-Fresenius-Stiftung, grant 2014_A92 (T.Z.); Ernst und Berta Grimmke-Stiftung, grant 8/18 (T.H.); German Research Foundation (DFG), Research Unit 2488 (F.J.K.); National Institute of Health (NIH), grant R01AI141787 (S.V.K.); Government of Rhineland-Palatinate (“Stiftung Rheinland-Pfalz für Innovation”), grant AZ 961-386261/733 (Gutenberg Health Study); “Wissen schafft Zukunft” (Gutenberg Health Study); Johannes Gutenberg-University of Mainz “Center for Translational Vascular Biology” (Gutenberg Health Study); Boehringer Ingelheimer, PHILIPS Medical Systems (Gutenberg Health Study); Swedish Heart and Lung Foundation (FIA2 study); Gustaf Sjölund Foundation (FIA2 study); Country councils of Northern Sweden (Visare Norr) (FIA2 study) and the Faculty of Medicine of Umeå University (FIA2 study). The funders had no role in the design of the study; in the collection, analyses or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Institutional Review Board Statement
The study was conducted in accordance with the Declaration of Helsinki and approved by the local ethics committee of the Medical Chamber of Rhineland-Palatinate, Germany (ethical approval codes 837.211.08 (6208) and 837.020.07 (5555)), by the local ethics committee of the University Medical Center Hamburg-Eppendorf, Germany (ethical approval code PV4137) and by the by the FIA2 local ethics committee (Dnr 05-142M, with additional review in 2009-01-19). All animal investigations conform to the Guide for the Care and Use of Laboratory Animals published by the US NIH. The study protocols were approved by the Hamburg Authority for Health and Consumer Protection (approval codes G060/15) and the Regional Office for Health and Social Affairs Berlin (approval code G0055/11).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The genotype, transcriptomics and phenotypic data are available under restricted access, as they contain identifying participant information. Deposition in online repositories or controlled access repositories is not authorized by the patients’ consent. Access requests, which must include a formal research proposal indicating the use of data and planned analyses, should be addressed to Tanja Zeller (t.zeller@uke.de).
Acknowledgments
The authors thank Caroline Röthemeier and Svenja Warnke for technical assistance, Andrej-Nikolai Spiess and René Riedel for statistical assistance, Julia Krause and Florian Weinberger for help with molecular techniques and constructive project-specific discussions and Simone Schnella for editorial assistance. The authors wish to acknowledge the Northern Sweden MONICA project, the Västerbotten Intervention Program, the Northern Sweden Medical Biobank and the funds supporting them.
Conflicts of Interest
The authors declare no conflict of interest.
References
- GBD 2015 DALYs and HALE Collaborators. Global, regional, and national disability-adjusted life-years (DALYs) for 315 diseases and injuries and healthy life expectancy (HALE), 1990–2015: A systematic analysis for the Global Burden of Disease Study 2015. Lancet 2016, 388, 1603–1658. [Google Scholar] [CrossRef]
- Roth, G.A.; Johnson, C.; Abajobir, A.; Abd-Allah, F.; Abera, S.F.; Abyu, G.; Ahmed, M.; Aksut, B.; Alam, T.; Alam, K.; et al. Global, Regional, and National Burden of Cardiovascular Diseases for 10 Causes, 1990 to 2015. J. Am. Coll. Cardiol. 2017, 70, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Abdolmaleki, F.; Gheibi Hayat, S.M.; Bianconi, V.; Johnston, T.P.; Sahebkar, A. Atherosclerosis and immunity: A perspective. Trends Cardiovasc. Med. 2019, 29, 363–371. [Google Scholar] [CrossRef] [PubMed]
