Research of Fluridone’s Effects on Growth and Pigment Accumulation of Haematococcus pluvialis Based on Transcriptome Sequencing
Abstract
:1. Introduction
2. Results
2.1. Effects of Fluridone on Cell Status and Biomass
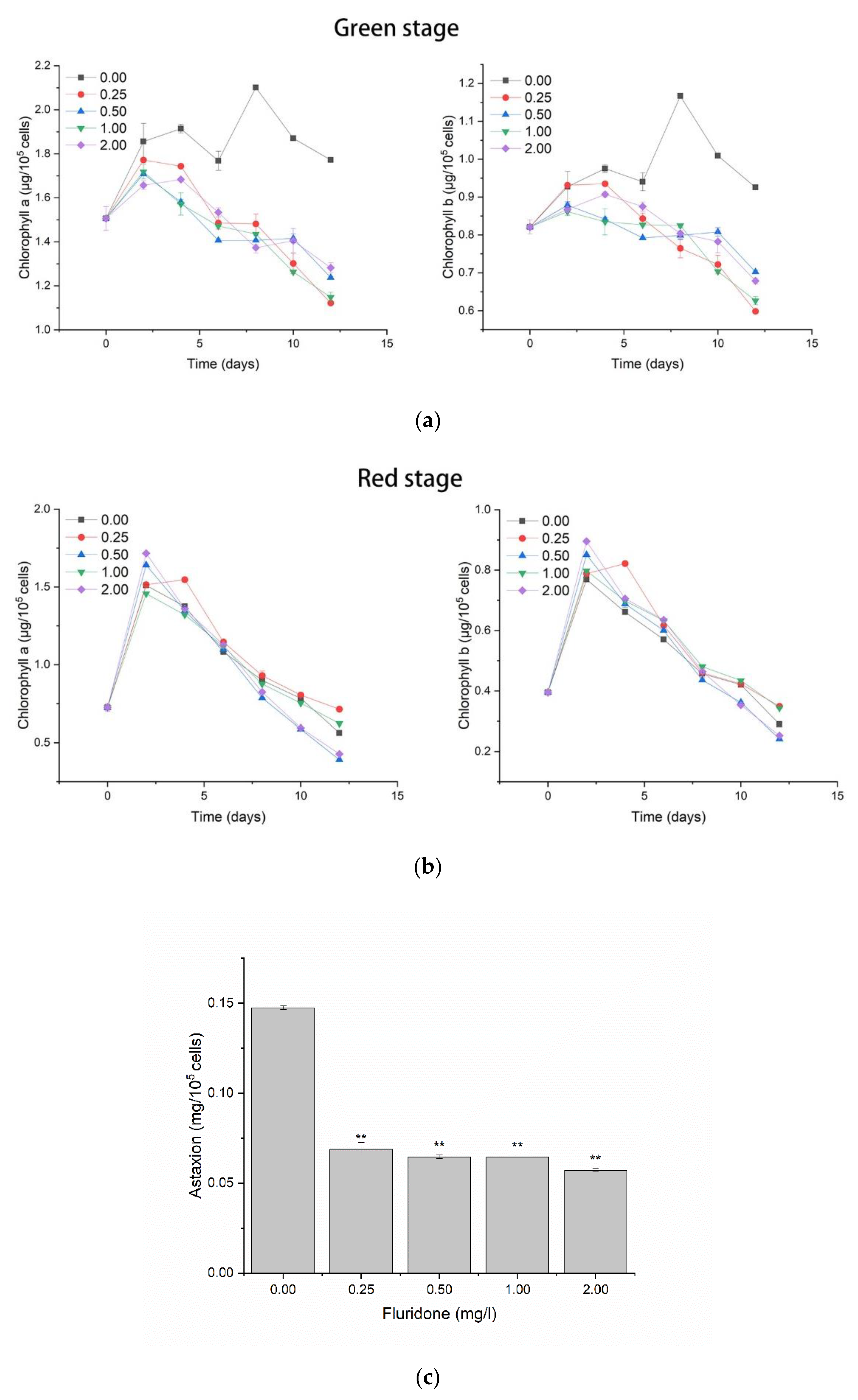
2.2. Effects of Pigments
2.3. Transcription Sequencing