Innovations in Genomics and Big Data Analytics for Personalized Medicine and Health Care: A Review
Abstract
:1. Introduction
- More than two and a half millennia ago, Hippocrates stated: “every human is distinct, and this affects both the disease prediction and the treatment”.
- In 1956, “favism”, the genetic basis for the selective toxicity of fava beans, was discovered to be due to a deficiency in the metabolic enzyme G6PD.
- In 1985, Renato Dulbecco realized that, in order to advance cancer research, it was necessary to sequence the human genome.
- In 1988, Genentech Inc. sequenced the entire human growth hormone locus (a world record), making evident the feasibility of sequencing the human genome.
- In 1990, the Human Genome Project (HGP) was launched, and the first draft was published in 2001, with its final version in 2003.
- Since the early 1990s, individualized treatments tailored to the genome of each patient have been envisioned but rarely realized.
- In 1994, a diagnostic test for the prediction of the success of rHGH replacement therapy was developed, being the earliest registry of a companion molecular diagnostics (CMDx) test ever invented.
- In 1998, when the FDA approved Herceptin (anti-EGFR mAb for EGFR+ breast tumors) and HerceptTest (to detect such tumors), it became the first “official” CMDx invented. Since then, a growing list of diagnostic packages/personalized medicine therapies has received, from the FDA, labels recognizing and recommending them.
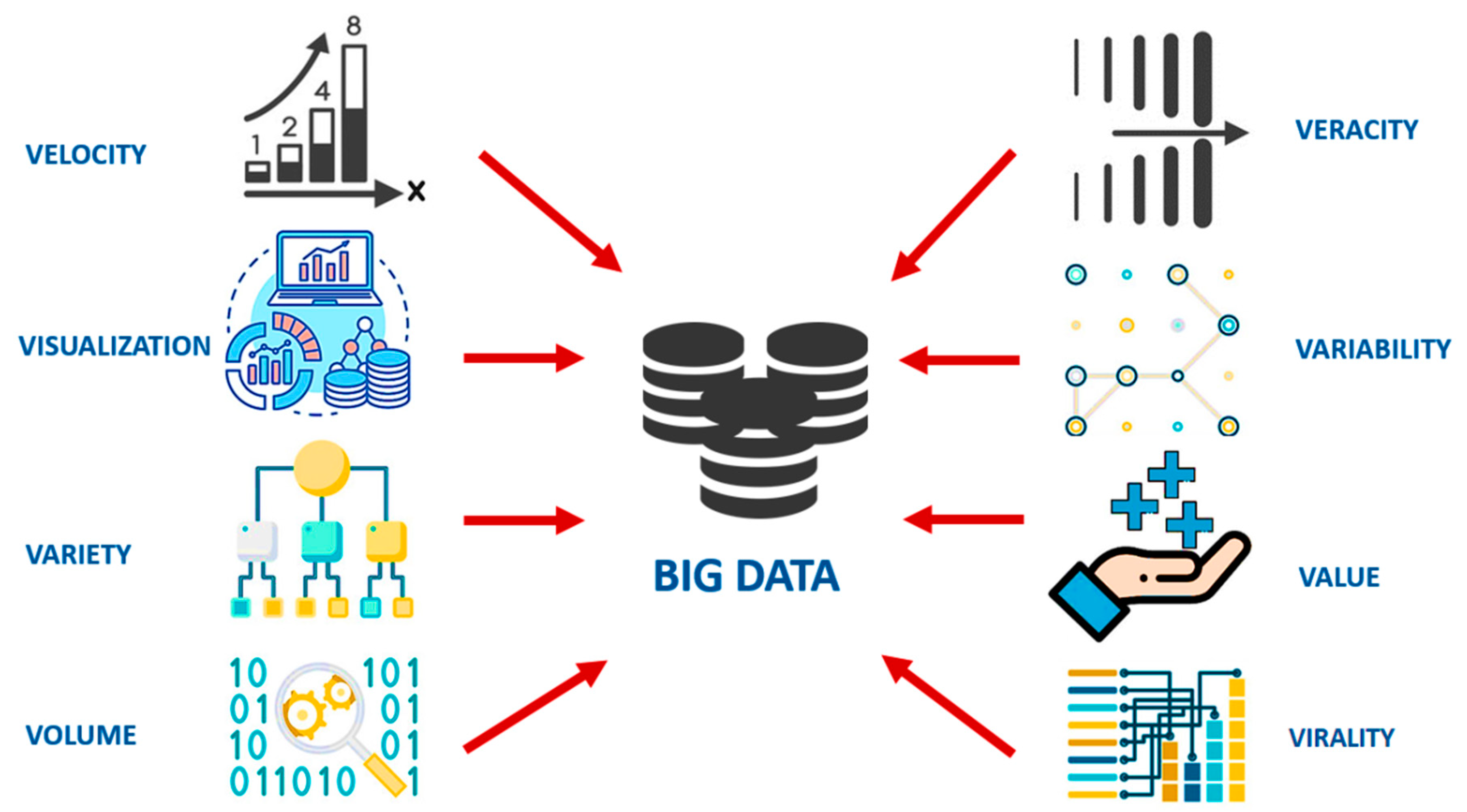
2. The Conceptualization of Big Data
3. Computational Approaches toward Personalized Medicine
3.1. Molecular Interaction Maps (MIMs)
3.2. Constraint-Based Models
3.3. Boolean Models (BMs)
3.4. Quantitative Models (QMs)
3.5. Pharmacokinetic Models
4. Machine Learning Perspectives on Personalized Medicine
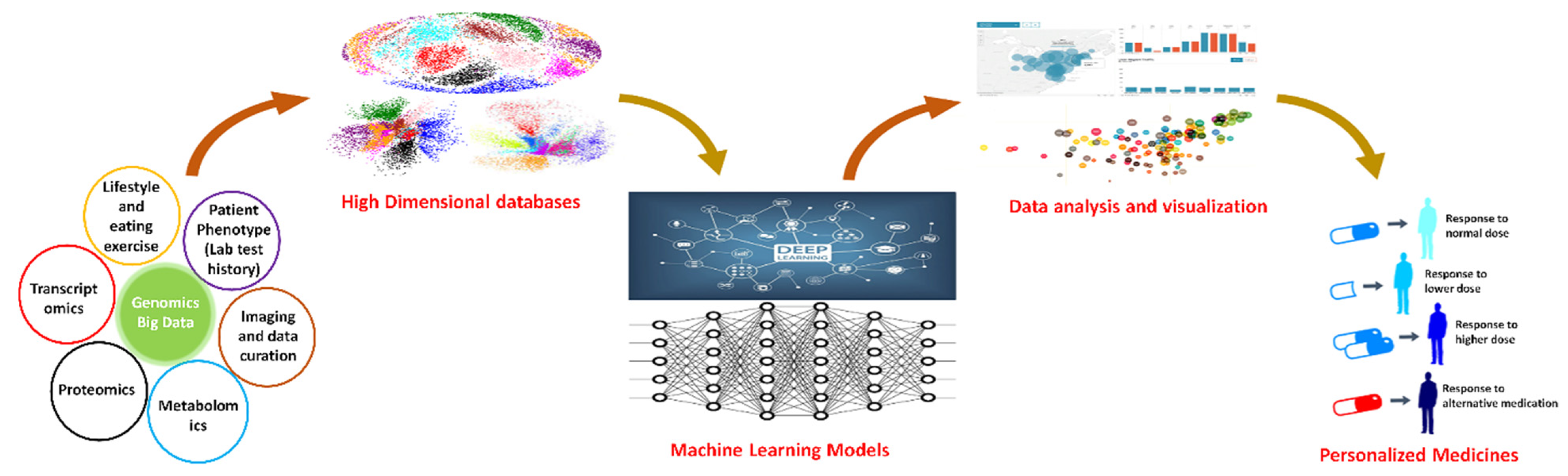
5. Modeling Genetic Data with Translational Purposes
- Replacing a mutated gene that causes disease with a healthy copy of the gene.
