Candidate Genes and Pathways in Rice Co-Responding to Drought and Salt Identified by gcHap Network
Abstract
:1. Introduction
2. Phenology, Physiological, and Biochemical Indicators of Drought and Salt Tolerance
3. Genetics of Drought Tolerance and Salt Tolerance in Rice
3.1. Collection of Cloned QTLs/Genes
3.2. Data Acquisition and Analysis by Public Databases
4. Gene–CDS–Haplotype and Functional Analysis of the DT and ST Genes in Rice
4.1. Gene Ontology and Pathway Analysis
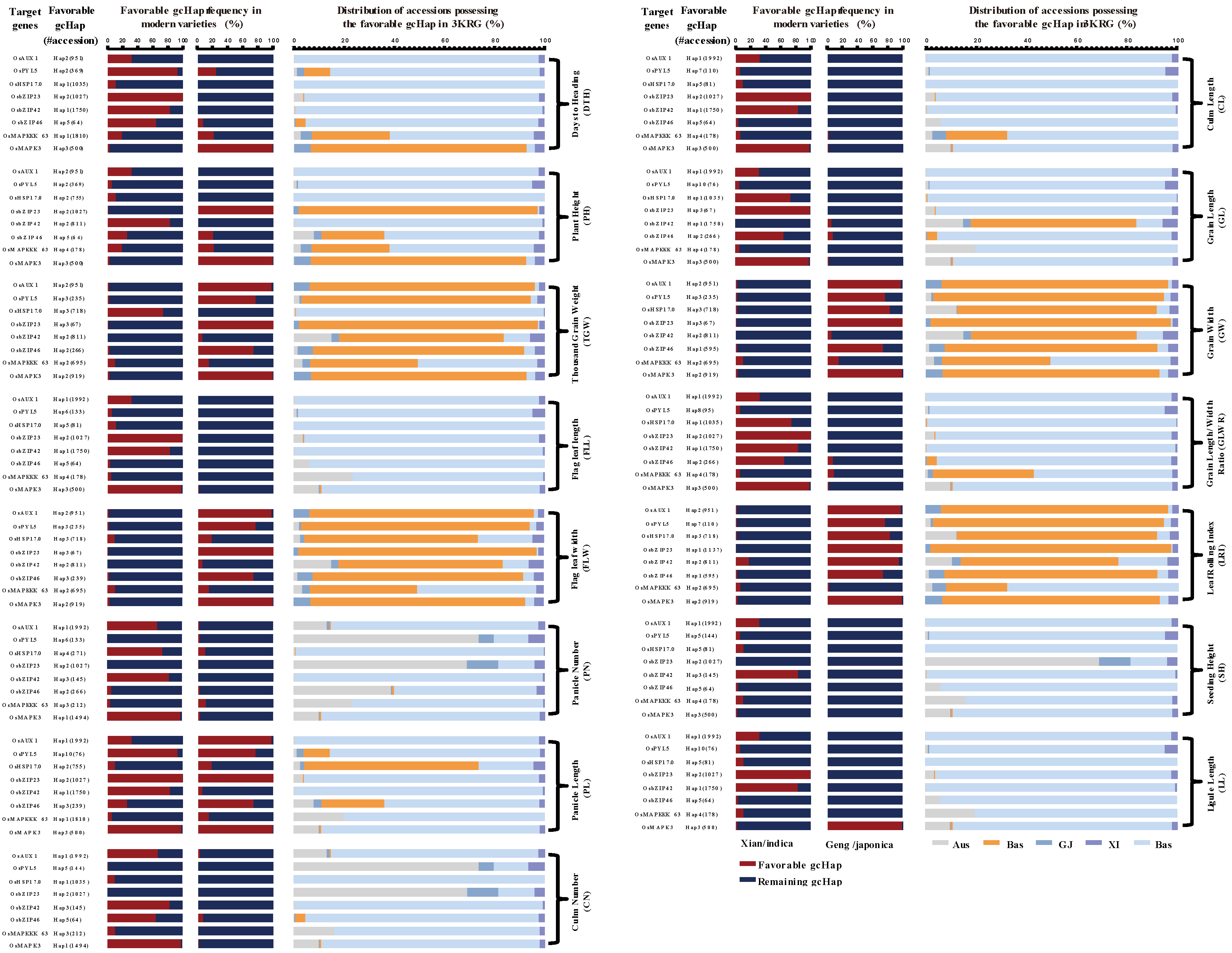
4.2. GcHap Diversity of Genes Positively Regulated by DT and ST
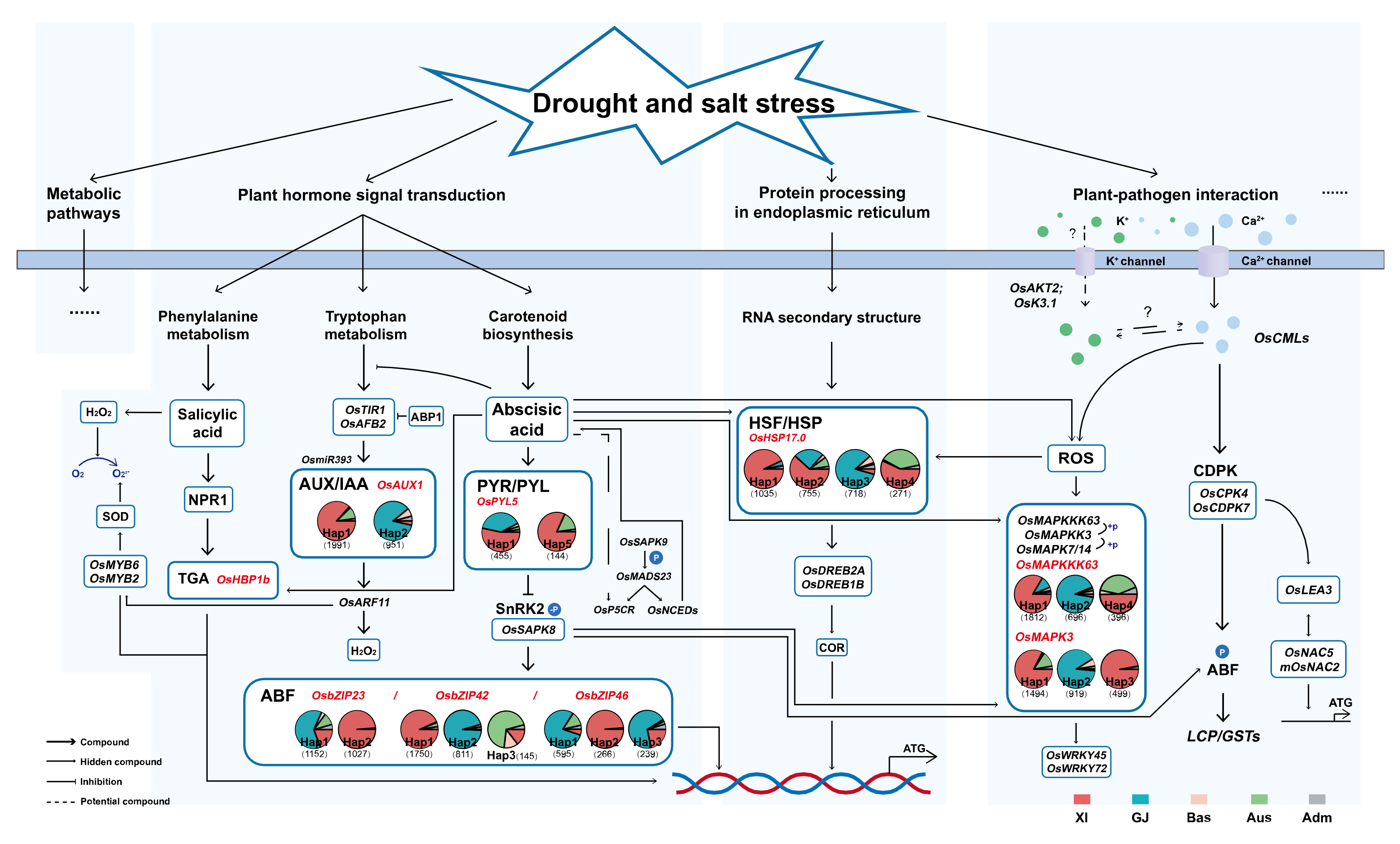
4.3. gcHap-Network Pathway Analysis
5. Breeding Approaches of Drought and Salt Tolerance
5.1. Improving DT and ST by Conventional Breeding Approaches
5.2. Improving DT and ST by Selective Introgression
6. Conclusions, Challenges, and Prospects
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef] [PubMed]
- Sircar, S.; Parekh, N. Meta-analysis of drought-tolerant genotypes in Oryza sativa: A network-based approach. PLoS ONE 2019, 14, e0216068. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jing, R.L. Advances of research on drought resistance and water use efficiency in crop plants. Rev. China Agric. Sci. Technol. 2007, 1, 1–5. [Google Scholar]
- Paik, S.; Le, D.T.P.; Nhu, L.T.; Mills, B.F. Salt-tolerant rice variety adoption in the Mekong River Delta: Farmer adaptation to sea-level rise. PLoS ONE 2020, 15, e0229464. [Google Scholar] [CrossRef] [PubMed]
- Vu, D.T.; Yamada, T.; Ishidaira, H. Assessing the impact of sea level rise due to climate change on seawater intrusion in mekong delta, Vietnam. Water Sci. Technol. 2018, 77, 1632–1639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neina, D. The Role of Soil pH in Plant Nutrition and Soil Remediation. Appl. Environ. Soil Sci. 2019, 2019, 1–9. [Google Scholar] [CrossRef]
- Kanno, I. Genesis and classification of humic allophane soils in Japan. Transact. Int. Soil Conf. Comm. IV 1962, 1962. [Google Scholar]
- Saychaudhuri, R. Agricultural land resources of india. Soil Sci. 1964, 97, 43–47. [Google Scholar] [CrossRef]
- Rao, P.S.; Mishra, B.; Gupta, S. Effects of Soil Salinity and Alkalinity on Grain Quality of Tolerant, Semi-Tolerant and Sensitive Rice Genotypes. Rice Sci. 2013, 20, 284–291. [Google Scholar] [CrossRef]
- Luo, L.J. Breeding for water-saving and drought-resistance rice (WDR) in China. J. Exp. Bot. 2010, 61, 3509–3517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yue, B.; Xiong, L.; Xue, W.; Xing, Y.; Luo, L.; Xu, C. Genetic analysis for drought resistance of rice at reproductive stage in field with different types of soil. Theor. Appl. Genet. 2005, 111, 1127–1136. [Google Scholar] [CrossRef] [PubMed]
- Torres, R.O.; McNally, K.; Cruz, C.V.; Serraj, R.; Henry, A. Screening of rice Genebank germplasm for yield and selection of new drought tolerance donors. Field Crop. Res. 2013, 147, 12–22. [Google Scholar] [CrossRef]
- Qin, H.; Li, Y.; Huang, R. Advances and Challenges in the Breeding of Salt-Tolerant Rice. Int. J. Mol. Sci. 2020, 21, 8385. [Google Scholar] [CrossRef] [PubMed]
- Uga, Y.; Okuno, K.; Yano, M. DRO1, a major QTL involved in deep rooting of rice under upland field conditions. J. Exp. Bot. 2011, 62, 2485–2494. [Google Scholar] [CrossRef] [Green Version]
