AutoCoEv—A High-Throughput In Silico Pipeline for Predicting Inter-Protein Coevolution
Abstract
1. Introduction
2. Implementation
2.1. Command Line Interface, Configuration, and Input
2.2. Identification of Orthologues
2.3. Multiple Sequence Alignment and Trees
2.4. Detection of Inter-Protein Coevolution by CAPS2
2.5. Post-Run Processing of the Results
3. Application
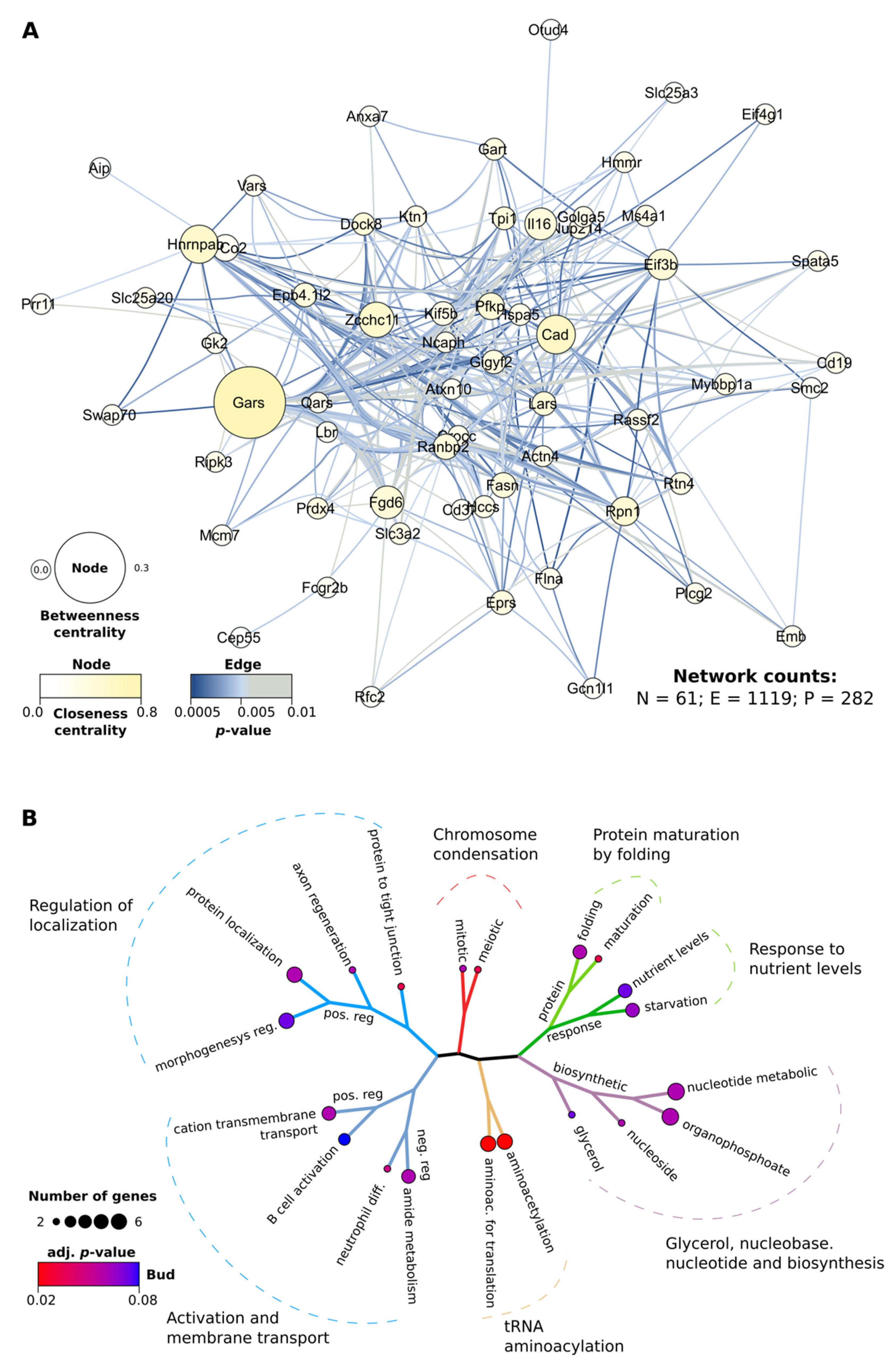
Large Dataset Analysis
4. Discussion
5. Materials and Methods
5.1. Availability and Required Software
5.2. Databases
5.3. Proximity Biotinylation
5.4. AutoCoEv Runtime Parameters
5.5. Protein Characterization
5.6. Data Analyses
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Baussand, J.; Carbone, A. A Combinatorial Approach to Detect Coevolved Amino Acid Networks in Protein Families of Variable Divergence. PLoS Comput Biol 2009, 5. [Google Scholar] [CrossRef]
- Kuriyan, J. Allostery and Coupled Sequence Variation in Nuclear Hormone Receptors. Cell 2004, 116, 354–356. [Google Scholar] [CrossRef][Green Version]
- Fares, M.A.; McNally, D. CAPS: Coevolution Analysis Using Protein Sequences. Bioinformatics 2006, 22, 2821–2822. [Google Scholar] [CrossRef]
- Oteri, F.; Nadalin, F.; Champeimont, R.; Carbone, A. BIS2Analyzer: A Server for Co-Evolution Analysis of Conserved Protein Families. Nucleic Acids Res. 2017, 45, W307–W314. [Google Scholar] [CrossRef]
- Wang, S.; Sun, S.; Li, Z.; Zhang, R.; Xu, J. Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model. PLoS Comput. Biol. 2017, 13, e1005324. [Google Scholar] [CrossRef]
- Morcos, F.; Pagnani, A.; Lunt, B.; Bertolino, A.; Marks, D.S.; Sander, C.; Zecchina, R.; Onuchic, J.N.; Hwa, T.; Weigt, M. Direct-Coupling Analysis of Residue Coevolution Captures Native Contacts across Many Protein Families. Proc. Natl. Acad. Sci. USA 2011, 108, E1293–E1301. [Google Scholar] [CrossRef]
- Hopf, T.A.; Green, A.G.; Schubert, B.; Mersmann, S.; Schärfe, C.P.I.; Ingraham, J.B.; Toth-Petroczy, A.; Brock, K.; Riesselman, A.J.; Palmedo, P.; et al. The EVcouplings Python Framework for Coevolutionary Sequence Analysis. Bioinformatics 2019, 35, 1582–1584. [Google Scholar] [CrossRef]
- Ovchinnikov, S.; Kamisetty, H.; Baker, D. Robust and Accurate Prediction of Residue–residue Interactions across Protein Interfaces Using Evolutionary Information. eLife 2014, 3, e02030. [Google Scholar] [CrossRef]
- Simonetti, F.L.; Teppa, E.; Chernomoretz, A.; Nielsen, M.; Marino Buslje, C. MISTIC: Mutual Information Server to Infer Coevolution. Nucleic Acids Res. 2013, 41, W8–W14. [Google Scholar] [CrossRef]
- Wang, Y.; Correa Marrero, M.; Medema, M.H.; van Dijk, A.D.J. Coevolution-Based Prediction of Protein–protein Interactions in Polyketide Biosynthetic Assembly Lines. Bioinformatics 2020, 36, 4846–4853. [Google Scholar] [CrossRef]
- Fares, M.A.; Travers, S.A.A. A Novel Method for Detecting Intramolecular Coevolution: Adding a Further Dimension to Selective Constraints Analyses. Genetics 2006, 173, 9–23. [Google Scholar] [CrossRef] [PubMed]