- Inactivating, or “knocking out,” a mutated gene that is functioning improperly.
- Introducing a new gene into the body to help fight a disease.
6. Data Mining Tools/Algorithms and Their Applications for Personalized Medicine
6.1. Pattern-Based Approaches in Data Mining for Analyzing Patient Data
6.2. Network Mining for Personalized Medicine and Health Care
6.3. Big Data Management Problems in Precision Medicine and Health Care
6.4. Significance of Next Generation Informatics for Big Data in Precision Medicine Era
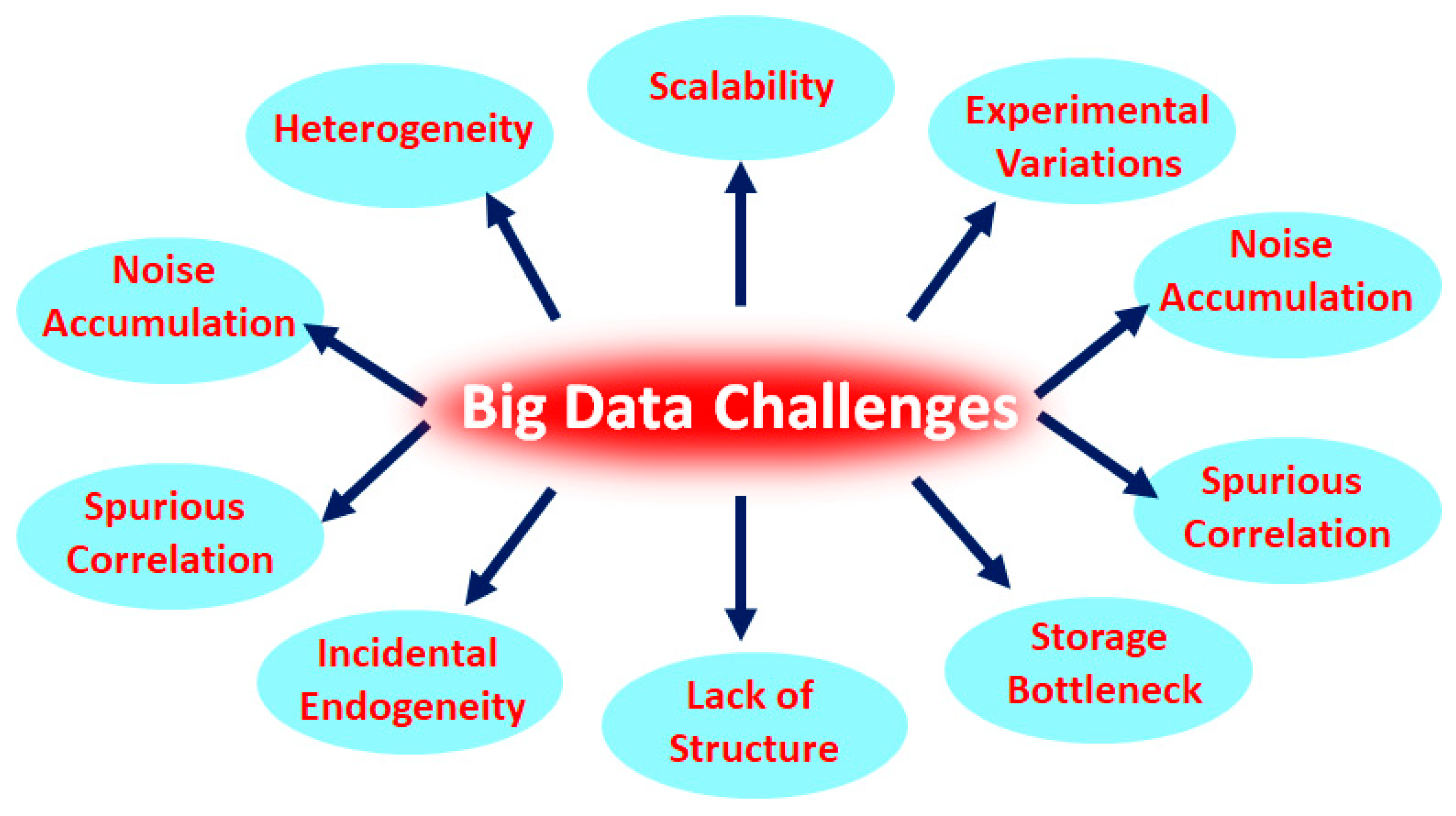
7. Heterogeneity, a Huge Challenge in Big Data Analysis
8. Role of Big Data in Accelerating Digital Healthcare
9. Big Data Applications in Health Care
10. Electronic Health Records
11. Health Big Data as a Key Player for Informed Strategic Planning
12. Advanced Risk and Disease Management through Big Data
13. Developing New Therapies and Big Data
14. Impediments of Big Data in Health Care
15. Conclusions and Future Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Iriart, J.A.B. Precision medicine/personalized medicine: A critical analysis of movements in the transformation of biomedicine in the early 21st century. Cadernos. Cad. De Saúde Publica 2019, 35. [Google Scholar] [CrossRef] [Green Version]
- Cirillo, D.; Valencia, A. Big data analytics for personalized medicine. Curr. Opin. Biotechnol. 2019, 58, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Ginsburg, G.S.; Willard, H.F. Genomic and personalized medicine: Foundations and applications. Transl. Res. 2009, 154, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Schaefer, G.O.; Tai, E.S.; Sun, S. Precision medicine and big data. Asian Bioeth. Rev. 2019, 11, 275–288. [Google Scholar] [CrossRef] [Green Version]
- Naqvi, M.R.; Jaffar, M.A.; Aslam, M.; Shahzad, S.K.; Iqbal, M.W.; Farooq, A. Importance of big data in precision and personalized medicine. In Proceedings of the 2020 International Congress on Human-Computer Interaction, Optimization and Robotic Applications (HORA), Ankara, Turkey, 30 July 2020; pp. 1–6. [Google Scholar]
- Beckmann, J.S.; Lew, D. Reconciling evidence-based medicine and precision medicine in the era of big data: Challenges and opportunities. Genome Med. 2016, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Espinal-Enríquez, J.; Mejía-Pedroza, R.; Hernández-Lemus, E. Computational approaches in precision medicine. In Progress and Challenges in Precision Medicine; Elsevier: Amsterdam, The Netherlands, 2017; pp. 233–250. [Google Scholar]
- Ashley, E.A. Towards precision medicine. Nat. Rev. Genet. 2016, 17, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Hulsen, T.; Jamuar, S.; Moody, A.; Karnes, J.; Varga, O.; Hedensted, S.; Spreafico, R.; Hafler, D.; McKinney, E. From big data to precision medicine. Front. Med. 2019, 6, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho, D.M.; Collins, K.M.; Powers, R.K.; Costello, J.C.; Collins, J.J. Next-generation machine learning for biological networks. Cell 2018, 173, 1581–1592. [Google Scholar] [CrossRef] [Green Version]
- Bibault, J.-E. Real-life clinical data mining: Generating hypotheses for evidence-based medicine. Ann. Transl. Med. 2020, 8, 69. [Google Scholar] [CrossRef]
- Normandeau, K. Beyond Volume, Variety and Velocity is the Issue of Big Data Veracity. Inside Big Data 2013. Available online: https://insidebigdata.com/2013/09/12/beyond-volume-variety-velocity-issue-big-data-veracity/ (accessed on 18 January 2022).
- Gandomi, A.; Haider, M. Beyond the hype: Big data concepts, methods, and analytics. Int. J. Inf. Manag. 2015, 35, 137–144. [Google Scholar] [CrossRef] [Green Version]
- Diebold, F.X.; Cheng, X.; Diebold, S.; Foster, D.; Halperin, M.; Lohr, S.; Mashey, J.; Nickolas, T.; Pai, M.; Pospiech, M. A Personal Perspective on the Origin (s) and Development of “Big Data”: The Phenomenon, the Term, and the Discipline*. CiteSeer 2012. [Google Scholar] [CrossRef] [Green Version]
- Auffray, C.; Balling, R.; Barroso, I.; Bencze, L.; Benson, M.; Bergeron, J.; Bernal-Delgado, E.; Blomberg, N.; Bock, C.; Conesa, A. Making sense of big data in health research: Towards an EU action plan. Genome Med. 2016, 8, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dash, S.; Shakyawar, S.K.; Sharma, M.; Kaushik, S. Big data in healthcare: Management, analysis and future prospects. J. Big Data 2019, 6, 54. [Google Scholar] [CrossRef] [Green Version]