- Ren, Z.-H.; Gao, J.-P.; Li, L.-G.; Cai, X.-L.; Huang, W.; Chao, D.-Y.; Zhu, M.-Z.; Wang, Z.-Y.; Luan, S.; Lin, H.-X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 37, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Chen, H.; Huang, W.; Chu, Y.; Shii, C.; Cheng, W. Ectopic expression of rice OsNCED3 in Arabidopsis increases ABA level and alters leaf morphology. Plant Sci. 2010, 178, 12–22. [Google Scholar] [CrossRef]
- Yamamuro, C.; Zhu, J.-K.; Yang, Z. Epigenetic Modifications and Plant Hormone Action. Mol. Plant. 2015, 9, 57–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajkumar, M.S.; Shankar, R.; Garg, R.; Jain, M. Bisulphite sequencing reveals dynamic DNA methylation under desiccation and salinity stresses in rice cultivars. Genomics 2020, 112, 3537–3548. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.; Duan, C. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Zhu, J.-K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought Resistance in Rice from Conventional to Molecular Breeding: A Review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhang, W.; da Silva, J.A.T.; Liu, X.; Duan, J. Rice histone deacetylase HDA704 positively regulates drought and salt tolerance by controlling stomatal aperture and density. Planta 2021, 254, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Partida, R.; Rosario, S.; Lozano-Juste, J. An Update on Crop ABA Receptors. Plants 2021, 10, 1087. [Google Scholar] [CrossRef]
- Huang, Y.; Zhou, J.; Li, Y.; Quan, R.; Wang, J.; Huang, R.; Qin, H. Salt Stress Promotes Abscisic Acid Accumulation to Affect Cell Proliferation and Expansion of Primary Roots in Rice. Int. J. Mol. Sci. 2021, 22, 10892. [Google Scholar] [CrossRef] [PubMed]
- Panda, D.; Mishra, S.S.; Behera, P.K. Drought Tolerance in Rice: Focus on Recent Mechanisms and Approaches. Rice Sci. 2021, 28, 119–132. [Google Scholar] [CrossRef]
- Cheng, S.; Zhou, D.-X.; Zhao, Y. WUSCHEL-related homeobox geneWOX11increases rice drought resistance by controlling root hair formation and root system development. Plant Signal. Behav. 2015, 11, e1130198. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Shen, J.; Xu, Y.; Lizhong, X.; Xiao, J.; Xiong, L. Ghd2, a CONSTANS-like gene, confers drought sensitivity through regulation of senescence in rice. J. Exp. Bot. 2016, 67, 5785–5798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Zhang, H.; Srivastava, A.K.; Pan, Y.; Bai, J.; Fang, J.; Shi, H.; Zhu, J.K. Knockdown of rice Mi-croRNA166 confers drought resistance by causing leaf rolling and altering stem xylem development. Plant Physiol. 2018, 176, 2082–2094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, D.; Sun, W.; Wang, D.; Dong, H.; Zhang, R.; Yu, S. A xylan glucuronosyltransferase gene exhibits pleiotropic effects on cellular composition and leaf development in rice. Sci. Rep. 2020, 10, 3726. [Google Scholar] [CrossRef] [Green Version]
- Mishra, A.; Panda, D. Novel Plant Growth Regulators and their Potential Uses in Agriculture. Int. J. Bio. Resour. Stress Manag. 2017, 8, 820–826. [Google Scholar] [CrossRef]