- Cong, Q.; Anishchenko, I.; Ovchinnikov, S.; Baker, D. Protein Interaction Networks Revealed by Proteome Coevolution. Science 2019, 365, 185–189. [Google Scholar] [CrossRef]
- Travers, S.A.A.; Fares, M.A. Functional Coevolutionary Networks of the Hsp70–Hop–Hsp90 System Revealed through Computational Analyses. Mol. Biol. Evol. 2007, 24, 1032–1044. [Google Scholar] [CrossRef]
- Huang, Y.; Temperley, N.D.; Ren, L.; Smith, J.; Li, N.; Burt, D.W. Molecular Evolution of the Vertebrate TLR1 Gene Family—A Complex History of Gene Duplication, Gene Conversion, Positive Selection and Co-Evolution. BMC Evol. Biol. 2011, 11, 149. [Google Scholar] [CrossRef]
- Ruiz-González, M.X.; Fares, M.A. Coevolution Analyses Illuminate the Dependencies between Amino Acid Sites in the Chaperonin System GroES-L. BMC Evol. Biol. 2013, 13, 156. [Google Scholar] [CrossRef]
- Petrov, P.; Syrjänen, R.; Smith, J.; Gutowska, M.W.; Uchida, T.; Vainio, O.; Burt, D.W. Characterization of the Avian Trojan Gene Family Reveals Contrasting Evolutionary Constraints. PLoS ONE 2015, 10, e0121672. [Google Scholar] [CrossRef]
- Champeimont, R.; Laine, E.; Hu, S.-W.; Penin, F.; Carbone, A. Coevolution Analysis of Hepatitis C Virus Genome to Identify the Structural and Functional Dependency Network of Viral Proteins. Sci. Rep. 2016, 6, 26401. [Google Scholar] [CrossRef]
- Awoniyi, L.O.; Runsala, M.; Hernández-Pérez, S.; Šuštar, V.; .Cunha, D.M.; Sarapulov, A.V.; Petrov, P.; Mattila, P.K. Novel players and large-scale protein dynamics of BCR activation revealed by APEX2 proximity labelling of lipid rafts. bioRxiv 2020. [Google Scholar] [CrossRef]
- Consortium, T.U. UniProt: A Worldwide Hub of Protein Knowledge. Nucleic Acids Res. 2019, 47, D506–D515. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Suleski, M.; Hedges, S.B. TimeTree: A Resource for Timelines, Timetrees, and Divergence Times. Mol. Biol. Evol. 2017, 34, 1812–1819. [Google Scholar] [CrossRef]
- Kriventseva, E.V.; Kuznetsov, D.; Tegenfeldt, F.; Manni, M.; Dias, R.; Simão, F.A.; Zdobnov, E.M. OrthoDB v10: Sampling the Diversity of Animal, Plant, Fungal, Protist, Bacterial and Viral Genomes for Evolutionary and Functional Annotations of Orthologs. Nucleic Acids Res. 2019, 47, D807–D811. [Google Scholar] [CrossRef] [PubMed]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and Applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [PubMed]
- Sela, I.; Ashkenazy, H.; Katoh, K.; Pupko, T. GUIDANCE2: Accurate Detection of Unreliable Alignment Regions Accounting for the Uncertainty of Multiple Parameters. Nucleic Acids Res. 2015, 43, W7–W14. [Google Scholar] [CrossRef] [PubMed]
