Diversity in Genetic Regulation of Bacterial Fimbriae Assembled by the Chaperone Usher Pathway
Abstract
1. Introduction
1.1. Fimbriae: Genetic Clusters and Assembly via the CU Pathway
1.2. Diversity in the CUP-Assembled Fimbriae
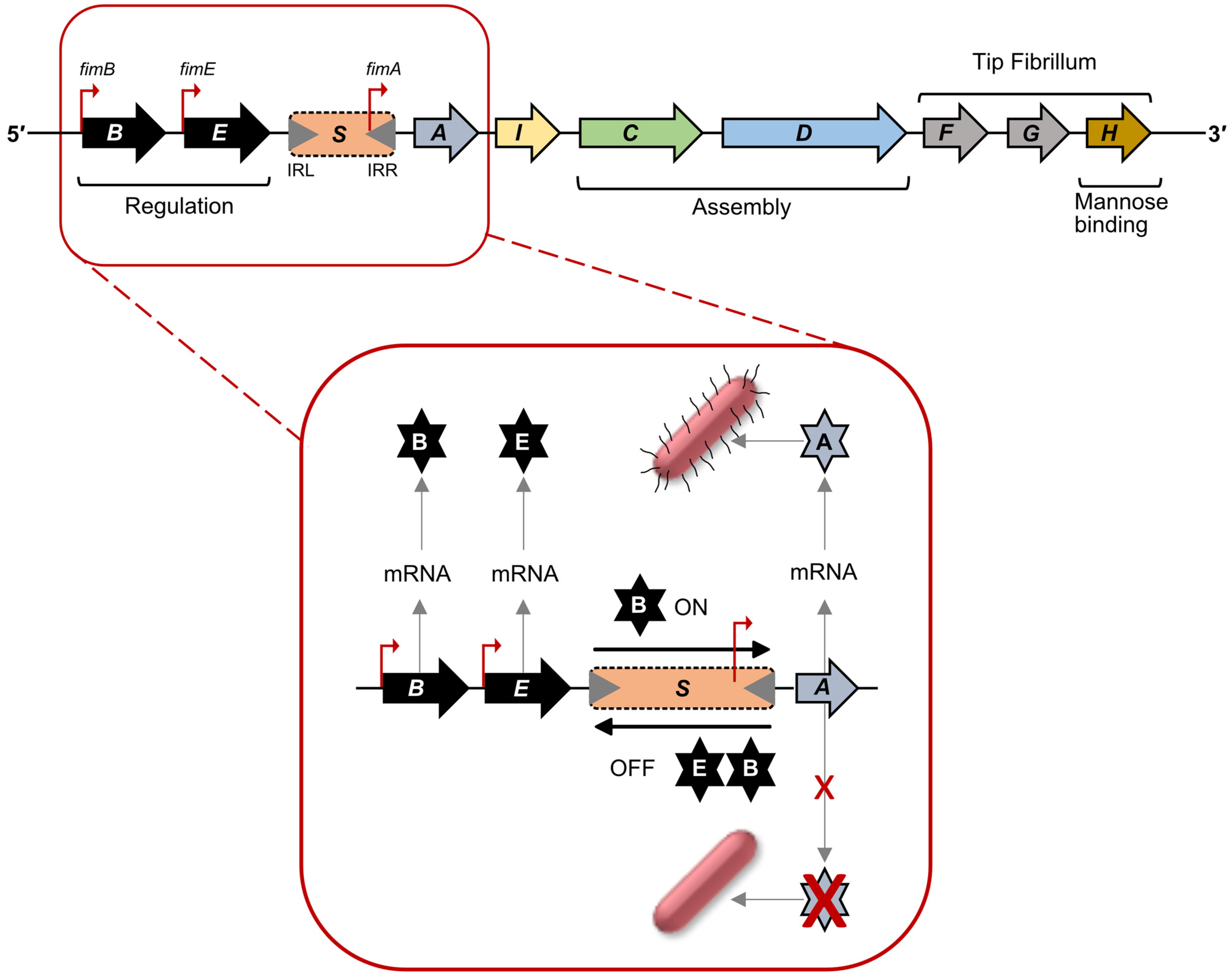
2. Diversity in Regulation: Phase Variation
2.1. Regulation of Type 1 Fimbriae Expression: Promoter Inversion
2.2. Regulation of Pap pili Expression: Methylation/Epigenetic Switch
3. Other DNA Binding Regulators, Dedicated and Global
3.1. pH 6 Antigen, Regulation by a Sensor-Regulator
3.2. Agg and Aaf Fimbriae, Regulation by A/X Family Regulator, AggR
3.3. CS and CfaI Fimbriae, Regulation by A/X Family Regulator, Rns
3.4. F1 Capsule, Regulation by A/X Family Regulator, Caf1R
4. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Proft, T.; Baker, E.N. Pili in Gram-negative and Gram-positive bacteriastructure, assembly and their role in disease. Cell Mol. Life Sci. 2009, 66, 613–635. [Google Scholar] [CrossRef] [PubMed]
- Rendon, M.A.; Saldana, Z.; Erdem, A.L.; Monteiro-Neto, V.; Vazquez, A.; Kaper, J.B.; Puente, J.L.; Giron, J.A. Commensal and pathogenic Escherichia coli use a common pilus adherence factor for epithelial cell colonization. Proc. Natl. Acad. Sci. USA 2007, 104, 10637–10642. [Google Scholar] [CrossRef]
- Lukaszczyk, M.; Pradhan, B.; Remaut, H. The Biosynthesis and Structures of Bacterial Pili. Subcell. Biochem. 2019, 92, 369–413. [Google Scholar] [PubMed]
- Berne, C.; Ducret, A.; Hardy, G.G.; Brun, Y.V. Adhesins Involved in Attachment to Abiotic Surfaces by Gram-Negative Bacteria. Microbiol. Spectr. 2015, 3, 163–199. [Google Scholar] [CrossRef] [PubMed]
- Pizarro-Cerda, J.; Cossart, P. Bacterial adhesion and entry into host cells. Cell 2006, 124, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Di Martino, P. Bacterial adherence: Much more than a bond. AIMS Microbiol. 2018, 4, 563–566. [Google Scholar] [CrossRef]
- Jin, X.; Marshall, J.S. Mechanics of biofilms formed of bacteria with fimbriae appendages. PLoS ONE 2020, 15, e0243280. [Google Scholar] [CrossRef]
- Werneburg, G.T.; Thanassi, D.G. Pili Assembled by the Chaperone/Usher Pathway in Escherichia coli and Salmonella. EcoSal Plus 2018, 8. [Google Scholar] [CrossRef]
- Pusz, P.; Bok, E.; Mazurek, J.; Stosik, M.; Baldy-Chudzik, K. Type 1 fimbriae in commensal Escherichia coli derived from healthy humans. Acta Biochim. Pol. 2014, 61, 389–392. [Google Scholar] [CrossRef]
- Muller, K.H.; Collinson, S.K.; Trust, T.J.; Kay, W.W. Type 1 fimbriae of Salmonella enteritidis. J. Bacteriol. 1991, 173, 4765–4772. [Google Scholar] [CrossRef][Green Version]
- Zhang, Y.; Tennent, J.M.; Ingham, A.; Beddome, G.; Prideaux, C.; Michalski, W.P. Identification of type-4 fimbriae in Actinobacillus pleuropneumoniae. FEMS Microbiol. Lett. 2000, 189, 15–18. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S.; Whitchurch, C.B.; Alm, R.A. The molecular genetics of type-4 fimbriae in Pseudomonas aeruginosa—A review. Gene 1996, 179, 147–155. [Google Scholar] [CrossRef] [PubMed]