Quality and Splicing Information
2.4. Feature Annotation Results
2.4.1. NR Annotated Species Classification
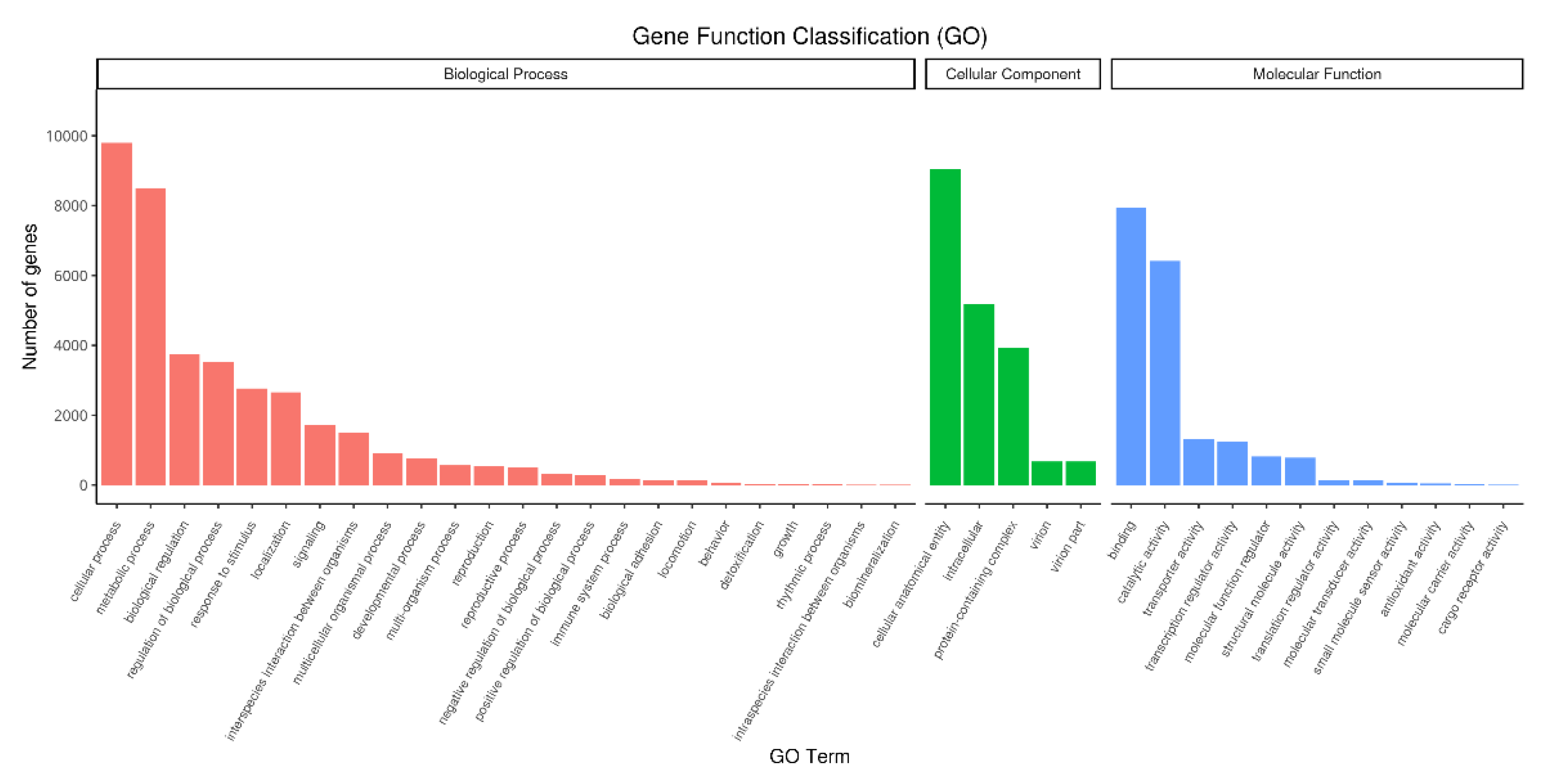
