The Development of Two High-Yield and High-Quality Functional Rice Cultivars Using Marker-Assisted Selection and Conventional Breeding Methods
Abstract
:1. Introduction
2. Results
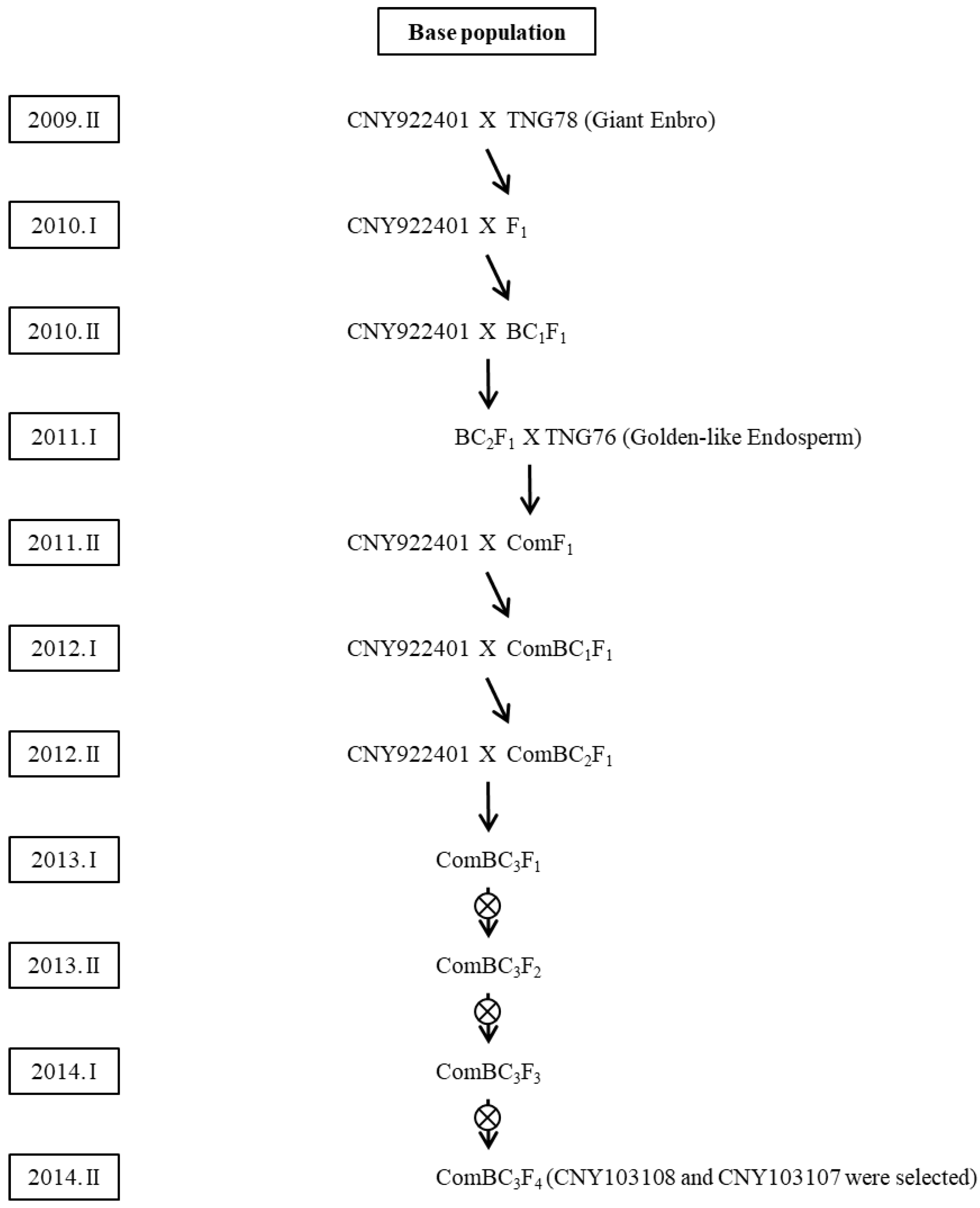
2.1. Base Population
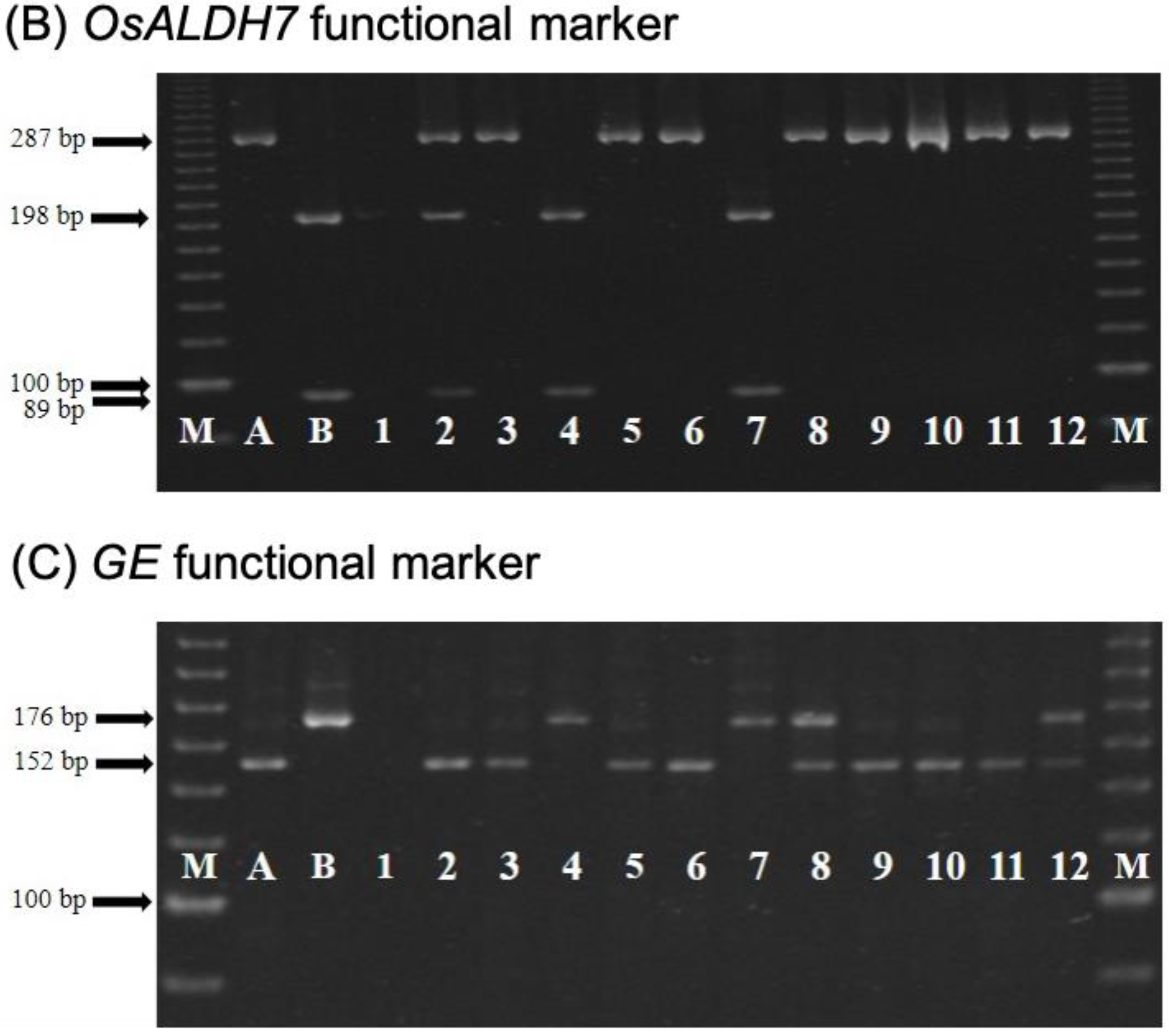
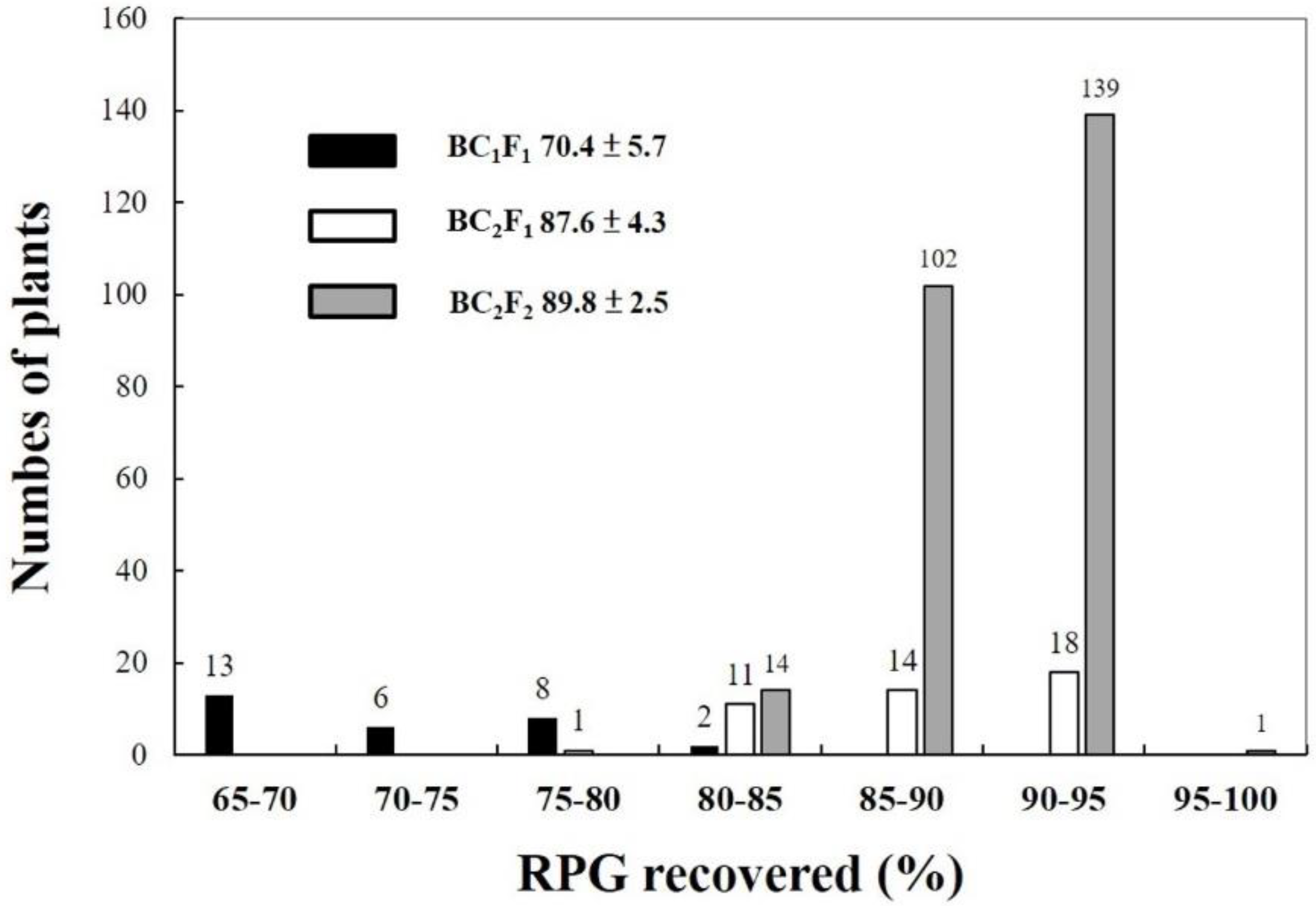
2.2. CNY922401/CNY103018 (PFR Population)
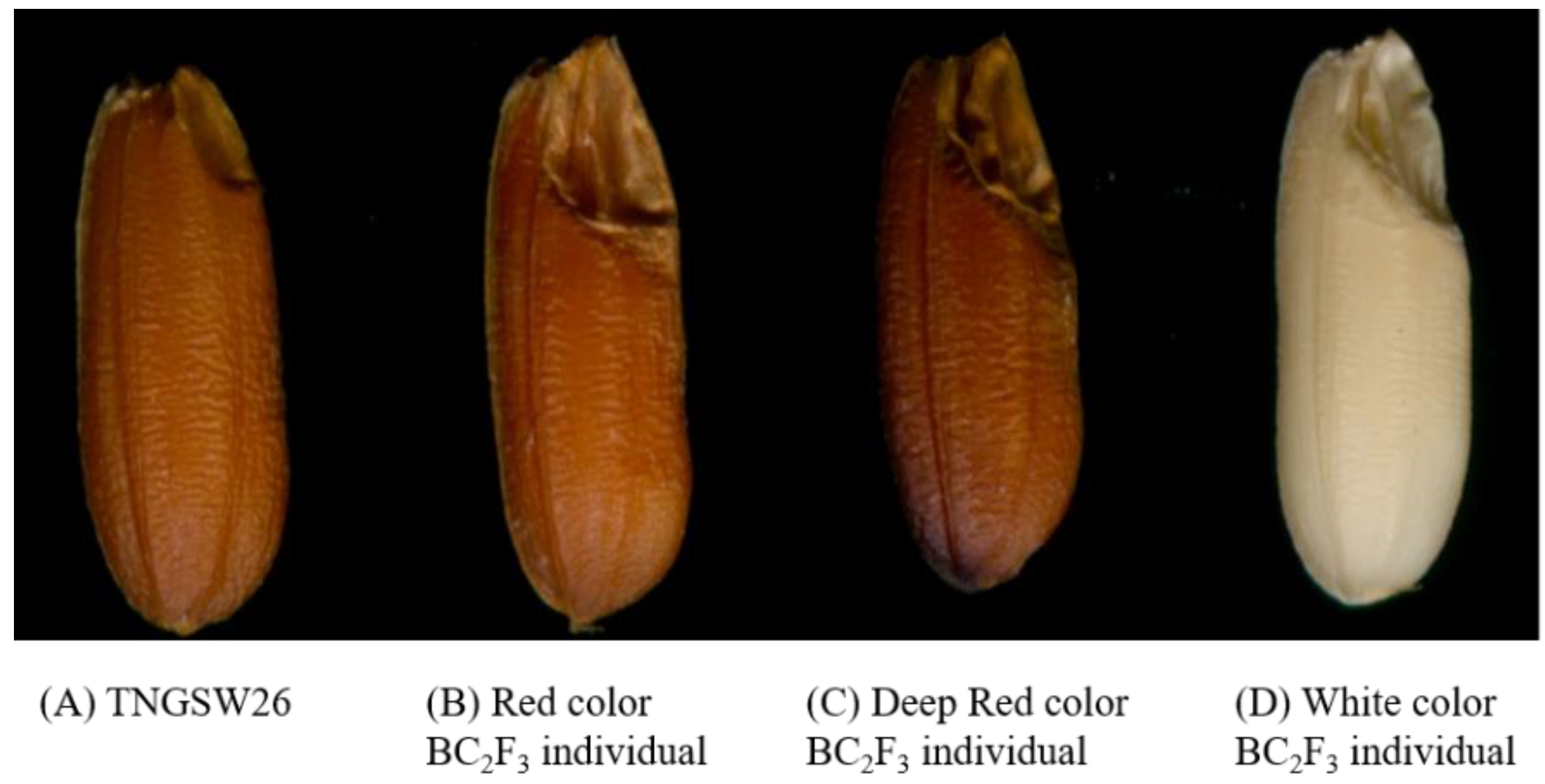
2.3. TNGSW26/CNY103017 (RFR Population)
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. Molecular Marker Design and Selection
4.3. DNA Isolation and PCR Amplification
4.4. Antioxidant Analysis
4.4.1. Total Phenolic Content
4.4.2. Flavonoid Contents
4.4.3. DPPH Scavenging Activity (%)
4.4.4. Monomeric Anthocyanin Pigment Content
4.5. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Con sent Statement
Acknowledgments
Conflicts of Interest
References
- Springmann, M.; Mason-D’Croz, D.; Robinson, S.; Garnett, T.; Godfray, H.C.J.; Gollin, D.; Rayner, M.; Ballon, P.; Scarborough, P. Global and regional health effects of future food production under climate change: A modelling study. Lancet 2016, 387, 1937–1946. [Google Scholar] [CrossRef] [Green Version]
- Juliano, B.O.; Duff, B. Rice grain quality as an emerging priority in National rice breeding programmes. In Rice Grain Marketing and Quality Issues: Selected Papers from the International Rice Research Conference, 27–31 August 1990, Seoul, Korea; IRRI: Manila, Philippines, 1991; pp. 55–64. [Google Scholar]
- Henderson, A.J.; Ollila, C.A.; Kumar, A.; Borresen, E.C.; Raina, K.; Agarwal, R.; Ryan, E.P. Chemopreventive properties of dietary rice bran: Current status and future prospects. Adv. Nutr. 2012, 3, 643–653. [Google Scholar] [CrossRef] [Green Version]
- Nestel, P.; Bouis, H.E.; Meenakshi, J.V.; Pfeiffer, W. Biofortification of staple food crops. J. Nutr. 2006, 136, 1064–1067. [Google Scholar] [CrossRef]
- Takahashi, S.; Ohtani, T.; Iida, S.; Sunohara, Y.; Matsushita, K.; Maeda, H.; Tanetani, Y.; Kawai, K.; Kawamukai, M.; Kadowaki, K.I. Development of CoQ10-enriched rice from giant embryo lines. Breed. Sci. 2009, 59, 321–326. [Google Scholar] [CrossRef] [Green Version]