- Bhoite, S.; van Gerven, N.; Chapman, M.R.; Remaut, H. Curli Biogenesis: Bacterial Amyloid Assembly by the Type VIII Secretion Pathway. EcoSal Plus 2019, 8. [Google Scholar] [CrossRef] [PubMed]
- Barnhart, M.M.; Chapman, M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006, 60, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.D.; Harb, L.; Khara, P.; Zeng, L.; Hu, B.; Christie, P.J. Type IV secretion systems: Advances in structure, function, and activation. Mol. Microbiol. 2021, 3, 436–452. [Google Scholar] [CrossRef]
- Thanassi, D.G.; Hultgren, S.J. Assembly of complex organelles: Pilus biogenesis in Gram-negative bacteria as a model system. Methods 2000, 20, 111–126. [Google Scholar] [CrossRef][Green Version]
- Thanassi, D.G.; Bliska, J.B.; Christie, P.J. Surface organelles assembled by secretion systems of Gram-negative bacteria: Diversity in structure and function. FEMS Microbiol. Rev. 2012, 36, 1046–1082. [Google Scholar] [CrossRef]
- Clegg, S.; Wilson, J.; Johnson, J. More than one way to control hair growth: Regulatory mechanisms in enterobacteria that affect fimbriae assembled by the chaperone/usher pathway. J. Bacteriol. 2011, 193, 2081–2088. [Google Scholar] [CrossRef][Green Version]
- Franca, F.L.; Wells, T.J.; Browning, D.F.; Nogueira, R.T.; Sarges, F.S.; Pereira, A.C.; Cunningham, A.F.; Lucheze, K.; Rosa, A.C.; Henderson, I.R.; et al. Genotypic and phenotypic characterisation of enteroaggregative Escherichia coli from children in Rio de Janeiro, Brazil. PLoS ONE. 2013, 8, e69971. [Google Scholar] [CrossRef]
- Thanassi, D.G.; Saulino, E.T.; Hultgren, S.J. The chaperone/usher pathway: A major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1998, 1, 223–231. [Google Scholar] [CrossRef]
- Perez-Rueda, E.; Tenorio-Salgado, S.; Huerta-Saquero, A.; Balderas-Martinez, Y.I.; Moreno-Hagelsieb, G. The functional landscape bound to the transcription factors of Escherichia coli K-12. Comput. Biol. Chem. 2015, 58, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Costa, T.R.; Felisberto-Rodrigues, C.; Meir, A.; Prevost, M.S.; Redzej, A.; Trokter, M.; Waksman, G. Secretion systems in Gram-negative bacteria: Structural and mechanistic insights. Nat. Rev. Microbiol. 2015, 13, 343–359. [Google Scholar] [CrossRef] [PubMed]
- Geibel, S.; Waksman, G. The molecular dissection of the chaperone-usher pathway. Biochim. Biophys. Acta. 2014, 1843, 1559–1567. [Google Scholar] [CrossRef]
- Zav’yalov, V.; Zavialov, A.; Zav’yalova, G.; Korpela, T. Adhesive organelles of Gram-negative pathogens assembled with the classical chaperone/usher machinery: Structure and function from a clinical standpoint. FEMS Microbiol. Rev. 2010, 34, 317–378. [Google Scholar] [CrossRef]
- Choudhury, D.; Thompson, A.; Stojanoff, V.; Langermann, S.; Pinkner, J.; Hultgren, S.J.; Knight, S.D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 1999, 285, 1061–1066. [Google Scholar] [CrossRef]
