clbP Gene, a Potential New Member of the β-Lactamase Family
Abstract
1. Introduction
2. Results
2.1. In Silico Visualisation of the clbP Gene and Enzyme
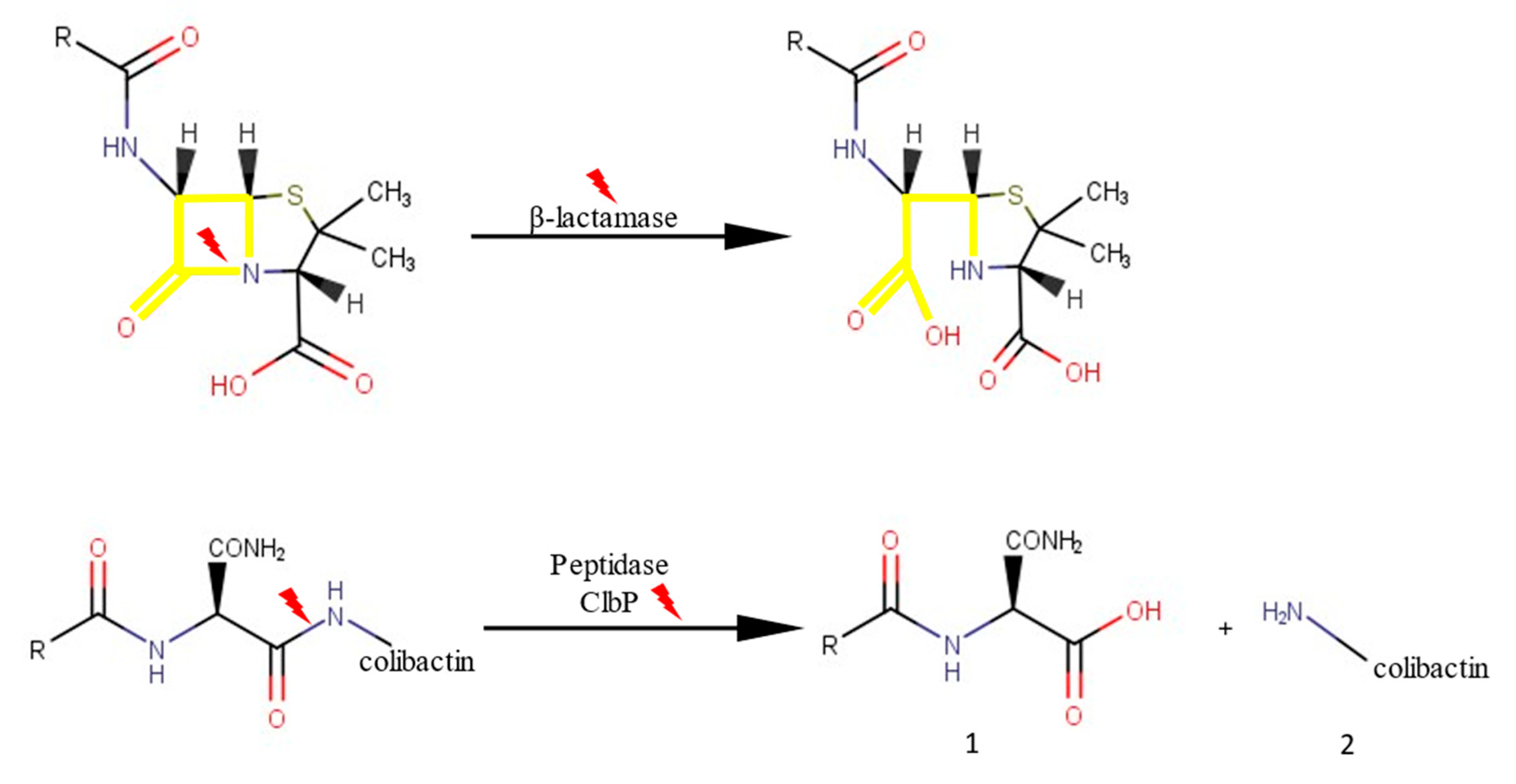
2.1.1. clbP Gene and Origin of the Enzyme
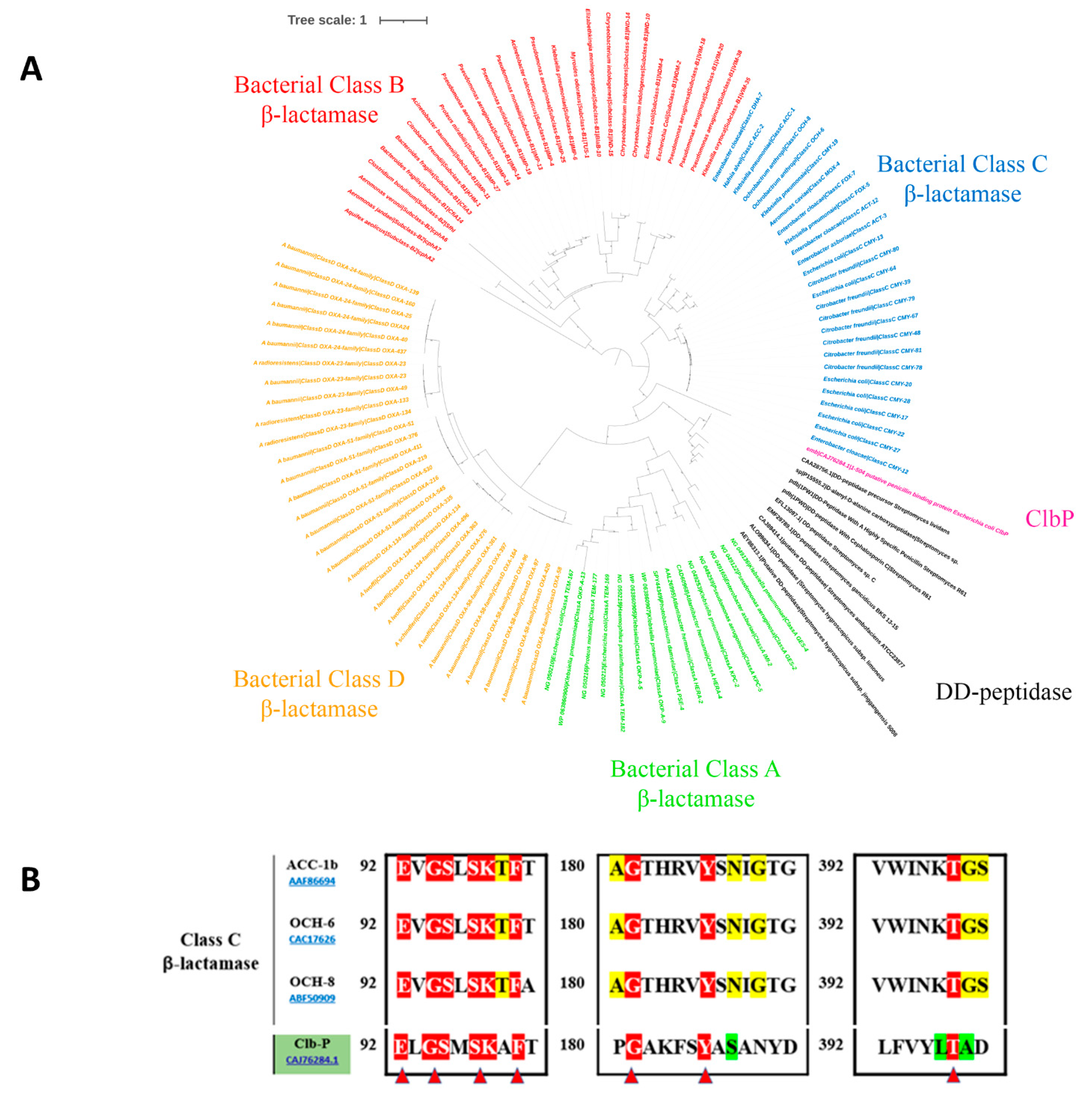
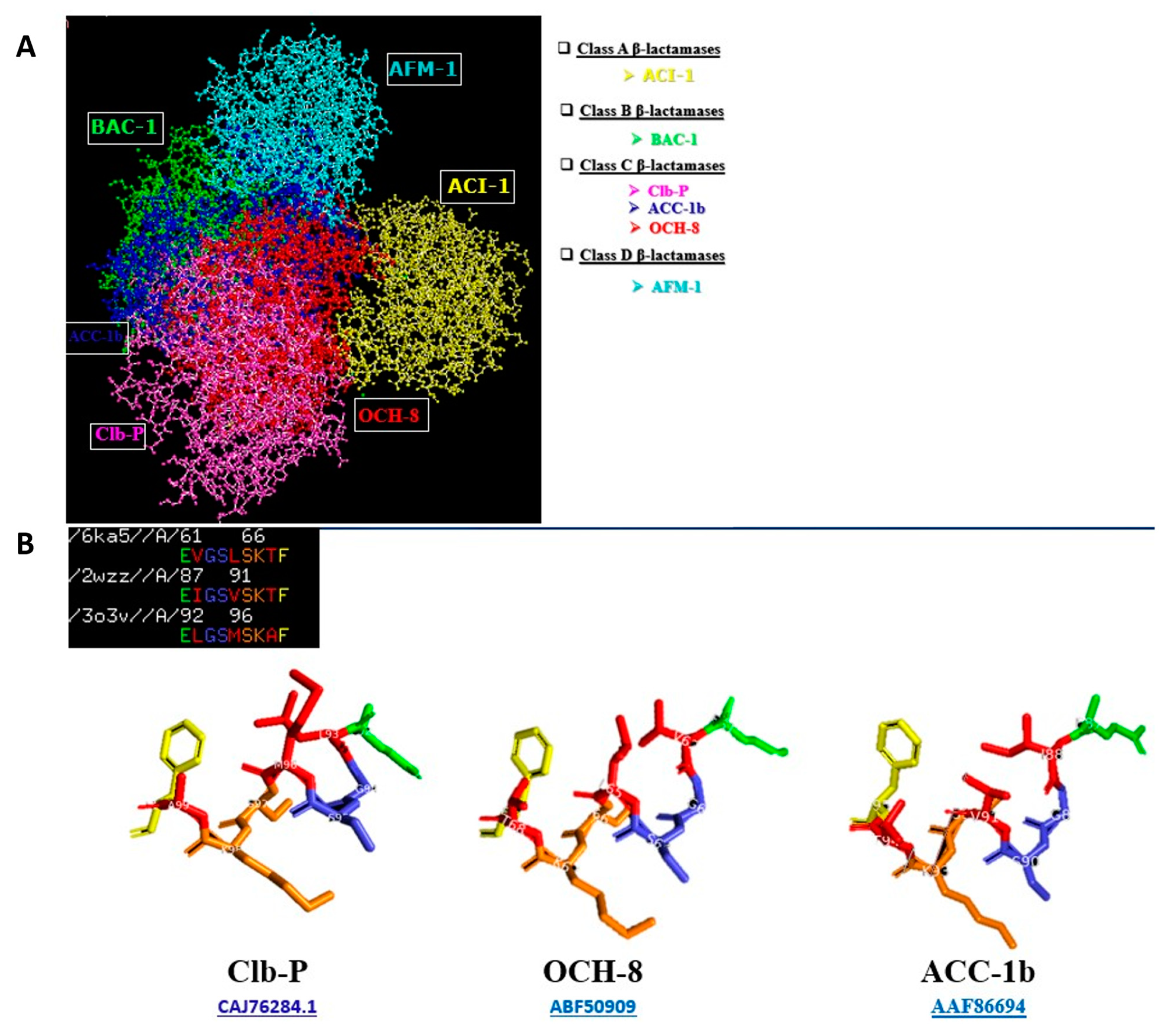
2.1.2. The ClbP Enzyme’s Phylogenetic Link to Bacterial β-Lactamases
2.2. In Vitro Expression of the clbP Gene
2.2.1. β-Lactam-Hydrolysing Activity
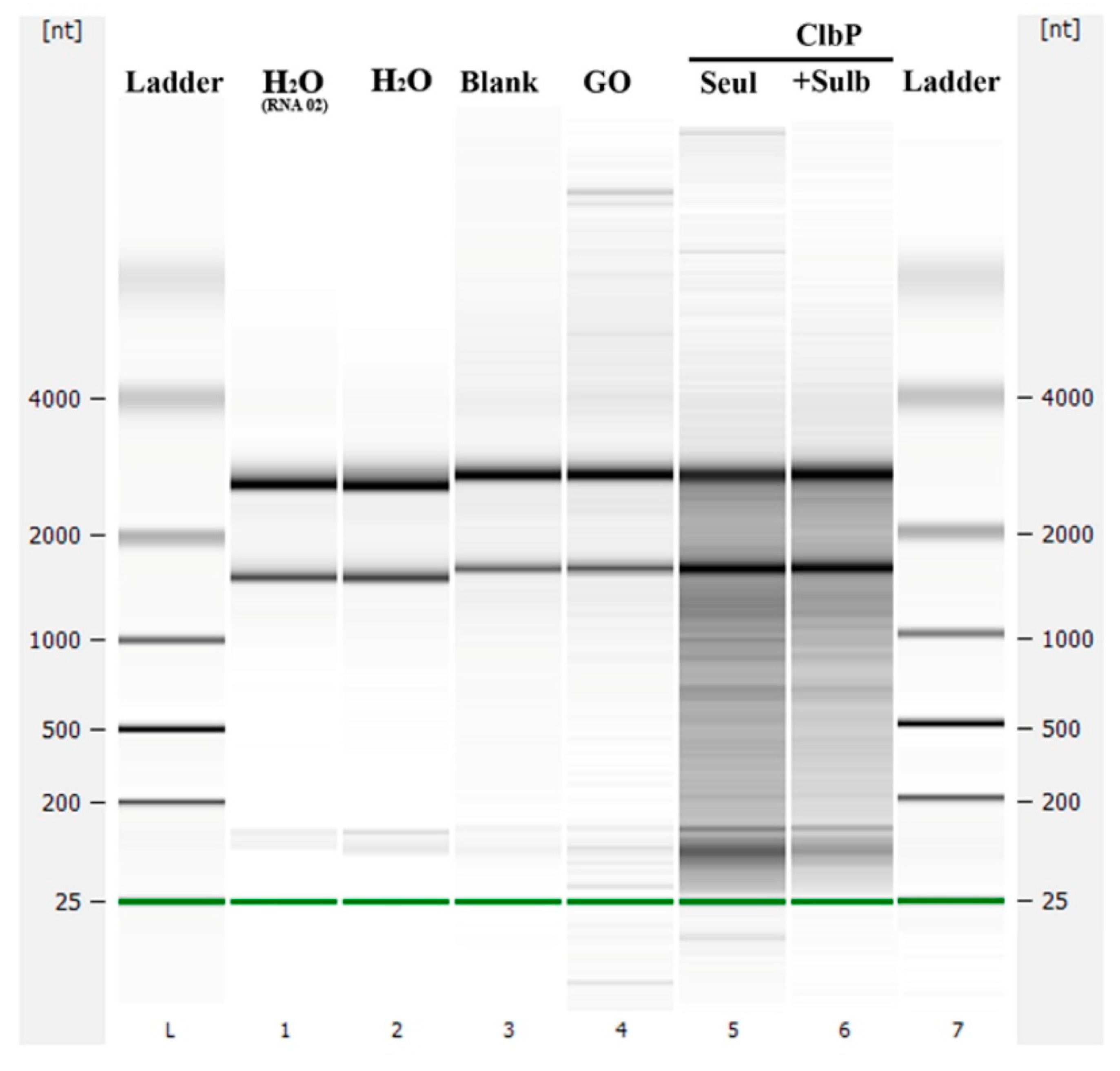
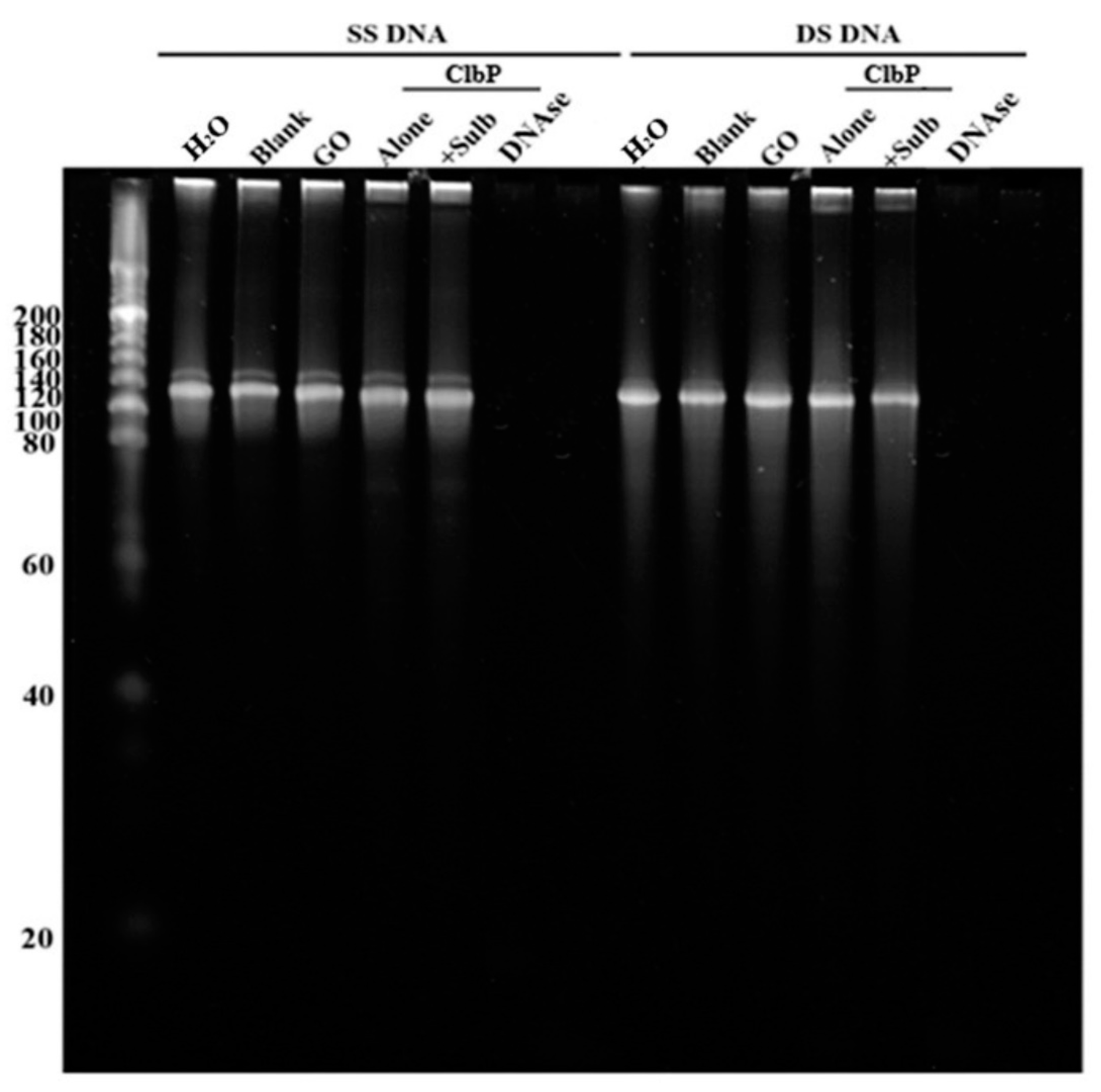
2.2.2. Nuclease and Ribonuclease Activity
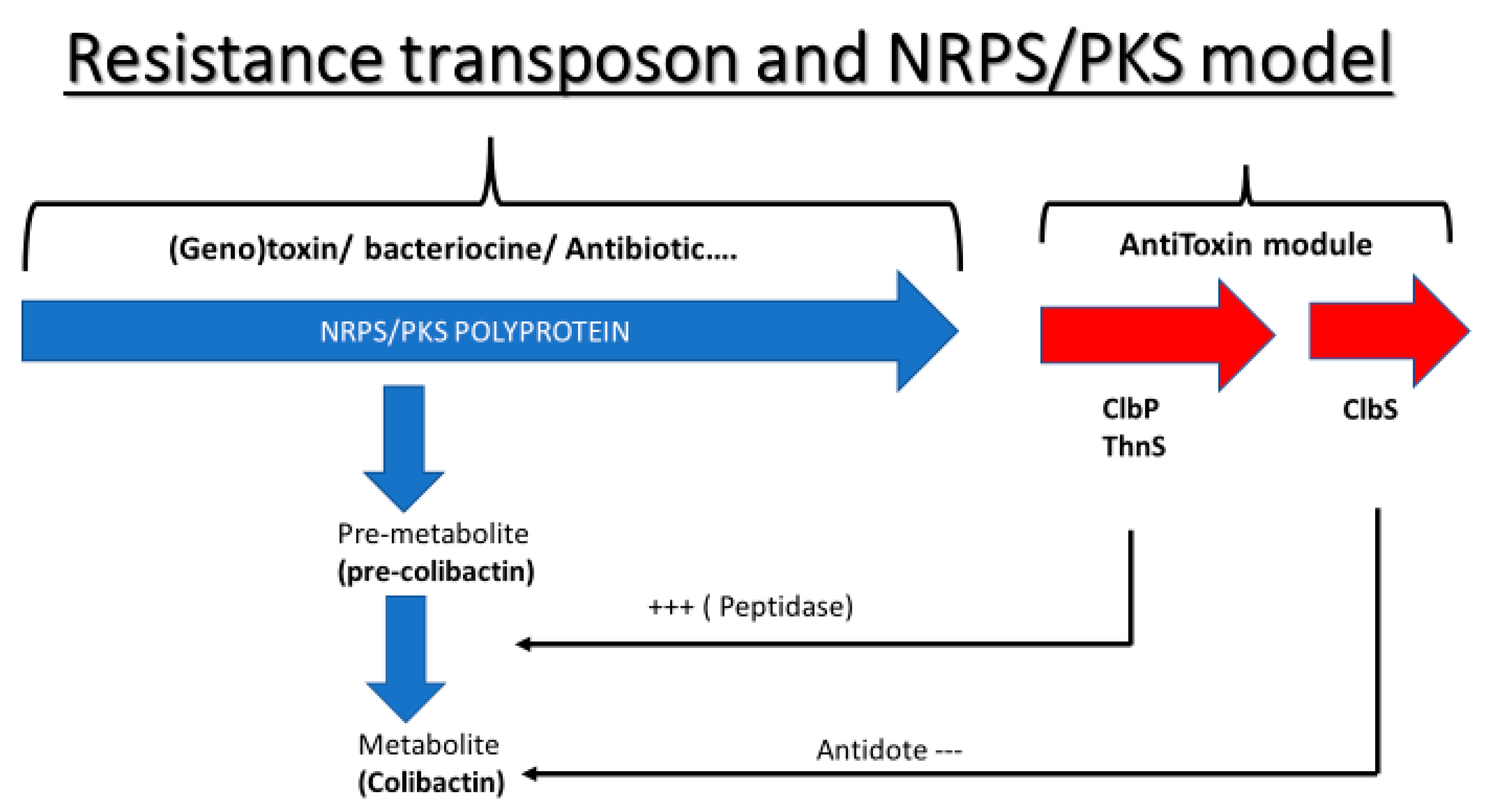
3. Discussion
4. Materials and Methods
4.1. Phylogeny of the ClbP Protein
4.2. Structural Prediction and Alignment of ClbP
4.3. Recombinant Protein Expression and Purification
4.4. Antibiotic Sensitivity Testing
4.5. Ribonuclease and Nuclease Activity
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Li, J.W.-H.; Vederas, J.C. Drug Discovery and Natural Products: End of an Era or an Endless Frontier? Science 2009, 325, 161–165. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, H.; Li, Y.; Hao, C. Association between Virulence Profile and Fluoroquinolone Resistance in Escherichia coli Isolated from Dogs and Cats in China. J. Infect. Dev. Ctries 2017, 11, 306–313. [Google Scholar] [CrossRef] [PubMed]