- Kawakatsu, T.; Wang, S.; Wakasa, Y.; Takaiwa, F. Increased lysine content in rice grains by over-accumulation of BiP in the endosperm. Biosci. Biotechnol. Biochem. 2010, 74, 2529–2531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.R.; Kim, C.E.; Kang, M.Y.; Nam, S.H. Cholesterol-lowering and antioxidant status-improving efficacy of germinated giant embryonic rice (Oryza sativa L.) in high cholesterol-fed rats. Ann. Nutr. Metab. 2007, 51, 519–526. [Google Scholar] [CrossRef] [PubMed]
- Min, B.; Gu, L.; McClung, A.M.; Bergman, C.J.; Chen, M.H. Free and bound total phenolic concentrations, antioxidant capacities, and profiles of proanthocyanidins and anthocyanins in whole grain rice (Oryza sativa L.) of different bran colours. Food Chem. 2012, 133, 715–722. [Google Scholar] [CrossRef]
- Zhu, L.; Gu, M.; Meng, X.; Cheung, S.C.K.; Yu, H.; Huang, J.; Sun, Y.; Shi, Y.; Liu, Q. High-amylose rice improves indices of animal health in normal and diabetic rats. Plant Biotechnol. J. 2012, 10, 353–362. [Google Scholar] [CrossRef]
- Friedman, M. Rice brans, rice bran oils, and rice hulls: Composition, food and industrial uses, and bioactivities in humans, animals, and cells. J. Agric. Food Chem. 2013, 61, 10626–10641. [Google Scholar] [CrossRef]
- Del Pozo-Insfran, D.; Brenes, C.H.; Saldivar, S.O.S.; Talcott, S.T. Polyphenolic and antioxidant content of white and blue corn (Zea mays L.) products. Food Res Int. 2006, 39, 696–703. [Google Scholar] [CrossRef]
- Urias-Lugo, D.A.; Heredia, J.B.; Serna-Saldivar, S.O.; Muy-Rangel, M.D.; Valdez-Torres, J.B. Total phenolics, total anthocyanins and antioxidant capacity of native and elite blue maize hybrids (Zea mays L.). CyTA-J. Food. 2015, 13, 336–339. [Google Scholar] [CrossRef]
- Urias-Peraldí, M.; Gutiérrez-Uribe, J.A.; Preciado-Ortiz, R.E.; Cruz-Morales, A.S.; Serna-Saldívar, S.O.; García-Lara, S. Nutraceutical profiles of improved blue maize (Zea mays) hybrids for subtropical regions. Field Crops Res. 2013, 141, 69–76. [Google Scholar] [CrossRef]