- MacIntyre, S.; Zyrianova, I.M.; Chernovskaya, T.V.; Leonard, M.; Rudenko, E.G.; Zav’Yalov, V.P.; Chapman, D.A. An extended hydrophobic interactive surface of Yersinia pestis Caf1M chaperone is essential for subunit binding and F1 capsule assembly. Mol. Microbiol. 2001, 39, 12–25. [Google Scholar] [CrossRef] [PubMed]
- Zavialov, A.V.; Kersley, J.; Korpela, T.; Zav’yalov, V.P.; MacIntyre, S.; Knight, S.D. Donor strand complementation mechanism in the biogenesis of non-pilus systems. Mol. Microbiol. 2002, 45, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Zavialov, A.V.; Berglund, J.; Pudney, A.F.; Fooks, L.J.; Ibrahim, T.M.; MacIntyre, S.; Knight, S.D. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: Preserved folding energy drives fiber formation. Cell 2003, 113, 587–596. [Google Scholar] [CrossRef]
- Dubnovitsky, A.P.; Duck, Z.; Kersley, J.E.; Härd, T.; MacIntyre, S.; Knight, S.D. Conserved hydrophobic clusters on the surface of the Caf1A usher C-terminal domain are important for F1 antigen assembly. J. Mol. Biol. 2010, 403, 243–259. [Google Scholar] [CrossRef]
- Yu, X.D.; Dubnovitsky, A.; Pudney, A.F.; Macintyre, S.; Knight, S.D.; Zavialov, A.V. Allosteric mechanism controls traffic in the chaperone/usher pathway. Structure 2012, 20, 1861–1871. [Google Scholar] [CrossRef]
- Barnhart, M.M.; Sauer, F.G.; Pinkner, J.S.; Hultgren, S.J. Chaperone-subunit-usher interactions required for donor strand exchange during bacterial pilus assembly. J. Bacteriol. 2003, 185, 2723–2730. [Google Scholar] [CrossRef] [PubMed]
- Le Trong, I.; Aprikian, P.; Kidd, B.A.; Thomas, W.E.; Sokurenko, E.V.; Stenkamp, R.E. Donor strand exchange and conformational changes during Escherichia coli fimbrial formation. J. Struct. Biol. 2010, 172, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Zyla, D.; Echeverria, B.; Glockshuber, R. Donor strand sequence, rather than donor strand orientation, determines the stability and non-equilibrium folding of the type 1 pilus subunit FimA. J. Biol. Chem. 2020, 295, 12437–12448. [Google Scholar] [CrossRef]
- Korea, C.G.; Badouraly, R.; Prevost, M.C.; Ghigo, J.M.; Beloin, C. Escherichia coli K-12 possesses multiple cryptic but functional chaperone-usher fimbriae with distinct surface specificities. Environ. Microbiol. 2010, 12, 1957–1977. [Google Scholar] [CrossRef] [PubMed]
- Wurpel, D.J.; Beatson, S.A.; Totsika, M.; Petty, N.K.; Schembri, M.A. Chaperone-usher fimbriae of Escherichia coli. PLoS ONE 2013, 8, e52835. [Google Scholar] [CrossRef]
- Schaffer, J.N.; Pearson, M.M. Proteus mirabilis and Urinary Tract Infections. Microbiol. Spectr. 2015, 3, 383–433. [Google Scholar] [CrossRef]
- Nuccio, S.P.; Baumler, A.J. Evolution of the Chaperone-Usher assembly pathway: Fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 2007, 71, 551–575. [Google Scholar] [CrossRef]
- Kumar, D. Caf1R-Mediated Regulation of the F1 Surface Antigen of Yersinia Pestis. Ph.D. Thesis, University of Reading, Reading, UK, 2016. Available online: http://centaur.reading.ac.uk/66018/ (accessed on 18 July 2022).