- Wong, K.M.; Suchard, M.A.; Huelsenbeck, J.P. Alignment Uncertainty and Genomic Analysis. Science 2008, 319, 473–476. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Löytynoja, A. Phylogeny-Aware Alignment with PRANK. Methods Mol. Biol. 2014, 1079, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Castresana, J. Selection of Conserved Blocks from Multiple Alignments for Their Use in Phylogenetic Analysis. Mol. Biol. Evol. 2000, 17, 540–552. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Vilella, A.J.; Severin, J.; Ureta-Vidal, A.; Heng, L.; Durbin, R.; Birney, E. EnsemblCompara GeneTrees: Complete, Duplication-Aware Phylogenetic Trees in Vertebrates. Genome Res. 2009, 19, 327–335. [Google Scholar] [CrossRef]
- Tange, O. GNU Parallel 20211122 (‘PengShuai’); Zenodo: Geneve, Switzerland, 2021. [Google Scholar] [CrossRef]
- Shen, W.; Le, S.; Li, Y.; Hu, F. SeqKit: A Cross-Platform and Ultrafast Toolkit for FASTA/Q File Manipulation. PLoS ONE 2016, 11, e0163962. [Google Scholar] [CrossRef] [PubMed]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 17 February 2022).
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Blohm, P.; Frishman, G.; Smialowski, P.; Goebels, F.; Wachinger, B.; Ruepp, A.; Frishman, D. Negatome 2.0: A Database of Non-Interacting Proteins Derived by Literature Mining, Manual Annotation and Protein Structure Analysis. Nucleic Acids Res. 2014, 42, D396–D400. [Google Scholar] [CrossRef] [PubMed]
- Giurgiu, M.; Reinhard, J.; Brauner, B.; Dunger-Kaltenbach, I.; Fobo, G.; Frishman, G.; Montrone, C.; Ruepp, A. CORUM: The Comprehensive Resource of Mammalian Protein Complexes-2019. Nucleic Acids Res. 2019, 47, D559–D563. [Google Scholar] [CrossRef] [PubMed]
- Lassmann, T.; Sonnhammer, E.L.L. Kalign, Kalignvu and Mumsa: Web Servers for Multiple Sequence Alignment. Nucleic Acids Research 2006, 34, W596–W599. [Google Scholar] [CrossRef]
- Moreno-Morcillo, M.; Grande-García, A.; Ruiz-Ramos, A.; del Caño-Ochoa, F.; Boskovic, J.; Ramón-Maiques, S. Structural Insight into the Core of CAD, the Multifunctional Protein Leading De Novo Pyrimidine Biosynthesis. Structure 2017, 25, 912–923.e5. [Google Scholar] [CrossRef]
- Thompson, J.D.; Linard, B.; Lecompte, O.; Poch, O. A Comprehensive Benchmark Study of Multiple Sequence Alignment Methods: Current Challenges and Future Perspectives. PLoS ONE 2011, 6, e18093. [Google Scholar] [CrossRef]
- Notredame, C.; Higgins, D.G.; Heringa, J. T-Coffee: A Novel Method for Fast and Accurate Multiple Sequence Alignment. J. Mol. Biol. 2000, 302, 205–217. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for Making Accurate Alignments of Many Protein Sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML Version 8: A Tool for Phylogenetic Analysis and Post-Analysis of Large Phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian Phylogenetic Inference and Model Choice across a Large Model Space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and Effective Stochastic Algorithm for Estimating Maximum-Likelihood Phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Anand, S.; Foot, N.; Ang, C.-S.; Gembus, K.M.; Keerthikumar, S.; Adda, C.G.; Mathivanan, S.; Kumar, S. Arrestin-Domain Containing Protein 1 (Arrdc1) Regulates the Protein Cargo and Release of Extracellular Vesicles. Proteomics 2018, 18, 1800266. [Google Scholar] [CrossRef] [PubMed]