- Han, K.H.; Sekikawa, M.; Shimada, K.I.; Hashimoto, M.; Hashimoto, N.; Noda, T.; Tanaka, H.; Fukushima, M. Anthocyanin-rich purple potato flake extract has antioxidant capacity and improves antioxidant potential in rats. Brit. J. Nutr. 2006, 96, 1125–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, Y.; Deng, L.; Chen, J.; Zhou, S.; Liu, S.; Fu, Y.; Yang, C.; Liao, Z.; Chen, M. An analytical pipeline to compare and characterise the anthocyanin antioxidant activities of purple sweet potato cultivars. Food Chem. 2016, 194, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, L.A.J.C.; Gomes, J.C.; Stringheta, P.C.; Gontijo, Á.M.; Padovani, C.R.; Ribeiro, L.R.; Salvadori, D.M.F. Black bean (Phaseolus vulgaris L.) as a protective agent against DNA damage in mice. Food Chem. Toxicol. 2003, 41, 1671–1676. [Google Scholar] [CrossRef]
- Tikkanen, M.J.; Adlercreutz, H. Dietary soy-derived isoflavone phytoestrogens: Could they have a role in coronary heart disease prevention? Biochem. Pharmacol. 2000, 60, 1–5. [Google Scholar] [CrossRef]
- Nishimura, M.; Yoshida, S.I.; Haramoto, M.; Mizuno, H.; Fukuda, T.; Kagami-Katsuyama, H.; Tanaka, A.; Ohkawara, T.; Sato, Y.; Nishihira, J. Effects of white rice containing enriched gamma-aminobutyric acid on blood pressure. J. Tradit. Complement. Med. 2016, 6, 66–71. [Google Scholar] [CrossRef] [Green Version]
- Hong, S.K.; Kitano, H.; Satoh, H.; Nagato, Y. How is embryo size genetically regulated in rice? Development 1996, 122, 2051–2058. [Google Scholar] [CrossRef] [PubMed]
- Koh, H.J.; Heu, M.H.; McCouch, S.R. Molecular mapping of the ges gene controlling the super-giant embryo character in rice (Oryza sativa L.). Theor. Appl. Genet. 1996, 93, 257–261. [Google Scholar] [CrossRef] [PubMed]
- Nagasawa, N.; Hibara, K.I.; Heppard, E.P.; Vander Velden, K.A.; Luck, S.; Beatty, M.; Nagato, Y.; Sakai, H. GIANT EMBRYO encodes CYP78A13, required for proper size balance between embryo and endosperm in rice. Plant J. 2013, 75, 592–605. [Google Scholar] [CrossRef]