- Fernandez Martinez, J.L.; Fernandez Muniz, M.Z.; Tompkins, M.J. On the topography of the cost functional in linear and nonlinear inverse problems. Geophysics 2012, 77, W1–W15. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.L.; Fernández-Muñiz, Z.; Pallero, J.; Pedruelo-González, L.M. From Bayes to Tarantola: New insights to understand uncertainty in inverse problems. J. Appl. Geophys. 2013, 98, 62–72. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.L.; Pallero, J.; Fernández-Muñiz, Z.; Pedruelo-González, L.M. The effect of noise and Tikhonov’s regularization in inverse problems. Part I: The linear case. J. Appl. Geophys. 2014, 108, 176–185. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.L.; Pallero, J.; Fernández-Muñiz, Z.; Pedruelo-González, L.M. The effect of noise and Tikhonov’s regularization in inverse problems. Part II: The nonlinear case. J. Appl. Geophys. 2014, 108, 186–193. [Google Scholar] [CrossRef]
- Zhang, H. Overview of sequence data formats. In Statistical Genomics; Springer: Berlin/Heidelberg, Germany, 2016; pp. 3–17. [Google Scholar]
- Lek, M.; Karczewski, K.; Minikel, E.; Samocha, K.; Banks, E.; Fennell, T.; O’Donnell-Luria, A.; Ware, J.; Hill, A.; Cummings, B.; et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 2016, 536, 285–291. [Google Scholar] [CrossRef] [Green Version]
- The Global Alliance for Genomics and Health. A federated ecosystem for sharing genomic, clinical data. Science 2016, 352, 1278–1280. [Google Scholar] [CrossRef] [Green Version]
- Wenger, A.M.; Guturu, H.; Bernstein, J.A.; Bejerano, G. Systematic reanalysis of clinical exome data yields additional diagnoses: Implications for providers. Genet. Med. 2017, 19, 209–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, C.F.; McRae, J.F.; Clayton, S.; Gallone, G.; Aitken, S.; FitzGerald, T.W.; Jones, P.; Prigmore, E.; Rajan, D.; Lord, J. Making new genetic diagnoses with old data: Iterative reanalysis and reporting from genome-wide data in 1133 families with developmental disorders. Genet. Med. 2018, 20, 1216–1223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, I.S.; Ginsburg, G.S. Personalized medicine: Progress and promise. Annu. Rev. Genom. Hum. Genet. 2011, 12, 217–244. [Google Scholar] [CrossRef] [PubMed]
- Masoudi-Nejad, A.; Wang, E. Cancer Modeling and Network Biology: Accelerating toward Personalized Medicine. Semin. Cancer Biol. 2015, 30, 1–3. [Google Scholar] [CrossRef]
- Meyer, U.A. Pharmacogenetics–five decades of therapeutic lessons from genetic diversity. Nat. Rev. Genet. 2004, 5, 669–676. [Google Scholar] [CrossRef]
- Janga, S.C.; Edupuganti, M.M.R. Systems and network-based approaches for personalized medicine. Curr. Synth. Syst. Biol. 2014, 2. [Google Scholar] [CrossRef] [Green Version]
- Tuena, C.; Semonella, M.; Fernández-Álvarez, J.; Colombo, D.; Cipresso, P. Predictive precision medicine: Towards the computational challenge. In P5 eHealth: An Agenda for the Health Technologies of the Future; Springer: Cham, Switzerland, 2020; pp. 71–86. [Google Scholar]
- Collin, C.B.; Gebhardt, T.; Golebiewski, M.; Karaderi, T.; Hillemanns, M.; Khan, F.M.; Salehzadeh-Yazdi, A.; Kirschner, M.; Krobitsch, S.; Consortium, E.-S.P. Computational Models for Clinical Applications in Personalized Medicine—Guidelines and Recommendations for Data Integration and Model Validation. J. Pers. Med. 2022, 12, 166. [Google Scholar] [CrossRef]
- Apweiler, R.; Beissbarth, T.; Berthold, M.R.; Blüthgen, N.; Burmeister, Y.; Dammann, O.; Deutsch, A.; Feuerhake, F.; Franke, A.; Hasenauer, J. Whither systems medicine? Exp. Mol. Med. 2018, 50, e453. [Google Scholar] [CrossRef] [Green Version]
- Pison, C.; Consortium, C. THE CASyM ROADMAP Implementation of Systems Medicine across Europe; Project Management Jülich, Forschungszentrum Jülich GmbH, Germany. 2014. Available online: https://hal.univ-grenoble-alpes.fr/hal-01969603 (accessed on 28 February 2022).
- Morrison, T.M.; Pathmanathan, P.; Adwan, M.; Margerrison, E. Advancing regulatory science with computational modeling for medical devices at the FDA’s office of science and engineering laboratories. Front. Med. 2018, 5, 241. [Google Scholar] [CrossRef] [Green Version]
- Musuamba, F.T.; Skottheim Rusten, I.; Lesage, R.; Russo, G.; Bursi, R.; Emili, L.; Wangorsch, G.; Manolis, E.; Karlsson, K.E.; Kulesza, A. Scientific and regulatory evaluation of mechanistic in silico drug and disease models in drug development: Building model credibility. CPT Pharmacomet. Syst. Pharmacol. 2021, 10, 804–825. [Google Scholar] [CrossRef]
- Arrieta, A.B.; Díaz-Rodríguez, N.; Del Ser, J.; Bennetot, A.; Tabik, S.; Barbado, A.; García, S.; Gil-López, S.; Molina, D.; Benjamins, R. Explainable Artificial Intelligence (XAI): Concepts, taxonomies, opportunities and challenges toward responsible AI. Inf. Fusion 2020, 58, 82–115. [Google Scholar] [CrossRef] [Green Version]
- Dimiduk, D.M.; Holm, E.A.; Niezgoda, S.R. Perspectives on the impact of machine learning, deep learning, and artificial intelligence on materials, processes, and structures engineering. Integr. Mater. Manuf. Innov. 2018, 7, 157–172. [Google Scholar] [CrossRef] [Green Version]