- Honarvar, S.; Choi, B.-K.; Schifferli, D.M. Phase variation of the 987P-like CS18 fimbriae of human enterotoxigenic Escherichia coli is regulated by site-specific recombinases. Mol. Microbiol. 2003, 48, 157–171. [Google Scholar] [CrossRef]
- Zhao, H.; Li, X.; Johnson, D.E.; Blomfield, I.; Mobley, H.L. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol. Microbiol. 1997, 23, 1009–1019. [Google Scholar] [CrossRef]
- Li, X.; Lockatell, C.V.; Johnson, D.E.; Mobley, H.L. Identification of MrpI as the sole recombinase that regulates the phase variation of MR/P fimbria, a bladder colonization factor of uropathogenic Proteus mirabilis. Mol. Microbiol. 2002, 45, 865–874. [Google Scholar] [CrossRef]
- Bode, N.J.; Debnath, I.; Kuan, L.; Schulfer, A.; Ty, M.; Pearson, M.M. Transcriptional analysis of the MrpJ network: Modulation of diverse virulence-associated genes and direct regulation of mrp fimbrial and flhDC flagellar operons in Proteus mirabilis. Infect. Immun. 2015, 83, 2542–2556. [Google Scholar] [CrossRef]
- Simms, A.N.; Mobley, H.L. PapX, a P fimbrial operon-encoded inhibitor of motility in uropathogenic Escherichia coli. Infect. Immun. 2008, 76, 4833–4841. [Google Scholar] [CrossRef] [PubMed][Green Version]
- White-Ziegler, C.A.; Villapakkam, A.; Ronaszeki, K.; Young, S. H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J. Bacteriol. 2000, 182, 6391–6400. [Google Scholar] [CrossRef] [PubMed]
- Midgett, C.R.; Talbot, K.M.; Day, J.L.; Munson, G.P.; Jon Kull, F. Structure of the master regulator Rns reveals an inhibitor of enterotoxigenic Escherichia coli virulence regulons. Sci. Rep. 2021, 11, 15663. [Google Scholar] [CrossRef] [PubMed]
- Hung, D.L.; Knight, S.D.; Woods, R.M.; Pinkner, J.S.; Hultgren, S.J. Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 1996, 15, 3792–3805. [Google Scholar] [CrossRef] [PubMed]
- Zavialov, A.; Zav’yalova, G.; Korpela, T.; Zav’yalov, V. FGL chaperone-assembled fimbrial polyadhesins: Anti-immune armament of Gram-negative bacterial pathogens. FEMS Microbiol. Rev. 2007, 31, 478–514. [Google Scholar] [CrossRef]
- MacIntyre, S.; Knight, S.D.; Fooks, L.J. Structure, assembly and applications of the polymeric F1 antigen of Yersinia pestis. In Yersinia: Molecular and Cellular Biology; Carniel, E., Hinnenbusch, B.J., Eds.; Horizon Bioscience: Norfolk, UK, 2004; pp. 363–407. [Google Scholar]
- Gahlot, D.K.; Ifill, G.; MacIntyre, S. Optimised Heterologous Expression and Functional Analysis of the Yersinia pestis F1-Capsular Antigen Regulator Caf1R. Int. J. Mol. Sci. 2021, 22, 9805. [Google Scholar] [CrossRef]
- Bao, R.; Nair, M.K.; Tang, W.K.; Esser, L.; Sadhukhan, A.; Holland, R.L.; Xia, D.; Schifferli, D.M. Structural basis for the specific recognition of dual receptors by the homopolymeric pH 6 antigen (Psa) fimbriae of Yersinia pestis. Proc. Natl. Acad. Sci. USA 2013, 110, 1065–1070. [Google Scholar] [CrossRef]
- Quinn, J.D.; Weening, E.H.; Miner, T.A.; Miller, V.L. Temperature Control of psaA Expression by PsaE and PsaF in Yersinia pestis. J. Bacteriol. 2019, 201, e00217-19. [Google Scholar] [CrossRef]
- Blomfield, I.C. The regulation of Pap and type 1 fimbriation in Escherichia coli. Adv. Microb. Physiol. 2001, 45, 1–49. [Google Scholar] [CrossRef]
- Katani, R.; Kudva, I.T.; Srinivasan, S.; Stasko, J.B.; Schilling, M.; Li, L.; Cote, R.; DebRoy, C.; Arthur, T.M.; Sokurenko, E.V.; et al. Strain and host-cell dependent role of type 1 fimbriae in the adherence phenotype of super-shed Escherichia coli O157:H7. Int. J. Med. Microbiol. 2021, 311, 151511. [Google Scholar] [CrossRef]
- Schwan, W.R. Regulation of fim genes in uropathogenic Escherichia coli. World J. Clin. Infect. Dis. 2011, 1, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Saldaña-Ahuactzi, Z.; Soria-Bustos, J.; Martínez-Santos, V.I.; Yañez-Santos, J.A.; Martínez-Laguna, Y.; Cedillo-Ramirez, M.L.; Puente, J.L.; Girón, J.A. The Fis Nucleoid Protein Negatively Regulates the Phase Variation fimS Switch of the type 1 Pilus Operon in Enteropathogenic Escherichia coli. Front. Microbiol. 2022, 13, 882563. [Google Scholar] [CrossRef] [PubMed]