- Paine, J.A.; Shipton, C.A.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adam, J.L.; Silverstone, A.L.; et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef] [PubMed]
- Burkhardt, P.K.; Beyer, P.; Wünn, J.; Klöti, A.; Armstrong, G.A.; Schledz, M.; von Lintig, J.; Potrykus, I. Transgenic rice (Oryza sativa) endosperm expressing daffodil (Narcissus pseudonarcissus) phytoene synthase accumulates phytoene, a key intermediate of provitamin A biosynthesis. Plant J. 1997, 11, 1071–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potrykus, I. Golden rice and beyond. Plant Physiol. 2001, 125, 1157–1161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, S.J. Analysis of Golden-Like Rice Pigment Compounds and ALDH Gene Expression. Master’s Thesis, Tunghai University, Taichung, Taiwan, 2009. (In Chinese with English Abstract). [Google Scholar]
- Gao, C.; Han, B. Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene 2009, 431, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.P.; Kuo, S.C.; Lin, B.N. Breeding of functional rice varieties. Agric. Biotechnol. Ind. Q. 2017, 49, 33–40. [Google Scholar]
- Nam, S.H.; Choi, S.P.; Kang, M.Y.; Koh, H.J.; Kozukue, N.; Friedman, M. Antioxidative activities of bran extracts from twenty one pigmented rice cultivars. Food Chem. 2006, 94, 613–620. [Google Scholar] [CrossRef]
- Qiu, Y.; Liu, Q.; Beta, T. Antioxidant activity of commercial wild rice and identification of flavonoid compounds in active fractions. J. Agric. Food Chem. 2009, 57, 7543–7551. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, M.; Kimura, T.; Yamagishi, K.; Shinmoto, H.; Yamaki, K. Comparison of mineral contents in 8 cultivars of pigmented brown rice. J. Jpn. Soc. Food. Sci. 2004, 51, 424–427. [Google Scholar] [CrossRef]
- Zhang, M.W.; Guo, B.J.; Peng, Z.M. Genetic effects on Fe, Zn, Mn and P contents in Indica black pericarp rice and their genetic correlations with grain characteristics. Euphytica 2004, 135, 315–323. [Google Scholar] [CrossRef]