- Ross, R. Atherosclerosis—An inflammatory disease. N. Engl. J. Med. 1999, 340, 115–126. [Google Scholar] [CrossRef]
- Yusuf, S.; Hawken, S.; Ounpuu, S.; Dans, T.; Avezum, A.; Lanas, F.; McQueen, M.; Budaj, A.; Pais, P.; Varigos, J.; et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): Case-control study. Lancet 2004, 364, 937–952. [Google Scholar] [CrossRef]
- Timmis, A.; Townsend, N.; Gale, C.; Grobbee, R.; Maniadakis, N.; Flather, M.; Wilkins, E.; Wright, L.; Vos, R.; Bax, J.; et al. European Society of Cardiology: Cardiovascular Disease Statistics 2017. Eur. Heart J. 2018, 39, 508–579. [Google Scholar] [CrossRef]
- Doughty, M.; Mehta, R.; Bruckman, D.; Das, S.; Karavite, D.; Tsai, T.; Eagle, K. Acute myocardial infarction in the young—The University of Michigan experience. Am. Heart J. 2002, 143, 56–62. [Google Scholar] [CrossRef]
- Brscic, E.; Bergerone, S.; Gagnor, A.; Colajanni, E.; Matullo, G.; Scaglione, L.; Cassader, M.; Gaschino, G.; Di Leo, M.; Brusca, A.; et al. Acute myocardial infarction in young adults: Prognostic role of angiotensin-converting enzyme, angiotensin II type I receptor, apolipoprotein E, endothelial constitutive nitric oxide synthase, and glycoprotein IIIa genetic polymorphisms at medium-term follow-up. Am. Heart J. 2000, 139, 979–984. [Google Scholar] [CrossRef]
- Marenberg, M.E.; Risch, N.; Berkman, L.F.; Floderus, B.; de Faire, U. Genetic susceptibility to death from coronary heart disease in a study of twins. N. Engl. J. Med. 1994, 330, 1041–1046. [Google Scholar] [CrossRef]
- Schunkert, H.; von Scheidt, M.; Kessler, T.; Stiller, B.; Zeng, L.; Vilne, B. Genetics of coronary artery disease in the light of genome-wide association studies. Clin. Res. Cardiol. 2018, 107, 2–9. [Google Scholar] [CrossRef]
- Myocardial Infarction Genetics Consortium; Kathiresan, S.; Voight, B.F.; Purcell, S.; Musunuru, K.; Ardissino, D.; Mannucci, P.M.; Anand, S.; Engert, J.C.; Samani, N.J.; et al. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009, 41, 334–341. [Google Scholar] [CrossRef] [PubMed]
- Erdmann, J.; Kessler, T.; Munoz Venegas, L.; Schunkert, H. A decade of genome-wide association studies for coronary artery disease: The challenges ahead. Cardiovasc. Res. 2018, 114, 1241–1257. [Google Scholar] [CrossRef] [PubMed]
- Tyndall, J.D.; Sandilya, R. GPCR agonists and antagonists in the clinic. Med. Chem. 2005, 1, 405–421. [Google Scholar] [CrossRef] [PubMed]
- Hendriks-Balk, M.C.; Peters, S.L.; Michel, M.C.; Alewijnse, A.E. Regulation of G protein-coupled receptor signalling: Focus on the cardiovascular system and regulator of G protein signalling proteins. Eur. J. Pharmacol. 2008, 585, 278–291. [Google Scholar] [CrossRef]
- Wang, J.; Gareri, C.; Rockman, H.A. G-Protein-Coupled Receptors in Heart Disease. Circ. Res. 2018, 123, 716–735. [Google Scholar] [CrossRef] [PubMed]
- Wan, E.S.; Qiu, W.; Baccarelli, A.; Carey, V.J.; Bacherman, H.; Rennard, S.I.; Agusti, A.; Anderson, W.; Lomas, D.A.; Demeo, D.L. Cigarette smoking behaviors and time since quitting are associated with differential DNA methylation across the human genome. Hum. Mol. Genet. 2012, 21, 3073–3082. [Google Scholar] [CrossRef]
- Haase, T.; Muller, C.; Krause, J.; Rothemeier, C.; Stenzig, J.; Kunze, S.; Waldenberger, M.; Munzel, T.; Pfeiffer, N.; Wild, P.S.; et al. Novel DNA Methylation Sites Influence GPR15 Expression in Relation to Smoking. Biomolecules 2018, 8, 74. [Google Scholar] [CrossRef]