- Burns, L.S.; Smith, S.G.; Dorman, C.J. Interaction of the FimB integrase with the fimS invertible DNA element in Escherichia coli in vivo and in vitro. J. Bacteriol. 2000, 182, 2953–2959. [Google Scholar] [CrossRef] [PubMed]
- Smith, S.G.; Dorman, C.J. Functional analysis of the FimE integrase of Escherichia coli K-12: Isolation of mutant derivatives with altered DNA inversion preferences. Mol. Microbiol. 1999, 34, 965–979. [Google Scholar] [CrossRef]
- Donato, G.M.; Lelivelt, M.J.; Kawula, T.H. Promoter-specific repression of fimB expression by the Escherichia coli nucleoid-associated protein H-NS. J. Bacteriol. 1997, 179, 6618–6625. [Google Scholar] [CrossRef][Green Version]
- Schwan, W.R.; Seifert, H.S.; Duncan, J.L. Analysis of the fimB promoter region involved in type 1 pilus phase variation in Escherichia coli. Mol. Gen. Genet. 1994, 242, 623–630. [Google Scholar] [CrossRef]
- Olsen, P.B.; Klemm, P. Localization of promoters in the fim gene cluster and the effect of H-NS on the transcription of fimB and fimE. FEMS Microbiol. Lett. 1994, 116, 95–100. [Google Scholar] [CrossRef][Green Version]
- Spears, P.A.; Schauer, D.; Orndorff, P.E. Metastable regulation of type 1 piliation in Escherichia coli and isolation and characterization of a phenotypically stable mutant. J. Bacteriol. 1986, 168, 179–185. [Google Scholar] [CrossRef]
- Rice, P.A.; Yang, S.; Mizuuchi, K.; Nash, H.A. Crystal structure of an IHF-DNA complex: A protein-induced DNA U-turn. Cell 1996, 87, 1295–1306. [Google Scholar] [CrossRef]
- Gally, D.L.; Bogan, J.A.; Eisenstein, B.I.; Blomfield, I.C. Environmental regulation of the fim switch controlling type 1 fimbrial phase variation in Escherichia coli K-12: Effects of temperature and media. J. Bacteriol. 1993, 175, 6186–6193. [Google Scholar] [CrossRef]
- Olsen, P.B.; Schembri, M.A.; Gally, D.L.; Klemm, P. Differential temperature modulation by H-NS of the fimB and fimE recombinase genes which control the orientation of the type 1 fimbrial phase switch. FEMS Microbiol. Lett. 1998, 162, 17–23. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Sauer, F.G.; Remaut, H.; Hultgren, S.J.; Waksman, G. Fiber assembly by the Chaperone-Usher pathway. Biochim. Biophys. Acta 2004, 1694, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Braaten, B.A.; Blyn, L.B.; Skinner, B.S.; Low, D.A. Evidence for a methylation-blocking factor (mbf) locus involved in Pap pilus expression and phase variation in Escherichia coli. J. Bacteriol. 1991, 173, 1789–1800. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Hernday, A.D.; Braaten, B.A.; Broitman-Maduro, G.; Engelberts, P.; Low, D.A. Regulation of the pap epigenetic switch by CpxAR: Phosphorylated CpxR inhibits transition to the phase ON state by competition with Lrp. Mol. Cell 2004, 16, 537–547. [Google Scholar] [CrossRef]
- Otto, K.; Silhavy, T.J. Surface sensing and adhesion of Escherichia coli controlled by the Cpx-signaling pathway. Proc. Natl. Acad. Sci. USA 2002, 99, 2287–2292. [Google Scholar] [CrossRef]
- Khandige, S.; Kronborg, T.; Uhlin, B.E.; Moller-Jensen, J. sRNA-Mediated Regulation of P-Fimbriae Phase Variation in Uropathogenic Escherichia coli. PLoS Pathog. 2015, 11, e1005109. [Google Scholar] [CrossRef]
- Madhavan, T.P.; Sakellaris, H. Colonization factors of enterotoxigenic Escherichia coli. Adv. Appl. Microbiol. 2015, 90, 155–197. [Google Scholar] [CrossRef]
- Alvarez-Fraga, L.; Phan, M.D.; Goh, K.G.K.; Nhu, N.T.K.; Hancock, S.J.; Allsopp, L.P.; Peters, K.M.; Forde, B.M.; Roberts, L.W.; Sullivan, M.J.; et al. Differential Afa/Dr Fimbriae Expression in the Multidrug-Resistant Escherichia coli ST131 Clone. mBio 2022, 13, e0351921. [Google Scholar] [CrossRef]