- Hussain, H.A.; Hussain, S.; Khaliq, A.; Ashraf, U.; Anjum, S.A.; Men, S.; Wang, L. Chilling and Drought Stresses in Crop Plants: Implications, Cross Talk, and Potential Management Opportunities. Front. Plant Sci. 2018, 9, 393. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.S.; Panda, D. Leaf Traits and Antioxidant Defense for Drought Tolerance During Early Growth Stage in Some Popular Traditional Rice Landraces from Koraput, India. Rice Sci. 2017, 24, 207–217. [Google Scholar] [CrossRef]
- Chen, T.; Shabala, S.; Niu, Y.; Chen, Z.-H.; Shabala, L.; Meinke, H.; Venkataraman, G.; Pareek, A.; Xu, J.; Zhou, M. Molecular mechanisms of salinity tolerance in rice. Crop J. 2021, 9, 506–520. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt Tolerance Mechanisms of Plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of Plant Responses to Salt Stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Jin, B.J.; Cho, H.M.; Park, M.S.; Lee, S.H.; Lim, L.H.; Cha, Y.J.; Bae, D.-W.; Kim, S.; et al. Microtubule Dynamics Plays a Vital Role in Plant Adaptation and Tolerance to Salt Stress. Int. J. Mol. Sci. 2021, 22, 5957. [Google Scholar] [CrossRef]
- Zhang, Y.; Fang, J.; Wu, X.; Dong, L. Na+/K+ balance and transport regulatory mechanisms in weedy and cultivated rice (Oryza sativa l.) under salt stress. BMC Plant Biol. 2018, 18, 375. [Google Scholar] [CrossRef]
- Chen, R.; Deng, Y.; Ding, Y.; Guo, J.; Qiu, J.; Wang, B.; Wang, C.; Xie, Y.; Zhang, Z.; Chen, J.; et al. Rice functional genomics: Decades’ efforts and roads ahead. Sci. China Life Sci. 2021, 65, 33–92. [Google Scholar] [CrossRef]
- Chen, Z.C.; Yamaji, N.; Horie, T.; Che, J.; Li, J.; An, G.; Ma, J.F. A magnesium transporter OsMGT1 plays a critical role in salt tolerance in rice. Plant Physiol. 2017, 174, 1837–1849. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Yamaji, N.; Costa, A.; Okuma, E.; Kobayashi, N.I.; Kashiwagi, T.; Katsuhara, M.; Wang, C.; Tanoi, K.; Murata, Y.; et al. OsHKT1;4-mediated Na+ transport in stems contributes to Na+ exclusion from leaf blades of rice at the reproductive growth stage upon salt stress. BMC Plant Biol. 2016, 16, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Ding, Y.; Yang, Y.; Song, C.; Wang, B.; Yang, S.; Guo, Y.; Gong, Z. Protein kinases in plant responses to drought, salt, and cold stress. J. Integr. Plant Biol. 2021, 63, 53–78. [Google Scholar] [CrossRef]
- Okada, A.; Okada, K.; Miyamoto, K.; Koga, J.; Shibuya, N.; Nojiri, H.; Yamane, H. OsTGAP1, a bZIP Transcription Factor, Coordinately Regulates the Inductive Production of Diterpenoid Phytoalexins in Rice. J. Biol. Chem. 2009, 284, 26510–26518. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Ma, Y.-Q.; Huang, X.-X.; Mu, T.-J.; Li, Y.-J.; Li, X.-K.; Liu, X.; Hou, B.-K. Overexpression of OsUGT3 enhances drought and salt tolerance through modulating ABA synthesis and scavenging ROS in rice. Environ. Exp. Bot. 2021, 192, 104653. [Google Scholar] [CrossRef]