- Gunaratne, A.; Wu, K.; Li, D.; Bentota, A.; Corke, H.; Cai, Y.Z. Antioxidant activity and nutritional quality of traditional red-grained rice varieties containing proanthocyanidins. Food Chem. 2013, 138, 1153–1161. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Shao, Y.; Bao, J.; Beta, T. Phenolic compounds and antioxidant properties of breeding lines between the white and black rice. Food Chem. 2015, 172, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Collard, B.; Vera Cruz, C.; McNally, K.L.; Virk, P.; Mackill, D. Rice molecular breeding laboratories in the genomics era: Current status and future considerations. Int. J. Plant Genom. 2008, 2008, 524847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, Y.R.; Chiang, S.H.; Wu, Y.P. Fine mapping of the giant embryo gene GE2 in rice. In International Symposium on Rice Research in the Era of Global Warming; Wu, M.T., Chen, J.R., Liu, D.J., Eds.; Taiwan Agricultural Research Institute: Taichung, Taiwan, 2009; pp. 23–36. [Google Scholar]
- Park, D.S.; Park, S.K.; Lee, B.C.; Song, S.Y.; Jun, N.S.; Manigbas, N.L.; Cho, J.H.; Nam, M.H.; Jeon, J.S.; Han, C.D.; et al. Molecular characterization and physico-chemical analysis of a new giant embryo mutant allele (get) in rice (Oryza sativa L.). Genes Genom. 2009, 31, 277–282. [Google Scholar] [CrossRef]
- Collard, B.C.; Mackill, D.J. Marker-assisted selection: An approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B 2008, 363, 557–572. [Google Scholar] [CrossRef] [Green Version]
- Welch, R.M. Breeding strategies for biofortified staple plant foods to reduce micronutrient malnutrition globally. J Nutr. 2002, 132, 495S–499S. [Google Scholar] [CrossRef] [PubMed]
- Connorton, J.M.; Balk, J. Iron biofortification of staple crops: Lessons and challenges in plant genetics. Plant Cell Physiol. 2019, 60, 1447–1456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lockyer, S.; White, A.; Buttriss, J.L. Biofortified crops for tackling micronutrient deficiencies–what impact are these having in developing countries and could they be of relevance within Europe? Nutr. Bull. 2018, 43, 319–357. [Google Scholar] [CrossRef]
- Dong, Y.; Tsuzuki, E.; Kamiunten, H.; Terao, H.; Lin, D. Mapping of QTL for embryo size in rice. Crop Sci. 2003, 43, 1068–1071. [Google Scholar] [CrossRef]