- Yang, Y.; Isberg, R.R. Transcriptional regulation of the Yersinia pseudotuberculosis pH 6 antigen adhesin by two envelope-associated components. Mol. Microbiol. 1997, 24, 499–510. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, L.; Fang, N.; Qu, S.; Tan, Y.; Guo, Z.; Qiu, J.; Zhou, D.; Yang, R. Reciprocal regulation of pH 6 antigen gene loci by PhoP and RovA in Yersinia pestis biovar Microtus. Future Microbiol. 2013, 8, 271–280. [Google Scholar] [CrossRef]
- Fang, H.; Liu, L.; Zhang, Y.; Yang, H.; Yan, Y.; Ding, X.; Han, Y.; Zhou, D.; Yang, R. BfvR, an AraC-Family Regulator, Controls Biofilm Formation and pH 6 Antigen Production in Opposite Ways in Yersinia pestis Biovar Microtus. Front. Cell. Infect. Microbiol. 2018, 8, 347. [Google Scholar] [CrossRef] [PubMed]
- Cathelyn, J.S.; Crosby, S.D.; Lathem, W.W.; Goldman, W.E.; Miller, V.L. RovA, a global regulator of Yersinia pestis, specifically required for bubonic plague. Proc. Natl. Acad. Sci. USA 2006, 103, 13514–13519. [Google Scholar] [CrossRef] [PubMed]
- Jonsson, R.; Struve, C.; Boisen, N.; Mateiu, R.V.; Santiago, A.E.; Jenssen, H.; Nataro, J.P.; Krogfelt, K.A. Novel aggregative adherence fimbria variant of enteroaggregative Escherichia coli. Infect. Immun. 2015, 83, 1396–1405. [Google Scholar] [CrossRef] [PubMed]
- Berry, A.A.; Yang, Y.; Pakharukova, N.; Garnett, J.A.; Lee, W.C.; Cota, E.; Marchant, J.; Roy, S.; Tuittila, M.; Liu, B.; et al. Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli. PLoS Pathog. 2014, 10, e1004404. [Google Scholar] [CrossRef]
- Boisen, N.; Struve, C.; Scheutz, F.; Krogfelt, K.A.; Nataro, J.P. New adhesin of enteroaggregative Escherichia coli related to the Afa/Dr/AAF family. Infect. Immun. 2008, 76, 3281–3292. [Google Scholar] [CrossRef]
- Nataro, J.P.; Yikang, D.; Yingkang, D.; Walker, K. AggR, a transcriptional activator of aggregative adherence fimbria I expression in enteroaggregative Escherichia coli. J. Bacteriol. 1994, 176, 4691–4699. [Google Scholar] [CrossRef]
- Yasir, M.; Icke, C.; Abdelwahab, R.; Haycocks, J.R.; Godfrey, R.E.; Sazinas, P.; Pallen, M.J.; Henderson, I.R.; Busby, S.J.W.; Browning, D.F. Organization and architecture of AggR-dependent promoters from enteroaggregative Escherichia coli. Mol. Microbiol. 2019, 111, 534–551. [Google Scholar] [CrossRef]
- Dudley, E.G.; Thomson, N.R.; Parkhill, J.; Morin, N.P.; Nataro, J.P. Proteomic and microarray characterization of the AggR regulon identifies a pheU pathogenicity island in enteroaggregative Escherichia coli. Mol. Microbiol. 2006, 61, 1267–1282. [Google Scholar] [CrossRef]
- Morin, N.; Santiago, A.E.; Ernst, R.K.; Guillot, S.J.; Nataro, J.P. Characterization of the AggR regulon in enteroaggregative Escherichia coli. Infect. Immun. 2013, 81, 122–132. [Google Scholar] [CrossRef]
- Jonsson, R.; Liu, B.; Struve, C.; Yang, Y.; Jorgensen, R.; Xu, Y.; Jenssen, H.; Krogfelt, K.A.; Matthews, S. Structural and functional studies of Escherichia coli aggregative adherence fimbriae (AAF/V) reveal a deficiency in extracellular matrix binding. Biochim. Biophys. Acta Proteins Proteom. 2017, 1865, 304–311. [Google Scholar] [CrossRef]
- Elias Jr, W.P.; Czeczulin, J.R.; Henderson, I.R.; Trabulsi, L.R.; Nataro, J.P. Organization of biogenesis genes for aggregative adherence fimbria II defines a virulence gene cluster in enteroaggregative Escherichia coli. J. Bacteriol. 1999, 181, 1779–1785. [Google Scholar] [CrossRef] [PubMed]
- Munson, G.P.; Scott, J.R. Binding site recognition by Rns, a virulence regulator in the AraC family. J. Bacteriol. 1999, 181, 2110–2117. [Google Scholar] [CrossRef] [PubMed]
- Munson, G.P.; Scott, J.R. Rns, a virulence regulator within the AraC family, requires binding sites upstream and downstream of its own promoter to function as an activator. Mol. Microbiol. 2000, 36, 1391–1402. [Google Scholar] [CrossRef] [PubMed]
- Caron, J.; Coffield, L.M.; Scott, J.R. A plasmid-encoded regulatory gene, rns, required for expression of the CS1 and CS2 adhesins of enterotoxigenic Escherichia coli. Proc. Natl. Acad. Sci. USA 1989, 86, 963–967. [Google Scholar] [CrossRef] [PubMed]