- Hussain, Q.; Asim, M.; Zhang, R.; Khan, R.; Farooq, S.; Wu, J. Transcription Factors Interact with ABA through Gene Expression and Signaling Pathways to Mitigate Drought and Salinity Stress. Biomolecules 2021, 11, 1159. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.C.; Chapagain, S.; Jang, C.S. A Negative Regulator in Response to Salinity in Rice: Oryza sativa Salt-, ABA- and Drought-Induced RING Finger Protein 1 (OsSADR1). Plant Cell Physiol. 2018, 59, 575–589. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.-K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, S.C.; Luan, S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2011, 35, 53–60. [Google Scholar] [CrossRef]
- June, M.K.; Izumi, C.M.; ZhenMing, P.; Nathalie, L.; Miguel, A.T.; Jeffery, L.D.; Rachel, E.B.; Sara, B.; Jonathan, D.G.J.; Julian, I.S. NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EBMO J. 2003, 22, 2623–2633. [Google Scholar]
- Hua, D.; Wang, C.; He, J.; Liao, H.; Duan, Y.; Zhu, Z.; Guo, Y.; Chen, Z.; Gong, Z. A Plasma Membrane Receptor Kinase, GHR1, Mediates Abscisic Acid- and Hydrogen Peroxide-Regulated Stomatal Movement in Arabidopsis. Plant Cell 2012, 24, 2546–2561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, T.; Wu, Y.; Zhang, Y.; Liu, L.; Ning, Y.; Wang, D.; Tong, H.; Chen, S.; Chu, C.; Xie, Q. OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Mol. Biol. 2011, 76, 145–156. [Google Scholar] [CrossRef]
- Selamat, N.; Nadarajah, K.K. Meta-Analysis of Quantitative Traits Loci (QTL) Identified in Drought Response in Rice (Oryza sativa L.). Plants 2021, 10, 716. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Zhong, H.; Gong, Z.; Fang, X.; Sun, T.; Deng, X.; Li, Y. Meta-analysis of salt stress transcriptome responses in different rice genotypes at the seedling stage. Plants 2019, 8, 3. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Lei, L.; Liu, H.; Wang, J.; Zheng, H.; Zou, D. Whole-genome mining of abiotic stress gene loci in rice. Planta 2020, 252, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Vikram, P.; Swamy, B.M.; Dixit, S.; Ahmed, H.U.; Cruz, M.T.S.; Singh, A.K.; Kumar, A. qDTY 1.1, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet. 2011, 12, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cui, Y. QTL Analysis of Highyield, Drought Tolerance and Salt Tolerance in Selected Population. Ph. D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2015. (In Chinese). [Google Scholar]
- Zang, J. Effect of Genetic Background on Expression of QTL for Drought and Salt Tolerance and Their Genetic Overlap in Rice. Ph. D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2008. (In Chinese). [Google Scholar]
- Wang, Y. Screening of High Yield, Drought and Salt Tolerance Plants from Backcross Introgression Lines and QTL Detection for Related Traits in Rice. Ph. D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2013. (In Chinese). [Google Scholar]
- Pang, Y. Breeding and Genetic Dissecting of Salinity Tolerance, Drought Tolerance, High Yield and High Grain Quality Rice Materials. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2017. (In Chinese). [Google Scholar]
- Zhong, R.; Lee, C.; McCarthy, R.L.; Reeves, C.K.; Jones, E.G.; Ye, Z.-H. Transcriptional Activation of Secondary Wall Biosynthesis by Rice and Maize NAC and MYB Transcription Factors. Plant Cell Physiol. 2011, 52, 1856–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar]