- Kitano, H.; Funahashi, A.; Matsuoka, Y.; Oda, K. Using process diagrams for the graphical representation of biological networks. Nat. Biotechnol. 2005, 23, 961–966. [Google Scholar] [CrossRef]
- Fujita, K.A.; Ostaszewski, M.; Matsuoka, Y.; Ghosh, S.; Glaab, E.; Trefois, C.; Crespo, I.; Perumal, T.M.; Jurkowski, W.; Antony, P. Integrating pathways of Parkinson’s disease in a molecular interaction map. Mol. Neurobiol. 2014, 49, 88–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuperstein, I.; Bonnet, E.; Nguyen, H.-A.; Cohen, D.; Viara, E.; Grieco, L.; Fourquet, S.; Calzone, L.; Russo, C.; Kondratova, M. Atlas of Cancer Signalling Network: A systems biology resource for integrative analysis of cancer data with Google Maps. Oncogenesis 2015, 4, e160. [Google Scholar] [CrossRef] [Green Version]
- Thiele, I.; Palsson, B.Ø. A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat. Protoc. 2010, 5, 93–121. [Google Scholar] [CrossRef] [Green Version]
- Uhlen, M.; Zhang, C.; Lee, S.; Sjöstedt, E.; Fagerberg, L.; Bidkhori, G.; Benfeitas, R.; Arif, M.; Liu, Z.; Edfors, F. A pathology atlas of the human cancer transcriptome. Science 2017, 357, e2507. [Google Scholar] [CrossRef] [Green Version]
- Mardinoglu, A.; Agren, R.; Kampf, C.; Asplund, A.; Nookaew, I.; Jacobson, P.; Walley, A.J.; Froguel, P.; Carlsson, L.M.; Uhlen, M. Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol. Syst. Biol. 2013, 9, 649. [Google Scholar] [CrossRef]
- Stempler, S.; Yizhak, K.; Ruppin, E. Integrating transcriptomics with metabolic modeling predicts biomarkers and drug targets for Alzheimer’s disease. PLoS ONE 2014, 9, e105383. [Google Scholar] [CrossRef] [Green Version]
- Wang, R.-S.; Saadatpour, A.; Albert, R. Boolean modeling in systems biology: An overview of methodology and applications. Phys. Biol. 2012, 9, 055001. [Google Scholar] [CrossRef] [Green Version]
- Eduati, F.; Jaaks, P.; Wappler, J.; Cramer, T.; Merten, C.A.; Garnett, M.J.; Saez-Rodriguez, J. Patient-specific logic models of signaling pathways from screenings on cancer biopsies to prioritize personalized combination therapies. Mol. Syst. Biol. 2020, 16, e8664. [Google Scholar] [CrossRef] [PubMed]
- Udyavar, A.R.; Wooten, D.J.; Hoeksema, M.; Bansal, M.; Califano, A.; Estrada, L.; Schnell, S.; Irish, J.M.; Massion, P.P.; Quaranta, V. Novel hybrid phenotype revealed in small cell lung cancer by a transcription factor network model that can explain tumor heterogeneity. Cancer Res. 2017, 77, 1063–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malik-Sheriff, R.S.; Glont, M.; Nguyen, T.V.; Tiwari, K.; Roberts, M.G.; Xavier, A.; Vu, M.T.; Men, J.; Maire, M.; Kananathan, S. BioModels—15 years of sharing computational models in life science. Nucleic Acids Res. 2020, 48, D407–D415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolch, W.; Fey, D. Personalized computational models as biomarkers. J. Pers. Med. 2017, 7, 9. [Google Scholar] [CrossRef] [Green Version]
- Hastings, J.F.; O’Donnell, Y.E.; Fey, D.; Croucher, D.R. Applications of personalised signalling network models in precision oncology. Pharmacol. Ther. 2020, 212, 107555. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Urizar, J.; Granados-Soto, V.; Flores-Murrieta, F.J.; Castañeda-Hernández, G. Pharmacokinetic-pharmacodynamic modeling: Why? Arch. Med. Res. 2000, 31, 539–545. [Google Scholar] [CrossRef]
- Edginton, A.N.; Willmann, S. Physiology-based simulations of a pathological condition. Clin. Pharmacokinet. 2008, 47, 743–752. [Google Scholar] [CrossRef]
- Mamoshina, P.; Vieira, A.; Putin, E.; Zhavoronkov, A. Applications of deep learning in biomedicine. Mol. Pharm. 2016, 13, 1445–1454. [Google Scholar] [CrossRef]
- Jordan, M.I.; Mitchell, T.M. Machine learning: Trends, perspectives, and prospects. Science 2015, 349, 255–260. [Google Scholar] [CrossRef]
- Fröhlich, H.; Balling, R.; Beerenwinkel, N.; Kohlbacher, O.; Kumar, S.; Lengauer, T.; Maathuis, M.H.; Moreau, Y.; Murphy, S.A.; Przytycka, T.M. From hype to reality: Data science enabling personalized medicine. BMC Med. 2018, 16, 1–15. [Google Scholar] [CrossRef]