- Caron, J.; Scott, J.R. A rns-like regulatory gene for colonization factor antigen I (CFA/I) that controls expression of CFA/I pilin. Infect. Immun. 1990, 58, 874–878. [Google Scholar] [CrossRef] [PubMed]
- Pichel, M.; Binsztein, N.; Viboud, G. CS22, a novel human enterotoxigenic Escherichia coli adhesin, is related to CS15. Infect. Immun. 2000, 68, 3280–3285. [Google Scholar] [CrossRef]
- Bodero, M.D.R.; Munson, G.P. The Virulence Regulator Rns Activates the Expression of CS14 Pili. Genes 2016, 7, 120. [Google Scholar] [CrossRef]
- Basturea, G.N.; Bodero, M.D.; Moreno, M.E.; Munson, G.P. Residues near the amino terminus of Rns are essential for positive autoregulation and DNA binding. J. Bacteriol. 2008, 190, 2279–2285. [Google Scholar] [CrossRef]
- Bodero, M.D.; Harden, E.A.; Munson, G.P. Transcriptional regulation of subclass 5b fimbriae. BMC Microbiol. 2008, 8, 180. [Google Scholar] [CrossRef]
- Favre, D.; Lüdi, S.; Stoffel, M.; Frey, J.; Horn, M.P.; Dietrich, G.; Spreng, S.; Viret, J.F. Expression of enterotoxigenic Escherichia coli colonization factors in Vibrio cholerae. Vaccine 2006, 24, 4354–4368. Available online: https://www.sciencedirect.com/science/article/pii/S0264410X06002489?via%3Dihub (accessed on 18 July 2022). [CrossRef]
- Froehlich, B.; Husmann, L.; Caron, J.; Scott, J.R. Regulation of rns, a positive regulatory factor for pili of enterotoxigenic Escherichia coli. J. Bacteriol. 1994, 176, 5385–5392. [Google Scholar] [CrossRef] [PubMed]
- Munson, G.P.; Holcomb, L.G.; Alexander, H.L.; Scott, J.R. In vitro identification of Rns-regulated genes. J. Bacteriol. 2002, 184, 1196–1199. [Google Scholar] [CrossRef] [PubMed]
- Pilonieta, M.C.; Bodero, M.D.; Munson, G.P. CfaD-dependent expression of a novel extracytoplasmic protein from enterotoxigenic Escherichia coli. J. Bacteriol. 2007, 189, 5060–5067. [Google Scholar] [CrossRef]
- de Haan, L.A.; Willshaw, G.A.; van der Zeijst, B.A.; Gaastra, W. The nucleotide sequence of a regulatory gene present on a plasmid in an enterotoxigenic Escherichia coli strain of serotype O167:H5. FEMS Microbiol. Lett. 1991, 83, 341–346. [Google Scholar] [CrossRef]
- Tobe, T.; Schoolnik, G.K.; Sohel, I.; Bustamante, V.H.; Puente, J.L. Cloning and characterization of bfpTVW, genes required for the transcriptional activation of bfpA in enteropathogenic Escherichia coli. Mol. Microbiol. 1996, 21, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Lowden, M.J.; Skorupski, K.; Pellegrini, M.; Chiorazzo, M.G.; Taylor, R.K.; Kull, F.J. Structure of Vibrio cholerae ToxT reveals a mechanism for fatty acid regulation of virulence genes. Proc. Natl. Acad. Sci. USA 2010, 107, 2860–2865. [Google Scholar] [CrossRef]
- Di Martino, M.L.; Falconi, M.; Micheli, G.; Colonna, B.; Prosseda, G. The Multifaceted Activity of the VirF Regulatory Protein in the Shigella Lifestyle. Front. Mol. Biosci. 2016, 3, 61. [Google Scholar] [CrossRef] [PubMed]
- Jordi, B.J.; Dagberg, B.; de Haan, L.A.; Hamers, A.M.; van der Zeijst, B.A.; Gaastra, W.; Uhlin, B.E. The positive regulator CfaD overcomes the repression mediated by histone-like protein H-NS (H1) in the CFA/I fimbrial operon of Escherichia coli. EMBO J. 1992, 11, 2627–2632. [Google Scholar] [CrossRef] [PubMed]
- Mahon, V.; Smyth, C.J.; Smith, S.G.J. Mutagenesis of the Rns regulator of enterotoxigenic Escherichia coli reveals roles for a linker sequence and two helix-turn-helix motifs. Microbiology 2010, 156, 2796–2806. [Google Scholar] [CrossRef]
- Mahon, V.; Fagan, R.P.; Smith, S.G. Snap denaturation reveals dimerization by AraC-like protein Rns. Biochimie 2012, 94, 2058–2061. [Google Scholar] [CrossRef][Green Version]
- Knight, S.D. Structure and assembly of Yersinia pestis F1 antigen. Adv. Exp. Med. Biol. 2007, 603, 74–87. [Google Scholar] [CrossRef] [PubMed]