- Weissman, K.J.; Leadlay, P.F. Combinatorial Biosynthesis of Reduced Polyketides. Nat. Rev. Microbiol. 2005, 3, 925–936. [Google Scholar] [CrossRef]
- Fechtner, J.L. Fungi and Bacteria Antibiotics: A Review. J. Am. Dent. Assoc. 1944, 31, 1217–1225. [Google Scholar] [CrossRef]
- Ogawara, H. Comparison of Antibiotic Resistance Mechanisms in Antibiotic-Producing and Pathogenic Bacteria. Molecules 2019, 24, 3430. [Google Scholar] [CrossRef]
- Antibiotics Classification Based on Mode of Its Action—BiokiMicroki. Available online: https://biokimicroki.com/antibiotics-classification-based-on-mode-of-action/ (accessed on 3 December 2022).
- Hayes, F.; Van Melderen, L. Toxins-antitoxins: Diversity, evolution and function. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 386–408. [Google Scholar] [CrossRef]
- Barbe, V.; Bouzon, M.; Mangenot, S.; Badet, B.; Poulain, J.; Segurens, B.; Vallenet, D.; Marlière, P.; Weissenbach, J. Complete Genome Sequence of Streptomyces cattleya NRRL 8057, a Producer of Antibiotics and Fluorometabolites. J. Bacteriol. 2011, 193, 5055–5056. [Google Scholar] [CrossRef]