- Cardoso, F.; van’t Veer, L.J.; Bogaerts, J.; Slaets, L.; Viale, G.; Delaloge, S.; Pierga, J.-Y.; Brain, E.; Causeret, S.; DeLorenzi, M. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 2016, 375, 717–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marchio, C.; Balmativola, D.; Castiglione, R.; Annaratone, L.; Sapino, A. Predictive diagnostic pathology in the target therapy era in breast cancer. Curr. Drug Targets 2017, 18, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Van’t Veer, L.J.; Dai, H.; Van De Vijver, M.J.; He, Y.D.; Hart, A.A.; Mao, M.; Peterse, H.L.; Van Der Kooy, K.; Marton, M.J.; Witteveen, A.T. Gene expression profiling predicts clinical outcome of breast cancer. Nature 2002, 415, 530–536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viale, G.; De Snoo, F.; Slaets, L.; Bogaerts, J.; Van’t Veer, L.; Rutgers, E.; Piccart-Gebhart, M.; Stork-Sloots, L.; Glas, A.; Russo, L. Immunohistochemical versus molecular (BluePrint and MammaPrint) subtyping of breast carcinoma. Outcome results from the EORTC 10041/BIG 3-04 MINDACT trial. Breast Cancer Res. Treat. 2018, 167, 123–131. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.; Hermsen, M.; Manson, Q.F.; Balkenhol, M. Diagnostic assessment of deep learning algorithms for detection of lymph node metastases in women with breast cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Madani, A.; Ong, J.R.; Tibrewal, A.; Mofrad, M.R. Deep echocardiography: Data-efficient supervised and semi-supervised deep learning towards automated diagnosis of cardiac disease. NPJ Digit. Med. 2018, 1, 1–11. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [Green Version]
- Rauschert, S.; Raubenheimer, K.; Melton, P.; Huang, R. Machine learning and clinical epigenetics: A review of challenges for diagnosis and classification. Clin. Epigenet. 2020, 12, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Bosco, G.L.; Rizzo, R.; Fiannaca, A.; La Rosa, M.; Urso, A. A deep learning model for epigenomic studies. In Proceedings of the 2016 12th International Conference on Signal-Image Technology & Internet-Based Systems (SITIS), Naples, Italy, 28 November–1 December 2016; Institute of Electrical and Electronics Engineers (IEEE): New York, NY, USA; pp. 688–692. [Google Scholar]
- Hamey, F.K.; Göttgens, B. Machine learning predicts putative hematopoietic stem cells within large single-cell transcriptomics data sets. Exp. Hematol. 2019, 78, 11–20. [Google Scholar] [CrossRef] [Green Version]
- Erban, A.; Fehrle, I.; Martinez-Seidel, F.; Brigante, F.; Más, A.L.; Baroni, V.; Wunderlin, D.; Kopka, J. Discovery of food identity markers by metabolomics and machine learning technology. Sci. Rep. 2019, 9, 9697. [Google Scholar] [CrossRef] [Green Version]
- Narita, A.; Ueki, M.; Tamiya, G. Artificial intelligence powered statistical genetics in biobanks. J. Hum. Genet. 2021, 66, 61–65. [Google Scholar] [CrossRef] [PubMed]
- Luz, C.F.; Vollmer, M.; Decruyenaere, J.; Nijsten, M.W.; Glasner, C.; Sinha, B. Machine learning in infection management using routine electronic health records: Tools, techniques, and reporting of future technologies. Clin. Microbiol. Infect. 2020, 26, 1291–1299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavrač, N. Machine learning for data mining in medicine. In Proceedings of the Joint European Conference on Artificial Intelligence in Medicine and Medical Decision Making, Aalborg, Denmark, 20–24 June 1999; Springer: Aalborg, Denmark; pp. 47–62. [Google Scholar]
- Cernea, A.; Fernández-Martínez, J.L.; deAndrés-Galiana, E.J.; Fernández-Ovies, F.J.; Alvarez-Machancoses, O.; Fernández-Muñiz, Z.; Saligan, L.N.; Sonis, S.T. Robust pathway sampling in phenotype prediction. Application to triple negative breast cancer. BMC Bioinform. 2020, 21, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bonder, M.J.; Kasela, S.; Kals, M.; Tamm, R.; Lokk, K.; Barragan, I.; Buurman, W.A.; Deelen, P.; Greve, J.-W.; Ivanov, M. Genetic and epigenetic regulation of gene expression in fetal and adult human livers. BMC Genom. 2014, 15, 860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niu, N.; Liu, T.; Cairns, J.; Ly, R.C.; Tan, X.; Deng, M.; Fridley, B.L.; Kalari, K.R.; Abo, R.P.; Jenkins, G. Metformin pharmacogenomics: A genome-wide association study to identify genetic and epigenetic biomarkers involved in metformin anticancer response using human lymphoblastoid cell lines. Hum. Mol. Genet. 2016, 25, 4819–4834. [Google Scholar] [CrossRef]
- Liou, S.-Y.; Stringer, F.; Hirayama, M. The impact of pharmacogenomics research on drug development. Drug Metab. Pharmacokinet. 2012, 27, 2–8. [Google Scholar] [CrossRef] [Green Version]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [Green Version]
- Hassan, M.; Raza, H.; Abbasi, M.A.; Moustafa, A.A.; Seo, S.-Y. The exploration of novel Alzheimer’s therapeutic agents from the pool of FDA approved medicines using drug repositioning, enzyme inhibition and kinetic mechanism approaches. Biomed. Pharmacother. 2019, 109, 2513–2526. [Google Scholar] [CrossRef]
- Cutter, G.R.; Liu, Y. Personalized medicine: The return of the house call? Neurol. Clin. Pract. 2012, 2, 343–351. [Google Scholar] [CrossRef] [Green Version]
- Hartenfeller, M.; Schneider, G. De novo drug design. Chem. Inform. Comput. Chem. Biol. 2010, 299–323. [Google Scholar]
- Álvarez-Machancoses, Ó.; Fernández-Martínez, J.L. Using artificial intelligence methods to speed up drug discovery. Expert Opin. Drug Discov. 2019, 14, 769–777. [Google Scholar] [CrossRef]