- Carrey, E.A.; Dietz, C.; Glubb, D.M.; Löffler, M.; Lucocq, J.M.; Watson, P.F. Detection and Location of the Enzymes of de Novo Pyrimidine Biosynthesis in Mammalian Spermatozoa. Reproduction 2002, 123, 757–768. [Google Scholar] [CrossRef] [PubMed]
- Puusepp, S.; Kovacs-Nagy, R.; Alhaddad, B.; Braunisch, M.; Hoffmann, G.F.; Kotzaeridou, U.; Lichvarova, L.; Liiv, M.; Makowski, C.; Mandel, M.; et al. Compound Heterozygous SPATA5 Variants in Four Families and Functional Studies of SPATA5 Deficiency. Eur. J. Hum. Genet. 2018, 26, 407–419. [Google Scholar] [CrossRef] [PubMed]
- Amstalden van Hove, E.R.; Smith, D.F.; Heeren, R.M.A. A Concise Review of Mass Spectrometry Imaging. J. Chromatogr. A 2010, 1217, 3946–3954. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. 20 Years of the SMART Protein Domain Annotation Resource. Nucleic Acids Res. 2018, 46, D493–D496. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The Protein Families Database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Erdős, G.; Pajkos, M.; Dosztányi, Z. IUPred3: Prediction of Protein Disorder Enhanced with Unambiguous Experimental Annotation and Visualization of Evolutionary Conservation. Nucleic Acids Res. 2021, 49, W297–W303. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.-G.; Han, Y.; He, Q.-Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Hulsen, T.; de Vlieg, J.; Alkema, W. BioVenn—A Web Application for the Comparison and Visualization of Biological Lists Using Area-Proportional Venn Diagrams. BMC Genom. 2008, 9, 488. [Google Scholar] [CrossRef]
- Li, M.; Li, D.; Tang, Y.; Wu, F.; Wang, J. CytoCluster: A Cytoscape Plugin for Cluster Analysis and Visualization of Biological Networks. Int. J. Mol. Sci. 2017, 18, 1880. [Google Scholar] [CrossRef] [PubMed]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petrov, P.B.; Awoniyi, L.O.; Šuštar, V.; Balci, M.Ö.; Mattila, P.K. AutoCoEv—A High-Throughput In Silico Pipeline for Predicting Inter-Protein Coevolution. Int. J. Mol. Sci. 2022, 23, 3351. https://doi.org/10.3390/ijms23063351
Petrov PB, Awoniyi LO, Šuštar V, Balci MÖ, Mattila PK. AutoCoEv—A High-Throughput In Silico Pipeline for Predicting Inter-Protein Coevolution. International Journal of Molecular Sciences. 2022; 23(6):3351. https://doi.org/10.3390/ijms23063351
Chicago/Turabian StylePetrov, Petar B., Luqman O. Awoniyi, Vid Šuštar, M. Özge Balci, and Pieta K. Mattila. 2022. "AutoCoEv—A High-Throughput In Silico Pipeline for Predicting Inter-Protein Coevolution" International Journal of Molecular Sciences 23, no. 6: 3351. https://doi.org/10.3390/ijms23063351
APA StylePetrov, P. B., Awoniyi, L. O., Šuštar, V., Balci, M. Ö., & Mattila, P. K. (2022). AutoCoEv—A High-Throughput In Silico Pipeline for Predicting Inter-Protein Coevolution. International Journal of Molecular Sciences, 23(6), 3351. https://doi.org/10.3390/ijms23063351