- Diene, S.M.; Pinault, L.; Baron, S.A.; Azza, S.; Armstrong, N.; Hadjadj, L.; Chabrière, E.; Rolain, J.-M.; Pontarotti, P.; Raoult, D. A metallo-β-lactamase enzyme for internal detoxification of the antibiotic thienamycin. Sci. Rep. 2021, 11, 1–8. [Google Scholar] [CrossRef]
- Pandey, N.; Cascella, M. Beta Lactam Antibiotics. In Antibiotic Discovery and Development; Springer: Berlin/Heidelberg, Germany, 2022; pp. 79–117. [Google Scholar] [CrossRef]
- Miyachiro, M.M.; Contreras-Martel, C.; Dessen, A. Penicillin-Binding Proteins (PBPs) and Bacterial Cell Wall Elongation Complexes. Subcell Biochem. 2019, 93, 273–289. [Google Scholar] [CrossRef]
- Nougayrède, J.-P.; Homburg, S.; Taieb, F.; Boury, M.; Brzuszkiewicz, E.; Gottschalk, G.; Buchrieser, C.; Hacker, J.; Dobrindt, U.; Oswald, E. Escherichia coli Induces DNA Double-Strand Breaks in Eukaryotic Cells. Science 2006, 313, 845–848. [Google Scholar] [CrossRef]
- Dubois, D.; Delmas, J.; Cady, A.; Robin, F.; Sivignon, A.; Oswald, E.; Bonnet, R. Cyclomodulins in Urosepsis Strains of Escherichia coli. J. Clin. Microbiol. 2010, 48, 2122–2129. [Google Scholar] [CrossRef] [PubMed]
- Johnson, J.R.; Johnston, B.; Kuskowski, M.A.; Nougayrede, J.P.; Oswald, E. Molecular Epidemiology and Phylogenetic Distribution of the Escherichia coli Pks Genomic Island. J. Clin. Microbiol. 2008, 46, 3906–3911. [Google Scholar] [CrossRef] [PubMed]