- Schneider, G. Automating drug discovery. Nat. Rev. Drug Discov. 2018, 17, 97–113. [Google Scholar] [CrossRef] [PubMed]
- Gediya, L.K.; Njar, V.C. Promise and challenges in drug discovery and development of hybrid anticancer drugs. Expert Opin. Drug Discov. 2009, 4, 1099–1111. [Google Scholar] [CrossRef] [PubMed]
- Gelb, M.H. Drug discovery for malaria: A very challenging and timely endeavor. Curr. Opin. Chem. Biol. 2007, 11, 440–445. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guha, R. On exploring structure–activity relationships. Silico Models Drug Discov. 2013, 81–94. [Google Scholar]
- Greene, N.; Fisk, L.; Naven, R.T.; Note, R.R.; Patel, M.L.; Pelletier, D.J. Developing structure− activity relationships for the prediction of hepatotoxicity. Chem. Res. Toxicol. 2010, 23, 1215–1222. [Google Scholar] [CrossRef]
- Patil, P.; Chaudhari, P.; Sahu, M.; Duragkar, N. Review article on gene therapy. Int. J. Genet. 2012, 4, 74. [Google Scholar]
- Cahan, E.M.; Hernandez-Boussard, T.; Thadaney-Israni, S.; Rubin, D.L. Putting the data before the algorithm in big data addressing personalized healthcare. NPJ Digit. Med. 2019, 2, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Lee, B.S.; McIntyre, R.S.; Gentle, J.E.; Park, N.S.; Chiriboga, D.A.; Lee, Y.; Singh, S.; McPherson, M.A. A computational algorithm for personalized medicine in schizophrenia. Schizophr. Res. 2018, 192, 131–136. [Google Scholar] [CrossRef]
- Ulyantsev, V.I.; Kazakov, S.V.; Dubinkina, V.B.; Tyakht, A.V.; Alexeev, D.G. MetaFast: Fast reference-free graph-based comparison of shotgun metagenomic data. Bioinformatics 2016, 32, 2760–2767. [Google Scholar] [CrossRef] [Green Version]
- Bellazzi, R.; Ferrazzi, F.; Sacchi, L. Predictive data mining in clinical medicine: A focus on selected methods and applications. WIREs Data Min. Knowl. Discov. 2011, 1, 416–430. [Google Scholar] [CrossRef]
- Ritchie, M.D.; Holzinger, E.R.; Li, R.; Pendergrass, S.A.; Kim, D. Methods of integrating data to uncover genotype–phenotype interactions. Nat. Rev. Genet. 2015, 16, 85–97. [Google Scholar] [CrossRef] [PubMed]
- Angermueller, C.; Clark, S.J.; Lee, H.J.; Macaulay, I.C.; Teng, M.J.; Hu, T.X.; Krueger, F.; Smallwood, S.A.; Ponting, C.P.; Voet, T. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 2016, 13, 229–232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLean, M.; Miles, C. Swift action needed to close the skills gap in bioinformatics. Nature 1999, 401, 10. [Google Scholar] [CrossRef] [Green Version]
- Hood, L. Systems biology and p4 medicine: Past, present, and future. Rambam Maimonides Med. J. 2013, 4. [Google Scholar] [CrossRef]
- Fernández-Martínez, J.L.; Fernández-Muñiz, Z.; Cernea, A.; Pallero, J.; DeAndrés-Galiana, E.J.; Pedruelo-González, L.M.; Álvarez, O.; Fernández-Ovies, F.J. How to deal with uncertainty in inverse and classification problems. In Advances in Modeling and Interpretation in Near Surface Geophysics; Springer: Berlin/Heidelberg, Germany, 2020; pp. 401–414. [Google Scholar]
- deAndrés-Galiana, E.J.; Fernández-Martínez, J.L.; Sonis, S.T. Design of biomedical robots for phenotype prediction problems. J. Comput. Biol. 2016, 23, 678–692. [Google Scholar] [CrossRef] [Green Version]
- Álvarez-Machancoses, Ó.; De Andrés-Galiana, E.J.; Fernández-Martínez, J.L.; Kloczkowski, A. Robust prediction of single and multiple point protein mutations stability changes. Biomolecules 2019, 10, 67. [Google Scholar] [CrossRef] [Green Version]
- deAndrés-Galiana, E.J.; Bea, G.; Fernández-Martínez, J.L.; Saligan, L.N. Analysis of defective pathways and drug repositioning in Multiple Sclerosis via machine learning approaches. Comput. Biol. Med. 2019, 115, 103492. [Google Scholar] [CrossRef]
- deAndrés-Galiana, E.J.; Fernández-Martínez, J.L.; Luaces, O.; del Coz, J.J.; Fernández, R.; Solano, J.; Nogués, E.A.; Zanabilli, Y.; Alonso, J.M.; Payer, A.R. On the prediction of Hodgkin lymphoma treatment response. Clin. Transl. Oncol. 2015, 17, 612–619. [Google Scholar] [CrossRef]
- Reinbolt, R.E.; Sonis, S.; Timmers, C.D.; Fernández-Martínez, J.L.; Cernea, A.; de Andrés-Galiana, E.J.; Hashemi, S.; Miller, K.; Pilarski, R.; Lustberg, M.B. Genomic risk prediction of aromatase inhibitor-related arthralgia in patients with breast cancer using a novel machine-learning algorithm. Cancer Med. 2018, 7, 240–253. [Google Scholar] [CrossRef] [Green Version]