- Teo, K.K.; Ounpuu, S.; Hawken, S.; Pandey, M.R.; Valentin, V.; Hunt, D.; Diaz, R.; Rashed, W.; Freeman, R.; Jiang, L.; et al. Tobacco use and risk of myocardial infarction in 52 countries in the INTERHEART study: A case-control study. Lancet 2006, 368, 647–658. [Google Scholar] [CrossRef]
- Kent, W.J.; Sugnet, C.W.; Furey, T.S.; Roskin, K.M.; Pringle, T.H.; Zahler, A.M.; Haussler, D. The human genome browser at UCSC. Genome Res. 2002, 12, 996–1006. [Google Scholar] [CrossRef]
- Kim, S.V.; Xiang, W.V.; Kwak, C.; Yang, Y.; Lin, X.W.; Ota, M.; Sarpel, U.; Rifkin, D.B.; Xu, R.; Littman, D.R. GPR15-mediated homing controls immune homeostasis in the large intestine mucosa. Science 2013, 340, 1456–1459. [Google Scholar] [CrossRef]
- Cartwright, A.; Schmutz, C.; Askari, A.; Kuiper, J.H.; Middleton, J. Orphan receptor GPR15/BOB is up-regulated in rheumatoid arthritis. Cytokine 2014, 67, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Zundler, S.; Atreya, R.; Rath, T.; Voskens, C.; Hirschmann, S.; Lopez-Posadas, R.; Watson, A.; Becker, C.; Schuler, G.; et al. Differential effects of alpha4beta7 and GPR15 on homing of effector and regulatory T cells from patients with UC to the inflamed gut in vivo. Gut 2016, 65, 1642–1664. [Google Scholar] [CrossRef] [PubMed]
- Adamczyk, A.; Gageik, D.; Frede, A.; Pastille, E.; Hansen, W.; Rueffer, A.; Buer, J.; Buning, J.; Langhorst, J.; Westendorf, A.M. Differential expression of GPR15 on T cells during ulcerative colitis. JCI Insight 2017, 2, e90585. [Google Scholar] [CrossRef] [PubMed]
- Ammitzboll, C.; von Essen, M.R.; Bornsen, L.; Petersen, E.R.; McWilliam, O.; Ratzer, R.; Jeppe, R.C.; Oturai, A.B.; Sondergaard, H.B.; Sellebjerg, F. GPR15(+) T cells are Th17 like, increased in smokers and associated with multiple sclerosis. J. Autoimmun. 2019, 97, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M. The Role of GPR15 Function in Blood and Vasculature. Int. J. Mol. Sci. 2021, 22, 824. [Google Scholar] [CrossRef]
- Frangogiannis, N.G. The inflammatory response in myocardial injury, repair, and remodelling. Nat. Rev. Cardiol. 2014, 11, 255–265. [Google Scholar] [CrossRef]
- Jjingo, D.; Conley, A.B.; Yi, S.V.; Lunyak, V.V.; Jordan, I.K. On the presence and role of human gene-body DNA methylation. Oncotarget 2012, 3, 462–474. [Google Scholar] [CrossRef]
- Teissandier, A.; Bourc’his, D. Gene body DNA methylation conspires with H3K36me3 to preclude aberrant transcription. EMBO J. 2017, 36, 1471–1473. [Google Scholar] [CrossRef]
- Bauer, M.; Hackermuller, J.; Schor, J.; Schreiber, S.; Fink, B.; Pierzchalski, A.; Herberth, G. Specific induction of the unique GPR15 expression in heterogeneous blood lymphocytes by tobacco smoking. Biomarkers 2019, 24, 217–224. [Google Scholar] [CrossRef]
- Dogan, M.V.; Xiang, J.; Beach, S.R.; Cutrona, C.; Gibbons, F.X.; Simons, R.L.; Brody, G.H.; Stapleton, J.T.; Philibert, R.A. Ethnicity and Smoking-Associated DNA Methylation Changes at HIV Co-Receptor GPR15. Front. Psychiatry 2015, 6, 132. [Google Scholar] [CrossRef]
- Bilsborough, J.; Viney, J.L. GPR15: A tale of two species. Nat. Immunol. 2015, 16, 137–139. [Google Scholar] [CrossRef] [PubMed]
- Ambroziak, M.; Niewczas-Wieprzowska, K.; Maicka, A.; Budaj, A. Younger age of patients with myocardial infarction is associated with a higher number of relatives with a history of premature atherosclerosis. BMC Cardiovasc. Disord. 2020, 20, 410. [Google Scholar] [CrossRef] [PubMed]