- Dougherty, M.W.; Jobin, C. Shining a Light on Colibactin Biology. Toxins 2021, 13, 346. [Google Scholar] [CrossRef] [PubMed]
- Chagneau, C.V.; Massip, C.; Bossuet-Greif, N.; Fremez, C.; Motta, J.P.; Shima, A.; Besson, C.; le Faouder, P.; Cénac, N.; Roth, M.P.; et al. Uropathogenic E. Coli Induces DNA Damage in the Bladder. PLoS Pathog. 2021, 17, e1009310. [Google Scholar] [CrossRef] [PubMed]
- Kharofa, J.; Apewokin, S.; Alenghat, T.; Ollberding, N.J. Metagenomic Analysis of the Fecal Microbiome in Colorectal Cancer Patients Compared to Healthy Controls as a Function of Age. Cancer Med. 2022; Online ahead of print. [Google Scholar] [CrossRef]
- Du, L.; Song, J. Delivery, Structure, and Function of Bacterial Genotoxins. Virulence 2022, 13, 1199. [Google Scholar] [CrossRef]
- Dubinsky, V.; Reshef, L.; Rabinowitz, K.; Wasserberg, N.; Dotan, I.; Gophna, U. Escherichia coli Strains from Patients with Inflammatory Bowel Diseases Have Disease-Specific Genomic Adaptations. J. Crohns Colitis 2022, 16, 1584–1597. [Google Scholar] [CrossRef]
- Bacterial Toxin Is Found in Patients with Urinary Tract Infections—ScienceDaily. Available online: https://www.sciencedaily.com/releases/2021/02/210225143648.htm (accessed on 3 December 2022).