- Cernea, A.; Fernández-Martínez, J.L.; de Andrés-Galiana, E.J.; Fernández-Muñiz, Z.; Bermejo-Millo, J.C.; González-Blanco, L.; Solano, J.J.; Abizanda, P.; Coto-Montes, A.; Caballero, B. Prognostic networks for unraveling the biological mechanisms of Sarcopenia. Mech. Ageing Dev. 2019, 182, 111129. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Martínez, J.L.; de Andrés-Galiana, E.J.; Fernández-Ovies, F.J.; Cernea, A.; Kloczkowski, A. Robust sampling of defective pathways in multiple myeloma. Int. J. Mol. Sci. 2019, 20, 4681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- deAndrés-Galiana, E.J.; Fernández-Ovies, F.J.; Cernea, A.; Fernández-Martínez, J.L.; Kloczkowski, A. Deep neural networks for phenotype prediction in rare diseases: Inclusion body myositis: A case study. In Artificial Intelligence in Precision Health; Elsevier: Amsterdam, The Netherlands, 2020; pp. 189–202. [Google Scholar]
- Álvarez-Machancoses, Ó.; deAndrés-Galiana, E.; Fernández-Martínez, J.L.; Kloczkowski, A. In The Utilization of Different Classifiers to Perform Drug Repositioning in Inclusion Body Myositis Supports the Concept of Biological Invariance. In Proceedings of the International Conference on Artificial Intelligence and Soft Computing, ICAISC 2020, Zakopane, Poland, 12–14 October 2020; Springer: Berlin/Heidelberg, Germany; pp. 589–598. [Google Scholar]
- Fan, J.; Han, F.; Liu, H. Challenges of big data analysis. Natl. Sci. Rev. 2014, 1, 293–314. [Google Scholar] [CrossRef] [Green Version]
- Wang, L. Heterogeneous data and big data analytics. Autom. Control. Inf. Sci. 2017, 3, 8–15. [Google Scholar] [CrossRef] [Green Version]
- Labrinidis, A.; Jagadish, H.V. Challenges and opportunities with big data. Proc. VLDB Endow. 2012, 5, 2032–2033. [Google Scholar] [CrossRef]
- Rahman, J.A. Knowledge Based Trade, Technical Change and Location Environment: The Case of Small and Medium Sized Enterprises Engaged in Advanced Producer Software Services in the South East Region. Ph.D. Thesis, University College London, London, UK, 2005. [Google Scholar]
- Alexander, C.A.; Wang, L. Big data analytics in heart attack prediction. J. Nurs. Care 2017, 6, 2167–2168. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, R.; Prabakaran, S. Big data in digital healthcare: Lessons learnt and recommendations for general practice. Heredity 2020, 124, 525–534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raghupathi, W.; Raghupathi, V. Big data analytics in healthcare: Promise and potential. Health Inf. Sci. Syst. 2014, 2, 3. [Google Scholar] [CrossRef]
- Cifuentes, C.; Romero, E.; Godoy, J. Design and implementation of a telepediatric primary-level and low-cost system to reduce unnecessary patient transfers. Telemed. e-Health 2017, 23, 521–526. [Google Scholar] [CrossRef]
- Danziger, J.; Ángel Armengol de la Hoz, M.; Li, W.; Komorowski, M.; Deliberato, R.O.; Rush, B.N.; Mukamal, K.J.; Celi, L.; Badawi, O. Temporal trends in critical care outcomes in US minority-serving hospitals. Am. J. Respir. Crit. Care Med. 2020, 201, 681–687. [Google Scholar] [CrossRef]
- Folchetti, L.G.D.; da Silva, I.T.; de Almeida-Pititto, B.; Ferreira, S.R.G. Nutritionists’ Health Study cohort: A web-based approach of life events, habits and health outcomes. BMJ Open 2016, 6, e012081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pastorino, R.; De Vito, C.; Migliara, G.; Glocker, K.; Binenbaum, I.; Ricciardi, W.; Boccia, S. Benefits and challenges of Big Data in healthcare: An overview of the European initiatives. Eur. J. Public Health 2019, 29 (Suppl. S3), 23–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zikopoulos, P.; Eaton, C. Understanding Big Data: Analytics for Enterprise Class Hadoop and Streaming Data; McGraw-Hill Osborne Media: New York, NY, USA, 2011. [Google Scholar]
- Murdoch, T.B.; Detsky, A.S. The inevitable application of big data to health care. JAMA 2013, 309, 1351–1352. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-Y.; Cheng, C.-W.; Kaddi, C.D.; Venugopalan, J.; Hoffman, R.; Wang, M.D. Omic and electronic health record big data analytics for precision medicine. IEEE Trans. Biomed. Eng. 2016, 64, 263–273. [Google Scholar]
- Khennou, F.; Khamlichi, Y.I.; Chaoui, N.E.H. Improving the use of big data analytics within electronic health records: A case study based OpenEHR. Procedia Comput. Sci. 2018, 127, 60–68. [Google Scholar] [CrossRef]
- Mazzei, M.J.; Noble, D. Big Data and Strategy: Theoretical Foundations and New Opportunities. In Strategy and Behaviors in the Digital Economy; IntechOpen: London, UK, 2019. [Google Scholar]
- Bates, D.W.; Saria, S.; Ohno-Machado, L.; Shah, A.; Escobar, G. Big data in health care: Using analytics to identify and manage high-risk and high-cost patients. Health Aff. 2014, 33, 1123–1131. [Google Scholar] [CrossRef] [Green Version]
- Dimitrov, D.V. Medical internet of things and big data in healthcare. Healthc. Inform. Res. 2016, 22, 156–163. [Google Scholar] [CrossRef]
- Razzak, M.I.; Imran, M.; Xu, G. Big data analytics for preventive medicine. Neural Comput. Appl. 2020, 32, 4417–4451. [Google Scholar] [CrossRef]