- Kedare, S.B.; Singh, R.P. Genesis and development of DPPH method of antioxidant assay. J. Food Sci. Technol. 2011, 48, 412–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shin, J.H.; Kim, S.R.; An, G. Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant Physiol. 2009, 149, 905–915. [Google Scholar] [CrossRef] [Green Version]
- Neff, M.M.; Turk, E.; Kalishman, M. Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 2002, 18, 613–615. [Google Scholar] [CrossRef]
- Hsu, J.H. The Platform of Polymorphic Markers and Genetic Mapping of a Drought Tolerance Gene at Seedling Stage in Rice. Master’s Thesis, National Chiayi University, Chiayi, Taiwan, 2012. (In Chinese with English Abstract). [Google Scholar]
- Shen, Y.J.; Jiang, H.; Jin, J.P.; Zhang, Z.B.; Xi, B.; He, Y.Y.; Wang, G.; Wang, C.; Qian, L.; Li, X.; et al. Development of genome-wide DNA polymorphism database for map-based cloning of rice genes. Plant Physiol. 2004, 135, 1198–1205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kazuhiro, K.; Niwa, Y.; Yamaguchi, T.; Sunohara, H.; Hirano, H.; Umeda, M. A rapid and easy-handling procedure for isolation of DNA from rice, Arabidopsis and tobacco. Plant Biotechnol. 1998, 15, 45–48. [Google Scholar]
- Bao, J.S.; Cai, Y.; Sun, M.; Wang, G.Y.; Corke, H. Anthocyanins, flavonols, and free radical scavenging activity of Chinese bayberry (Myrica rubra) extracts and their color properties and stability. J. Agric. Food Chem. 2005, 53, 2327–2332. [Google Scholar] [CrossRef] [PubMed]
- Hsieh, K.J.; Wu, Y.P.; Lin, S.J. Investigation of the functional compositions of turmeric collected from Cishan District of Kaohsiung City, Taiwan. Crop Environ. Bioinform. 2014, 11, 158–168. [Google Scholar]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Berloo, R. GGT: Software for the display of graphical genotypes. J. Hered. 1999, 90, 328–330. [Google Scholar] [CrossRef] [Green Version]





| Lines | 2017-1 | 2017-2 | 2018-1 | 2018-2 | 2019-1 | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| CNY922401 | 7377.0 | abcd | 5088.4 | e | 8532.5 | a | 1422.1 | abc | 8221.4 | ab |