- Auvray, F.; Perrat, A.; Arimizu, Y.; Chagneau, C.V.; Bossuet-Greif, N.; Massip, C.; Brugère, H.; Nougayrède, J.-P.; Hayashi, T.; Branchu, P.; et al. Insights into the Acquisition of the Pks Island and Production of Colibactin in the Escherichia coli Population. bioRxiv 2021. [Google Scholar] [CrossRef]
- Nougayrède, J.-P.; Chagneau, C.V.; Motta, J.-P.; Bossuet-Greif, N.; Belloy, M.; Taieb, F.; Gratadoux, J.-J.; Thomas, M.; Langella, P.; Oswald, E. A Toxic Friend: Genotoxic and Mutagenic Activity of the Probiotic Strain Escherichia coli Nissle 1917. mSphere 2021, 6, e00624-21. [Google Scholar] [CrossRef]
- Suresh, A.; Shaik, S.; Baddam, R.; Ranjan, A.; Qumar, S.; Jadhav, S.; Ghazi, I.A.; Wieler, L.H.; Ahmed, N.; Semmler, T. Evolutionary Dynamics Based on Comparative Genomics of Pathogenic Escherichia coli Lineages Harboring Polyketide Synthase ( Pks) Island. mBio 2021, 12, e03634-20. [Google Scholar] [CrossRef]
- Tang, J.W.; Liu, X.; Ye, W.; Li, Z.R.; Qian, P.Y. Biosynthesis and Bioactivities of Microbial Genotoxin Colibactins. Nat. Prod. Rep. 2022, 39, 991–1014. [Google Scholar] [CrossRef]
- Faïs, T.; Delmas, J.; Barnich, N.; Bonnet, R.; Dalmasso, G. Colibactin: More Than a New Bacterial Toxin. Toxins 2018, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Parker, W.L.; Rathnum, M.L.; Scott Wells, J.; Trejo, W.H.; Principe, P.A.; Sykes, R.B. SQ 27, 860, a simple carbapenem produced by species of serratia and erwinia. J. Antibiot. 