- Chen, M.; Zhao, Y.; Zhuo, C.; Lu, S.; Guo, Z. Overexpression of a nf-yc transcription factor from bermudagrass confers tolerance to drought and salinity in transgenic rice. Plant Biotechnol. J. 2015, 13, 482–491. [Google Scholar] [CrossRef] [PubMed]
- Lakra, N.; Nutan, K.K.; Das, P.; Anwar, K.; Singla-Pareek, S.L.; Pareek, A. A nuclear-localized histone-gene binding protein from rice (oshbp1b) functions in salinity and drought stress tolerance by maintaining chlorophyll content and improving the antioxidant machinery. J. Plant Physiol. 2015, 176, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Saijo, Y.; Kinoshita, N.; Ishiyama, K.; Hata, S.; Kyozuka, J.; Hayakawa, T.; Nakamura, T.; Shimamoto, K.; Yamaya, T.; Izui, K. A Ca2+-Dependent Protein Kinase that Endows Rice Plants with Cold- and Salt-Stress Tolerance Functions in Vascular Bundles. Plant Cell Physiol. 2001, 42, 1228–1233. [Google Scholar] [CrossRef] [Green Version]
- Yi, X.; Du, Z.; Su, Z. PlantGSEA: A gene set enrichment analysis toolkit for plant community. Nucleic Acids Res. 2013, 41, W98–W103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanehisa, M.S.G. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Yu, H.; Huang, J.; Wang, W.; Faruquee, M.; Zhang, F.; Zhao, X.; Fu, B.; Chen, K.; Zhang, H.; et al. Towards a deeper haplotype mining of complex traits in rice with RFGB v2.0. Plant Biotechnol. J. 2019, 18, 14–16. [Google Scholar] [CrossRef] [PubMed]
- Paradis, E. pegas: An R package for population genetics with an integrated-modular approach. Bioinformatics 2010, 26, 419–420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mahmood ur, R.; Ijaz, M.; Qamar, S.; Bukhari, S.A.; Malik, K. Abiotic Stress Signaling in Rice Crop. In Advances in Rice Research for Abiotic Stress Tolerance; Mirza, H., Masayuki, F., Kamrun, N., Jiban, B., Eds.; Woodhead Publishing: Duxford, UK, 2019; pp. 551–569. [Google Scholar]
- Nghi, K.N.; Tagliani, A.; Mariotti, L.; Weits, D.A.; Perata, P.; Pucciariello, C. Auxin is required for the long coleoptile trait in japonica rice under submergence. New Phytol. 2021, 229, 85–93. [Google Scholar] [CrossRef] [PubMed]
- Xia, K.F.; Wang, R.; Ou, X.J.; Fang, Z.M.; Tian, C.G.; Duan, J.; Wang, Y.Q.; Zhang, M.Y. OsTIR1 and OsAFB2 Downregulation via OsmiR393 Overexpression Leads to More Tillers, Early Flowering and Less Tolerance to Salt and Drought in Rice. PLoS ONE 2012, 7, e30039. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Huang, Y.; Qi, P.; Lian, G.; Hu, X.; Han, N.; Wang, J.; Zhu, M.; Qian, Q.; Bian, H. Functional analysis of auxin receptor OsTIR1/osafb family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol. 2021, 229, 2676–2692. [Google Scholar] [CrossRef] [PubMed]
- Sims, K.; Abedi-Samakush, F.; Szulc, N.; Honti, M.M.; Mattsson, J. OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. Int. J. Mol. Sci. 2021, 22, 4089. [Google Scholar] [CrossRef]