- Wild, P.S.; Zeller, T.; Beutel, M.; Blettner, M.; Dugi, K.A.; Lackner, K.J.; Pfeiffer, N.; Munzel, T.; Blankenberg, S. The Gutenberg Health Study. Bundesgesundheitsblatt Gesundh. Gesundh. 2012, 55, 824–829. [Google Scholar] [CrossRef] [PubMed]
- Wild, P.S.; Sinning, C.R.; Roth, A.; Wilde, S.; Schnabel, R.B.; Lubos, E.; Zeller, T.; Keller, T.; Lackner, K.J.; Blettner, M.; et al. Distribution and categorization of left ventricular measurements in the general population: Results from the population-based Gutenberg Heart Study. Circ. Cardiovasc. Imaging 2010, 3, 604–613. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic. Acids. Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Zeller, T.; Wild, P.; Szymczak, S.; Rotival, M.; Schillert, A.; Castagne, R.; Maouche, S.; Germain, M.; Lackner, K.; Rossmann, H.; et al. Genetics and beyond—The transcriptome of human monocytes and disease susceptibility. PLoS ONE 2010, 5, e10693. [Google Scholar] [CrossRef]
- Ekblom, K.; Marklund, S.L.; Jansson, J.H.; Hallmans, G.; Weinehall, L.; Hultdin, J. Iron stores and HFE genotypes are not related to increased risk of first-time myocardial infarction: A prospective nested case-referent study. Int. J. Cardiol. 2011, 150, 169–172. [Google Scholar] [CrossRef]
- Crawford, A.A.; Soderberg, S.; Kirschbaum, C.; Murphy, L.; Eliasson, M.; Ebrahim, S.; Davey Smith, G.; Olsson, T.; Sattar, N.; Lawlor, D.A.; et al. Morning plasma cortisol as a cardiovascular risk factor: Findings from prospective cohort and Mendelian randomization studies. Eur. J. Endocrinol. 2019, 181, 429–438. [Google Scholar] [CrossRef]
- Bayne, K. Revised Guide for the Care and Use of Laboratory Animals available. American Physiological Society. Physiologist 1996, 39, 208–211. [Google Scholar]
- Hinrichs, S.; Scherschel, K.; Kruger, S.; Neumann, J.T.; Schwarzl, M.; Yan, I.; Warnke, S.; Ojeda, F.M.; Zeller, T.; Karakas, M.; et al. Precursor proadrenomedullin influences cardiomyocyte survival and local inflammation related to myocardial infarction. Proc. Natl. Acad. Sci. USA 2018, 115, E8727–E8736. [Google Scholar] [CrossRef]
- Bacmeister, L.; Schwarzl, M.; Warnke, S.; Stoffers, B.; Blankenberg, S.; Westermann, D.; Lindner, D. Inflammation and fibrosis in murine models of heart failure. Basic Res. Cardiol. 2019, 114, 19. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Kunze, S. Quantitative Region-Specific DNA Methylation Analysis by the EpiTYPER Technology. Methods Mol. Biol. 2018, 1708, 515–535. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- R Core Team. nlme: Linear and Nonlinear Mixed Effects Models; R Core Team: Vienna, Austria, 2013. [Google Scholar]
- Storey, J.D.; Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. USA 2003, 100, 9440–9445. [Google Scholar] [CrossRef] [PubMed]
- Storey, J.D.; Bass, A.J. Qvalue: Q-Value Estimation for False Discovery Rate Control; Bioconductor: Boston, MA, USA, 2013. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Supek, F.; Bosnjak, M.; Skunca, N.; Smuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef]
- Wickham, H. Elegant graphics for data analysis. Media 2009, 35, 10–1007. [Google Scholar]
- Tingley, D.; Yamamoto, T.; Hirose, K.; Keele, L.; Imai, K. Mediation: R package for causal mediation analysis. JSS J. Stat. Softw. 2014, 59. Available online: http://hdl.handle.net/1721.1/91154 (accessed on 4 November 2021).
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).