1982, 35, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Aoki, H.; Okuhara, M. Natural Beta-Lactam Antibiotics. Annu. Rev. Microbiol. 2003, 34, 159–181. [Google Scholar] [CrossRef]
- Cougnoux, A.; Gibold, L.; Robin, F.; Dubois, D.; Pradel, N.; Darfeuille-Michaud, A.; Dalmasso, G.; Delmas, J.; Bonnet, R. Analysis of Structure-Function Relationships in the Colibactin-Maturating Enzyme ClbP. J. Mol. Biol. 2012, 424, 203–214. [Google Scholar] [CrossRef] [PubMed]
- Morales-Lozada, Y.; Gómez-Moreno, R.; Báez-Bravo, G.; Robledo, I.E.; Suazo-Dávila, D.; Cabrera, C.R.; Baerga-Ortiz, A.; Juan, S.; Rico, P. The Accumulation of Colibactin Intermediates Does Not Affect the Growth and Morphology of Pks+ Escherichia coli. bioRxiv 2020. [Google Scholar] [CrossRef]
- Massip, C.; Branchu, P.; Bossuet-Greif, N.; Chagneau, C.V.; Gaillard, D.; Martin, P.; Boury, M.; Sécher, T.; Dubois, D.; Nougayrède, J.-P.; et al. Deciphering the Interplay between the Genotoxic and Probiotic Activities of Escherichia coli Nissle 1917. PLoS Pathog. 2019, 15, e1008029. [Google Scholar] [CrossRef]