| PFR4 | 6354.9 | cdef | - | - | - | - | ||||
| PFR5 | 6510.5 | bcdef | 4355.1 | f | - | - | - | |||
| PFR20 | 6043.8 | ef | - | - | - | - | ||||
| PFR24 | 5955.0 | f | - | - | - | - | ||||
| PFR32 | 7599.2 | ab | 5066.2 | e | 7599.2 | ab | 911.0 | c | 7999.2 | b |
| TNGSW26 | 7910.3 | a | 6621.6 | a | 6666.0 | ab | 1177.7 | bc | 9065.8 | a |
| RFR1 | 7732.6 | ab | - | - | - | - | ||||
| RFR2 | 7221.5 | abcde | 5466.1 | cde | - | - | - | |||
| RFR13 | 7510.4 | abc | 5466.1 | cde | 7799.2 | ab | 2733.1 | ab | 7799.2 | b |
| RFR16 | 6354.9 | cdef | 5732.8 | cd | - | - | - | |||
| RFR21 | 5755.0 | f | - | - | - | - | ||||
| RFR22 | 6577.1 | bcdef | - | - | - | - | ||||
| RFR34 | 6754.9 | abcdef | - | - | - | - | ||||
| RFR36 | 6754.9 | abcdef | 5288.4 | de | 6043.8 | b | 2955.3 | a | - | |
| RFR39 | 6132.7 | def | - | - | - | - | ||||
| RFR44 | 6621.6 | bcdef | 6199.4 | ab | 6377.1 | b | 1133.2 | c | - | |
| RFR46 | 6488.2 | bcdef | - | - | - | - | ||||
| RFR48 | 6354.9 | cdef | - | - | - | - | ||||
| RFR51 | 7621.5 | ab | 5910.5 | bc | 6821.5 | ab | 1799.8 | abc | 6710.4 | c |
| Line | DPPH (%) x | Anthocyanin Content y | ||
|---|---|---|---|---|
| CNY922401 | 88.89 | ab | 1.48 | d |
| PFR4 | 87.58 | abc | 2.25 | a |
| PFR5 | 86.32 | bc | 2.09 | abc |
| PFR20 | 86.83 | abc | 2.15 | ab |
| PFR24 | 85.52 | bc | 1.84 | bc |
| PFR32 | 87.63 | abc | 1.81 | c |
| TNGSW26 | 82.4 | bc | nd | |
| RFR1 | 85.72 | bc | nd | |
| RFR2 | 87.63 | abc | nd | |
| RFR13 | 81.25 | c | nd | |
| RFR16 | 37.31 | g | nd | |
| RFR21 | 27.9 | h | nd | |
| RFR22 | 27.45 | h | nd | e |
| RFR34 | 86.07 | bc | 0.11 | e |
| RFR36 | 93.06 | a | 0.22 | e |
| RFR39 | 88.24 | ab | 0.34 | e |
| RFR44 | 86.17 | bc | 0.05 | |
| RFR46 | 67.17 | e | nd | |
| RFR48 | 50.18 | f | nd | |
| RFR51 | 74.16 | d | nd | |
| Line | Total Phenolic Content w | Flavonoid Content x | DPPH (%) y | Anthocyanin Content z | ||||
|---|---|---|---|---|---|---|---|---|
| CNY922401 | 5.41 | a | 1.94 | a | 84.36 | a | 2.34 | a |