- Yadav, S.K.; Kumar, V.V.S.; Verma, R.K.; Yadav, P.; Saroha, A.; Wankhede, D.P.; Chaudhary, B.; Chinnusamy, V. Genome-wide identification and characterization of ABA receptor PYL gene family in rice. BMC Genom. 2020, 21, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Verma, R.K.; Kumar, V.V.S.; Yadav, S.K.; Pushkar, S.; Rao, M.V.; Chinnusamy, V. Overexpression of ABA Receptor PYL10 Gene Confers Drought and Cold Tolerance to Indica Rice. Front. Plant Sci. 2019, 10, 1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, J.; Wang, L.; Yuan, M. Update on the Roles of Rice MAPK Cascades. Int. J. Mol. Sci. 2021, 22, 1679. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Zheng, R.; He, J.; Zhou, Z.; Wang, J.; Xiong, Y.; Xu, T. Noncanonical auxin signaling regulates cell di-vision pattern during lateral root development. Proc. Natl. Acad. Sci. USA 2019, 116, 21285–21290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ponce, K.; Guo, L.; Leng, Y.; Meng, L.; Ye, G. Advances in Sensing, Response and Regulation Mechanism of Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 2254. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yu, B.; Wu, Q.; Min, Q.; Zeng, R.; Xie, Z.; Huang, J. OsMADS23 phosphorylated by SAPK9 confers drought and salt tolerance by regulating ABA biosynthesis in rice. PLoS Genet. 2021, 17, e1009699. [Google Scholar] [CrossRef]
- Chang, Y.; Nguyen, B.H.; Xie, Y.; Xiao, B.; Tang, N.; Zhu, W.; Mou, T.; Xiong, L. Co-overexpression of the constitutively active form of osbzip46 and ABA-activated protein kinase sapk6 improves drought and temperature stress resistance in rice. Front Plant Sci. 2017, 8, 1102. [Google Scholar] [CrossRef]
- Kim, N.; Moon, S.-J.; Min, M.K.; Choi, E.-H.; Kim, J.-A.; Koh, E.Y.; Yoon, I.; Byun, M.-O.; Yoo, S.-D.; Kim, B.-G. Functional characterization and reconstitution of ABA signaling components using transient gene expression in rice protoplasts. Front. Plant Sci. 2015, 6, 614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, S.; Dong, X. Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 2014, 20, 64–68. [Google Scholar] [CrossRef] [Green Version]
- Durga, K.; Soumen, B. Redox regulation of adventitious root formation through downstream changes in hormonal system in mung bean [vigna radiata (l.) R. Wilczek]. Ann. Syst. Biol. 2021, 4, 5–12. [Google Scholar] [CrossRef]
- Kora, D.; Bhattacharjee, S. The interaction of reactive oxygen species and antioxidants at the metabolic inter-face in salicylic acid-induced adventitious root formation in mung bean [vigna radiata (l.) R. Wilczek]. J. Plant Physiol. 2020, 248, 153152. [Google Scholar] [CrossRef]
- Zinati, Z.; Barati, V. Unveiling the molecular mechanisms of drought stress tolerance in rice (Oryza sativa L.) using computational approaches. BioTechnologia 2018, 99, 385–400. [Google Scholar] [CrossRef]
- Kan, Y.; Lin, H.-X. Molecular regulation and genetic control of rice thermal response. Crop J. 2021, 9, 497–505. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef]
- Ma, H.; Chen, J.; Zhang, Z.; Ma, L.; Yang, Z.; Zhang, Q.; Li, X.; Xiao, J.; Wang, S. MAPK kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 2017, 92, 557–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Na, Y.-J.; Choi, H.-K.; Park, M.Y.; Choi, S.-W.; Vo, K.T.X.; Jeon, J.-S.; Kim, S.Y. OsMAPKKK63 is involved in salt stress response and seed dormancy control. Plant Signal. Behav. 2019, 14, e1578633. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Zhu, J.; Gong, Z.; Zhu, J.K. Abiotic stress responses in plants. Nat. Rev. Genet. 2021, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Pittman, J.K.; Hirschi, K.D. Phylogenetic analysis and protein structure modelling identifies distinct Ca2+/cation antiporters and conservation of gene family structure within Arabidopsis and rice species. Rice 2016, 9, 3. [Google Scholar] [CrossRef] [Green Version]