- Volpe, M.R.; Wilson, M.R.; Brotherton, C.A.; Winter, E.S.; Johnson, S.E.; Balskus, E.P. In Vitro Characterization of the Colibactin-Activating Peptidase ClbP Enables Development of a Fluorogenic Activity Probe. ACS Chem. Biol. 2019, 14, 1097–1101. [Google Scholar] [CrossRef]
- Dubois, D.; Baron, O.; Cougnoux, A.; Delmas, J.; Pradel, N.; Boury, M.; Bouchon, B.; Bringer, M.A.; Nougayrède, J.P.; Oswald, E.; et al. ClbP Is a Prototype of a Peptidase Subgroup Involved in Biosynthesis of Nonribosomal Peptides. J. Biol. Chem. 2011, 286, 35562–35570. [Google Scholar] [CrossRef]
- Pope, J.L.; Yang, Y.; Newsome, R.C.; Sun, W.; Sun, X.; Ukhanova, M.; Neu, J.; Issa, J.P.; Mai, V.; Jobin, C. Microbial Colonization Coordinates the Pathogenesis of a Klebsiella Pneumoniae Infant Isolate. Sci. Rep. 2019, 9, 3380. [Google Scholar] [CrossRef]
- Gupta, S.K.; Padmanabhan, B.R.; Diene, S.M.; Lopez-Rojas, R.; Kempf, M.; Landraud, L.; Rolain, J.M. ARG-Annot, a New Bioinformatic Tool to Discover Antibiotic Resistance Genes in Bacterial Genomes. Antimicrob. Agents Chemother. 2014, 58, 212–220. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. TrimAl: A Tool for Automated Alignment Trimming in Large-Scale Phylogenetic Analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2 Approximately Maximum-Likelihood Trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef] [PubMed]