- Karlyshev, A.V.; Galyov, E.E.; Abramov, V.M.; Zav’yalov, V.P. Caf1R gene and its role in the regulation of capsule formation of Y. pestis. FEBS Lett. 1992, 305, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Porter, M.E.; Mitchell, P.; Roe, A.J.; Free, A.; Smith, D.G.; Gally, D.L. Direct and indirect transcriptional activation of virulence genes by an AraC-like protein, PerA from enteropathogenic Escherichia coli. Mol. Microbiol. 2004, 54, 1117–1133. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.G.; Jair, K.W.; Wolf, R.E., Jr.; Rosner, J.L. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 1996, 178, 2216–2223. [Google Scholar] [CrossRef][Green Version]
- Schleif, R. AraC protein, regulation of the L-arabinose operon in Escherichia coli, and the light switch mechanism of AraC action. FEMS Microbiol. Rev. 2010, 34, 779–796. [Google Scholar] [CrossRef]
- Kolin, A.; Balasubramaniam, V.; Skredenske, J.M.; Wickstrum, J.R.; Egan, S.M. Differences in the mechanism of the allosteric l-rhamnose responses of the AraC/XylS family transcription activators RhaS and RhaR. Mol. Microbiol. 2008, 68, 448–461. [Google Scholar] [CrossRef]
- Dominguez-Cuevas, P.; Marin, P.; Busby, S.; Ramos, J.L.; Marques, S. Roles of effectors in XylS-dependent transcription activation: Intramolecular domain derepression and DNA binding. J. Bacteriol. 2008, 190, 3118–3128. [Google Scholar] [CrossRef]
- Ni, L.; Tonthat, N.K.; Chinnam, N.; Schumacher, M.A. Structures of the Escherichia coli transcription activator and regulator of diauxie, XylR: An AraC DNA-binding family member with a LacI/GalR ligand-binding domain. Nucleic Acids Res. 2013, 41, 1998–2008. [Google Scholar] [CrossRef]
- Schuller, A.; Slater, A.W.; Norambuena, T.; Cifuentes, J.J.; Almonacid, L.I.; Melo, F. Computer-based annotation of putative AraC/XylS-family transcription factors of known structure but unknown function. J. Biomed. Biotechnol. 2012, 2012, 103132. [Google Scholar] [CrossRef]
- Derbise, A.; Cerda Marin, A.; Ave, P.; Blisnick, T.; Huerre, M.; Carniel, E.; Demeure, C.E. An encapsulated Yersinia pseudotuberculosis is a highly efficient vaccine against pneumonic plague. PLoS Negl. Trop. Dis. 2012, 6, e1528. [Google Scholar] [CrossRef]
- Gallagher, T.B.; Mellado-Sanchez, G.; Jorgensen, A.L.; Moore, S.; Nataro, J.P.; Pasetti, M.F.; Baillie, L.W. Development of a multiple-antigen protein fusion vaccine candidate that confers protection against Bacillus anthracis and Yersinia pestis. PLoS Negl. Trop. Dis. 2019, 13, e0007644. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Singh, A.K. Plague vaccine: Recent progress and prospects. NPJ Vaccines 2019, 4, 11. [Google Scholar] [CrossRef] [PubMed]
- van Ham, S.M.; van Alphen, L.; Mooi, F.R.; van Putten, J.P. Phase variation of H. influenzae fimbriae: Transcriptional control of two divergent genes through a variable combined promoter region. Cell 1993, 73, 1187–1196. [Google Scholar] [CrossRef] [PubMed]
- Kinch, L.N.; Cong, Q.; Jaishankar, J.; Orth, K. Co-component signal transduction systems: Fast-evolving virulence regulation cassettes discovered in enteric bacteria. Proc. Natl. Acad. Sci. USA 2022, 119, e2203176119. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gahlot, D.K.; Taheri, N.; MacIntyre, S. Diversity in Genetic Regulation of Bacterial Fimbriae Assembled by the Chaperone Usher Pathway. Int. J. Mol. Sci. 2023, 24, 161. https://doi.org/10.3390/ijms24010161
Gahlot DK, Taheri N, MacIntyre S. Diversity in Genetic Regulation of Bacterial Fimbriae Assembled by the Chaperone Usher Pathway. International Journal of Molecular Sciences. 2023; 24(1):161. https://doi.org/10.3390/ijms24010161
Chicago/Turabian StyleGahlot, Dharmender K., Nayyer Taheri, and Sheila MacIntyre. 2023. "Diversity in Genetic Regulation of Bacterial Fimbriae Assembled by the Chaperone Usher Pathway" International Journal of Molecular Sciences 24, no. 1: 161. https://doi.org/10.3390/ijms24010161
APA StyleGahlot, D. K., Taheri, N., & MacIntyre, S. (2023). Diversity in Genetic Regulation of Bacterial Fimbriae Assembled by the Chaperone Usher Pathway. International Journal of Molecular Sciences, 24(1), 161. https://doi.org/10.3390/ijms24010161