- Marcec, M.J.; Gilroy, S.; Poovaiah, B.W.; Tanaka, K. Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 2019, 283, 343–354. [Google Scholar] [CrossRef] [PubMed]
- Sharma, D.; Verma, N.; Pandey, C.; Verma, D.; Bhagat, P.K.; Noryang, S.; Singh, K.; Tayyeba, S.; Banerjee, G.; Sinha, A.K. Map kinase as regulators for stress responses in plants: An over-view. In Protein Kinases and Stress Signaling in Plants: Functional Genomic Perspective; Pandey, G.K., Ed.; Wiley: Hoboken, NJ, USA, 2021. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed]
- Musavizadeh, Z.; Najafi-Zarrini, H.; Kazemitabar, S.K.; Hashemi, S.H.; Faraji, S.; Barcaccia, G.; Heidari, P. Genome-wide analysis of potassium channel genes in rice: Expression of the osakt and oskat genes under salt stress. Genes 2021, 12, 5. [Google Scholar] [CrossRef]
- Zhang, A.; Yang, X.; Lu, J.; Song, F.; Sun, J.; Wang, C.; Lian, J.; Zhao, L.; Zhao, B. OsIAA20, an Aux/IAA protein, mediates abiotic stress tolerance in rice through an ABA pathway. Plant Sci. 2021, 308, 110903. [Google Scholar] [CrossRef] [PubMed]
- Santos, A.P.; Serra, T.; Figueiredo, D.D.; Barros, P.; Lourenco, T.; Chander, S.; Oliveira, M.M.; Saibo, N.J. Transcription regulation of abiotic stress responses in rice: A combined action of transcription factors and epigenetic mechanisms. OMICS 2011, 15, 839–857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, L.; Wu, J.; Jiang, M.; Wang, Y. Plant Mitogen-Activated Protein Kinase Cascades in Environmental Stresses. Int. J. Mol. Sci. 2021, 22, 1543. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.; Kumar, I.S. Drought Response in Rice: The miRNA Story. Int. J. Mol. Sci. 2019, 20, 3766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.; Chang, Y.; Qin, Y.; Chen, D.; Zhu, T.; Peng, K.; Wang, H.; Tang, N.; Li, X.; Wang, Y.; et al. A lamin-like protein OsNMCP1 regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeller OsSWI3C in rice. New Phytol. 2020, 227, 65–83. [Google Scholar] [CrossRef] [PubMed]
- Frisch, M.; Bohn, M.; Melchinger, A.E. Comparison of Selection Strategies for Marker-Assisted Backcrossing of a Gene. Crop Sci. 1999, 39, 1295–1301. [Google Scholar] [CrossRef] [Green Version]
- Sandhu, N.; Dixit, S.; Swamy, B.P.M.; Vikram, P.; Venkateshwarlu, C.; Catolos, M.; Kumar, A. Positive interactions of major-effect QTLs with genetic background that enhances rice yield under drought. Sci. Rep. 2018, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Muthu, V.; Abbai, R.; Nallathambi, J.; Rahman, H.; Ramasamy, S.; Kambale, R.; Thulasinathan, T.; Ayyenar, B.; Muthurajan, R. Pyramiding QTLs controlling tolerance against drought, salinity, and submergence in rice through marker assisted breeding. PLoS ONE 2020, 15, e0227421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, X.; Jiang, H.; Meng, L.; Chen, J. Gene Mapping, Cloning and Association Analysis for Salt Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 11674. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.R.; Gregorio, G.B. Progress of salinity tolerant rice variety development in bangladesh. SABRAO J. Breed. Genet. 2013, 45, 21–30. [Google Scholar]