- Diene, S.M.; Pinault, L.; Keshri, V.; Armstrong, N.; Khelaifia, S.; Chabrière, E.; Caetano-Anolles, G.; Colson, P.; la Scola, B.; Rolain, J.M.; et al. Human Metallo-β-Lactamase Enzymes Degrade Penicillin. Sci. Rep. 2019, 9, 12173. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azour, A.; Al-Bayssari, C.; Pinault, L.; Azza, S.; Rolain, J.-M.; Diene, S.M. clbP Gene, a Potential New Member of the β-Lactamase Family. Int. J. Mol. Sci. 2022, 23, 15642. https://doi.org/10.3390/ijms232415642
Azour A, Al-Bayssari C, Pinault L, Azza S, Rolain J-M, Diene SM. clbP Gene, a Potential New Member of the β-Lactamase Family. International Journal of Molecular Sciences. 2022; 23(24):15642. https://doi.org/10.3390/ijms232415642
Chicago/Turabian StyleAzour, Adel, Charbel Al-Bayssari, Lucile Pinault, Saïd Azza, Jean-Marc Rolain, and Seydina M. Diene. 2022. "clbP Gene, a Potential New Member of the β-Lactamase Family" International Journal of Molecular Sciences 23, no. 24: 15642. https://doi.org/10.3390/ijms232415642
APA StyleAzour, A., Al-Bayssari, C., Pinault, L., Azza, S., Rolain, J.-M., & Diene, S. M. (2022). clbP Gene, a Potential New Member of the β-Lactamase Family. International Journal of Molecular Sciences, 23(24), 15642. https://doi.org/10.3390/ijms232415642