| PFR32 | 4.51 | a | 1.59 | b | 75.73 | ab | 1.66 | b |
| TNGSW26 | 0.73 | c | 0.89 | c | 58.98 | c | 0.03 | d |
| RFR13 | 2.44 | b | 0.85 | c | 64.00 | bc | 0.08 | d |
| RFR36 | 1.62 | bc | 0.81 | c | 45.46 | d | 0.19 | c |
| RFR44 | 0.70 | c | 0.59 | d | 36.44 | d | 0.04 | d |
| RFR51 | 2.08 | b | 0.82 | c | 70.35 | bc | 0.05 | d |
| Line | Total Phenolic Content w | Flavonoid Content x | DPPH (%) y | Anthocyanin Content z | ||||
|---|---|---|---|---|---|---|---|---|
| CNY922401 | 4.58 | a | 1.38 | a | 84.79 | b | 1.64 | a |
| PFR32 | 2.37 | bc | 1.40 | a | 86.30 | ab | 1.06 | b |
| TNGSW26 | 3.55 | ab | 0.82 | b | 90.59 | a | 0.06 | c |
| RFR13 | 1.58 | c | 0.63 | c | 88.63 | ab | 0.03 | c |
| TK9 | 1.15 | c | 0.20 | d | 34.47 | c | 0.01 | c |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, Y.-P.; Chang, Y.-C.; Kuo, H.-I.; Lin, B.-N.; Wang, S.-M.; Tseng, Y.-C. The Development of Two High-Yield and High-Quality Functional Rice Cultivars Using Marker-Assisted Selection and Conventional Breeding Methods. Int. J. Mol. Sci. 2022, 23, 4678. https://doi.org/10.3390/ijms23094678
Wu Y-P, Chang Y-C, Kuo H-I, Lin B-N, Wang S-M, Tseng Y-C. The Development of Two High-Yield and High-Quality Functional Rice Cultivars Using Marker-Assisted Selection and Conventional Breeding Methods. International Journal of Molecular Sciences. 2022; 23(9):4678. https://doi.org/10.3390/ijms23094678
Chicago/Turabian StyleWu, Yong-Pei, Yu-Chi Chang, Hsin-I Kuo, Bing-Nan Lin, Shu-Mei Wang, and Yu-Chien Tseng. 2022. "The Development of Two High-Yield and High-Quality Functional Rice Cultivars Using Marker-Assisted Selection and Conventional Breeding Methods" International Journal of Molecular Sciences 23, no. 9: 4678. https://doi.org/10.3390/ijms23094678
APA StyleWu, Y.-P., Chang, Y.-C., Kuo, H.-I., Lin, B.-N., Wang, S.-M., & Tseng, Y.-C. (2022). The Development of Two High-Yield and High-Quality Functional Rice Cultivars Using Marker-Assisted Selection and Conventional Breeding Methods. International Journal of Molecular Sciences, 23(9), 4678. https://doi.org/10.3390/ijms23094678