- Lang, N.T.; Buu, B.C.; Ismail, A. Molecular mapping and marker-assisted selection for salt tolerance in rice (Oryza sativa l.). Pak. J. Bot. 2008, 16, 50–56. [Google Scholar]
- Vu, H.T.T.; Le, D.D.; Ismail, A.M.; Le, H.H. Marker-assisted backcrossing (mabc) for improved salinity tolerance in rice (‘Oryza sativa’ l.) to cope with climate change in Vietnam. Aust. J. Crop Sci. 2012, 6, 1649–1654. [Google Scholar]
- Linh le, H.; Linh, T.H.; Xuan, T.D.; Ham le, H.; Ismail, A.M.; Khanh, T.D. Molecular breeding to improve salt tolerance of rice (Oryza sativa l.) in the red river delta of Vietnam. Int. J. Plant Genom. 2012, 2012, 949038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patwary, M.A.K.; Moniruzzaman, M.; Islam, M.M.; Begum, S.N.; Alam, M.S. Marker-assisted foreground se-lection for identification of salt tolerant rice genotypes. Agriculturists 2012, 10, 1–8. [Google Scholar]
- Huyen, L.T.N.; Cuc, L.M.; Abdelbagi, M.I.; Ham, L.H. Introgression of the Saltol into as996, the elite variety of Vietnam, using marker assisted backcrossing. J. Sci. Nat. Sci. Technol. 2012, 28, 37–46. [Google Scholar]
- Huyen, L.T.N. Introgression the saltol QTL into q5db, the elite variety of Vietnam using marker- assisted—se-lection (mas). Am. J. BioSci. 2013, 1, 80–84. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.; Ali, J.; Zhou, S.; Ren, G.; Xie, H.; Xu, J.; Yu, X.; Zhou, F.; Peng, S.; Ma, L.; et al. From Green Super Rice to green agriculture: Reaping the promise of functional genomics research. Mol. Plant 2022, 15, 9–26. [Google Scholar] [CrossRef]
- Yu, S.; Ali, J.; Zhang, C.; Li, Z.; Zhang, Q. Genomic breeding of Green Super Rice varieties and their deployment in Asia and africa. Theor. Appl. Genet. 2020, 133, 1427–1442. [Google Scholar] [CrossRef] [PubMed] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, Z.; Ma, S.; Liang, L.; Feng, T.; Xiong, M.; Lian, S.; Zhu, J.; Chen, Y.; Meng, L.; Li, M. Candidate Genes and Pathways in Rice Co-Responding to Drought and Salt Identified by gcHap Network. Int. J. Mol. Sci. 2022, 23, 4016. https://doi.org/10.3390/ijms23074016
Hao Z, Ma S, Liang L, Feng T, Xiong M, Lian S, Zhu J, Chen Y, Meng L, Li M. Candidate Genes and Pathways in Rice Co-Responding to Drought and Salt Identified by gcHap Network. International Journal of Molecular Sciences. 2022; 23(7):4016. https://doi.org/10.3390/ijms23074016
Chicago/Turabian StyleHao, Zhiqi, Sai Ma, Lunping Liang, Ting Feng, Mengyuan Xiong, Shangshu Lian, Jingyan Zhu, Yanjun Chen, Lijun Meng, and Min Li. 2022. "Candidate Genes and Pathways in Rice Co-Responding to Drought and Salt Identified by gcHap Network" International Journal of Molecular Sciences 23, no. 7: 4016. https://doi.org/10.3390/ijms23074016
APA StyleHao, Z., Ma, S., Liang, L., Feng, T., Xiong, M., Lian, S., Zhu, J., Chen, Y., Meng, L., & Li, M. (2022). Candidate Genes and Pathways in Rice Co-Responding to Drought and Salt Identified by gcHap Network. International Journal of Molecular Sciences, 23(7), 4016. https://doi.org/10.3390/ijms23074016