- Leff, D.; Yang, G. Big data for precision medicine. Engineering 2015, 1, 277–279. [Google Scholar] [CrossRef] [Green Version]
- Wooden, B.; Goossens, N.; Hoshida, Y.; Friedman, S.L. Using big data to discover diagnostics and therapeutics for gastrointestinal and liver diseases. Gastroenterology 2017, 152, 53–67.e3. [Google Scholar] [CrossRef] [Green Version]
- Podlesny, N.J.; Kayem, A.V.; Meinel, C. Towards identifying de-anonymisation risks in distributed health data silos. In Proceedings of the International Conference on Database and Expert Systems Applications, Linz, Austria, 26–29 August 2019; Springer: Berlin/Heidelberg, Germany; pp. 33–43. [Google Scholar]
- Belle, A.; Thiagarajan, R.; Soroushmehr, S.; Navidi, F.; Beard, D.A.; Najarian, K. Big data analytics in healthcare. BioMed Res. Int. 2015, 2015, 370194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alemayehu, D.; Berger, M.L. Big Data: Transforming drug development and health policy decision making. Health Serv. Outcomes Res. Methodol. 2016, 16, 92–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wielki, J. Implementation of the big data concept in organizations-possibilities, impediments and challenges. In Proceedings of the 2013 Federated Conference on Computer Science and Information Systems, Kraków, Poland, 8–11 September 2013; pp. 985–989. [Google Scholar]
- Furda, R.; Gregus, M. Impediments in healthcare digital transformation. Int. J. Appl. Res. Public Health Manag. (IJARPHM) 2019, 4, 21–34. [Google Scholar] [CrossRef]
- Mathew, P.S.; Pillai, A.S. Big Data solutions in Healthcare: Problems and perspectives. In Proceedings of the International Conference on Innovations in Information, Embedded and Communication Systems (ICIIECS), Coimbatore, India, 19–20 March 2015; pp. 1–6. [Google Scholar]
- Strang, K.D.; Sun, Z. Hidden big data analytics issues in the healthcare industry. Health Inform. J. 2020, 26, 981–998. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, X.; Kambourakis, G. Special issue on Security, Privacy and Trust in network-based Big Data. Inf. Sci.—Inform. Comput. Sci. Intell. Syst. Appl. Int. J. 2015, 318, 48–50. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, Y. Transmission protocol for secure big data in two-hop wireless networks with cooperative jamming. Inf. Sci. 2014, 281, 201–210. [Google Scholar] [CrossRef]
- Shull, F. The true cost of mobility? IEEE Softw. 2014, 31, 5–9. [Google Scholar] [CrossRef] [Green Version]
- Brown, B. HIPAA Beyond HIPAA: ONCHIT, ONC, AHIC, HITSP, and CCHIT. J. Health Care Compliance 2008, 10, 41–44. [Google Scholar]
- van Loenen, B.; Kulk, S.; Ploeger, H. Data protection legislation: A very hungry caterpillar: The case of mapping data in the European Union. Gov. Inf. Q. 2016, 33, 338–345. [Google Scholar] [CrossRef]
- Stephens, Z.D.; Lee, S.Y.; Faghri, F.; Campbell, R.H.; Zhai, C.; Efron, M.J.; Iyer, R.; Schatz, M.C.; Sinha, S.; Robinson, G.E. Big data: Astronomical or genomical? PLoS Biol. 2015, 13, e1002195. [Google Scholar] [CrossRef]
- Patil, H.K.; Seshadri, R. Big data security and privacy issues in healthcare. In Proceedings of the 2014 IEEE International Congress on Big Data, Washington, DC, USA, 27–30 October 2014; pp. 762–765. [Google Scholar]
- Jimenez-Sanchez, G. Genomics innovation: Transforming healthcare, business, and the global economy. Genome 2015, 58, 511–517. [Google Scholar] [CrossRef] [PubMed]
- Martin-Sanchez, F.; Verspoor, K. Big data in medicine is driving big changes. Yearb. Med. Inform. 2014, 23, 14–20. [Google Scholar]
- Wang, Y.; Kung, L.; Byrd, T.A. Big data analytics: Understanding its capabilities and potential benefits for healthcare organizations. Technol. Forecast. Soc. Chang. 2018, 126, 3–13. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hassan, M.; Awan, F.M.; Naz, A.; deAndrés-Galiana, E.J.; Alvarez, O.; Cernea, A.; Fernández-Brillet, L.; Fernández-Martínez, J.L.; Kloczkowski, A. Innovations in Genomics and Big Data Analytics for Personalized Medicine and Health Care: A Review. Int. J. Mol. Sci. 2022, 23, 4645. https://doi.org/10.3390/ijms23094645
Hassan M, Awan FM, Naz A, deAndrés-Galiana EJ, Alvarez O, Cernea A, Fernández-Brillet L, Fernández-Martínez JL, Kloczkowski A. Innovations in Genomics and Big Data Analytics for Personalized Medicine and Health Care: A Review. International Journal of Molecular Sciences. 2022; 23(9):4645. https://doi.org/10.3390/ijms23094645
Chicago/Turabian StyleHassan, Mubashir, Faryal Mehwish Awan, Anam Naz, Enrique J. deAndrés-Galiana, Oscar Alvarez, Ana Cernea, Lucas Fernández-Brillet, Juan Luis Fernández-Martínez, and Andrzej Kloczkowski. 2022. "Innovations in Genomics and Big Data Analytics for Personalized Medicine and Health Care: A Review" International Journal of Molecular Sciences 23, no. 9: 4645. https://doi.org/10.3390/ijms23094645