2.4.2. GO Classification
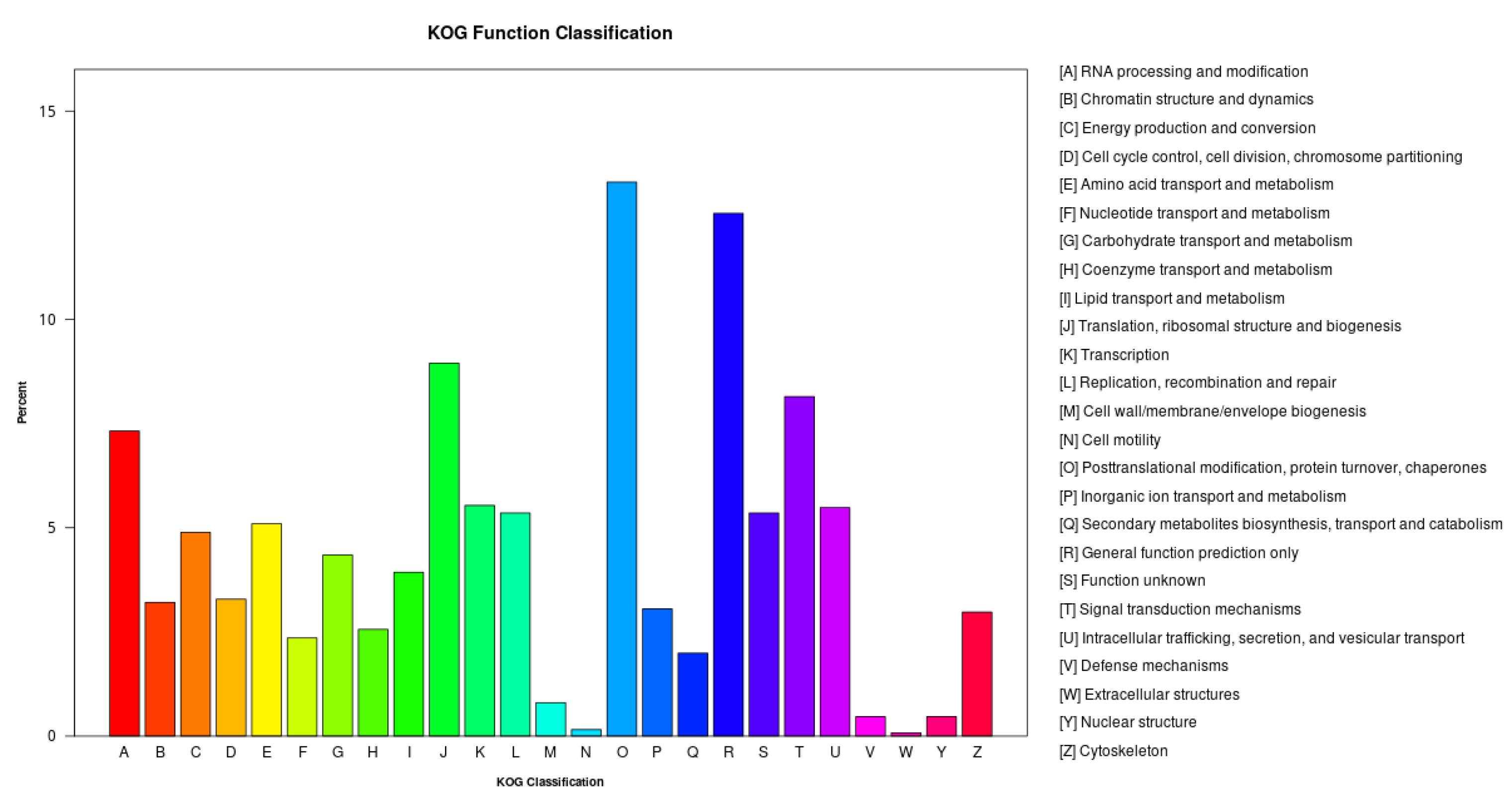
2.4.3. KOG Classification
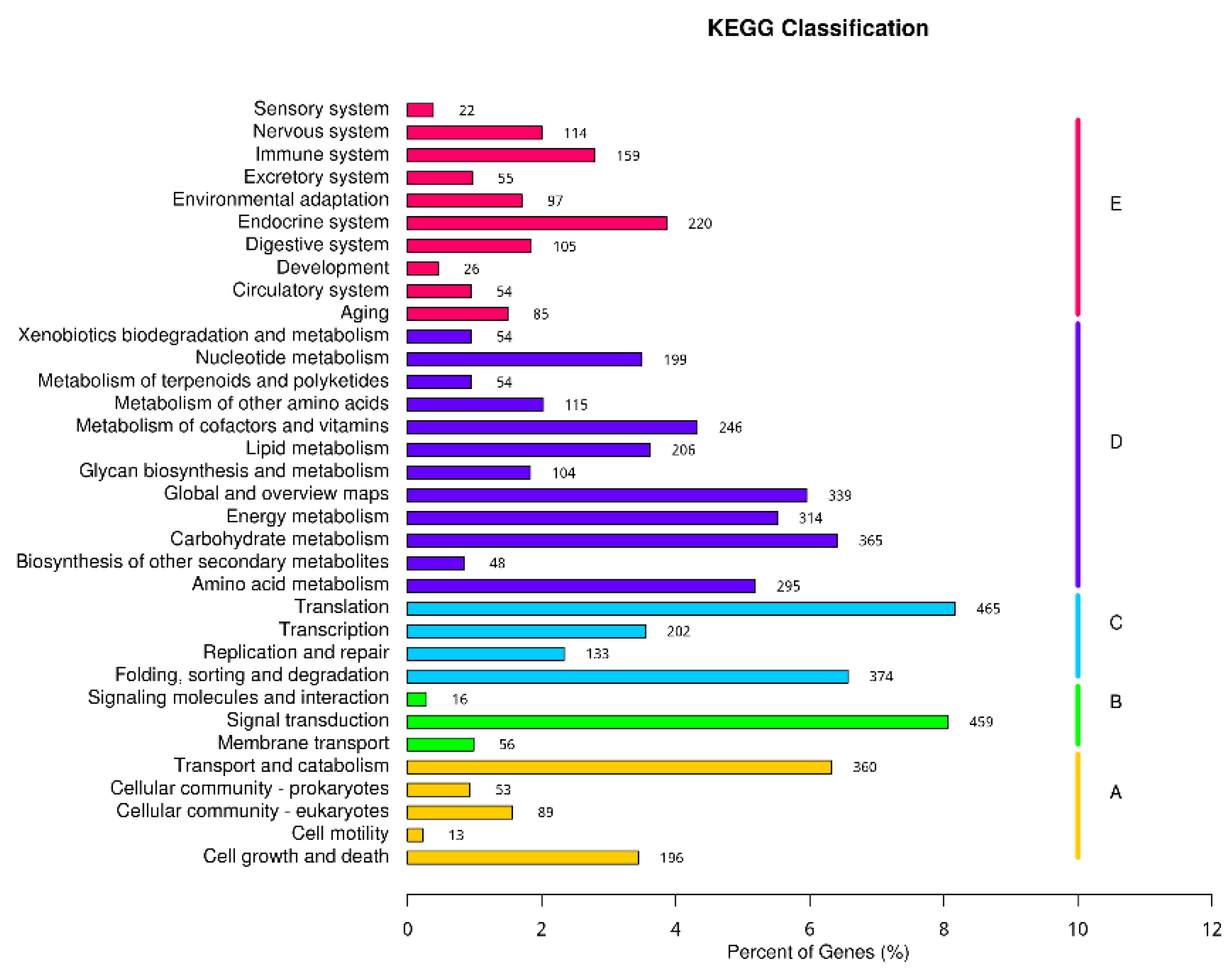
2.4.4. KEGG Classification
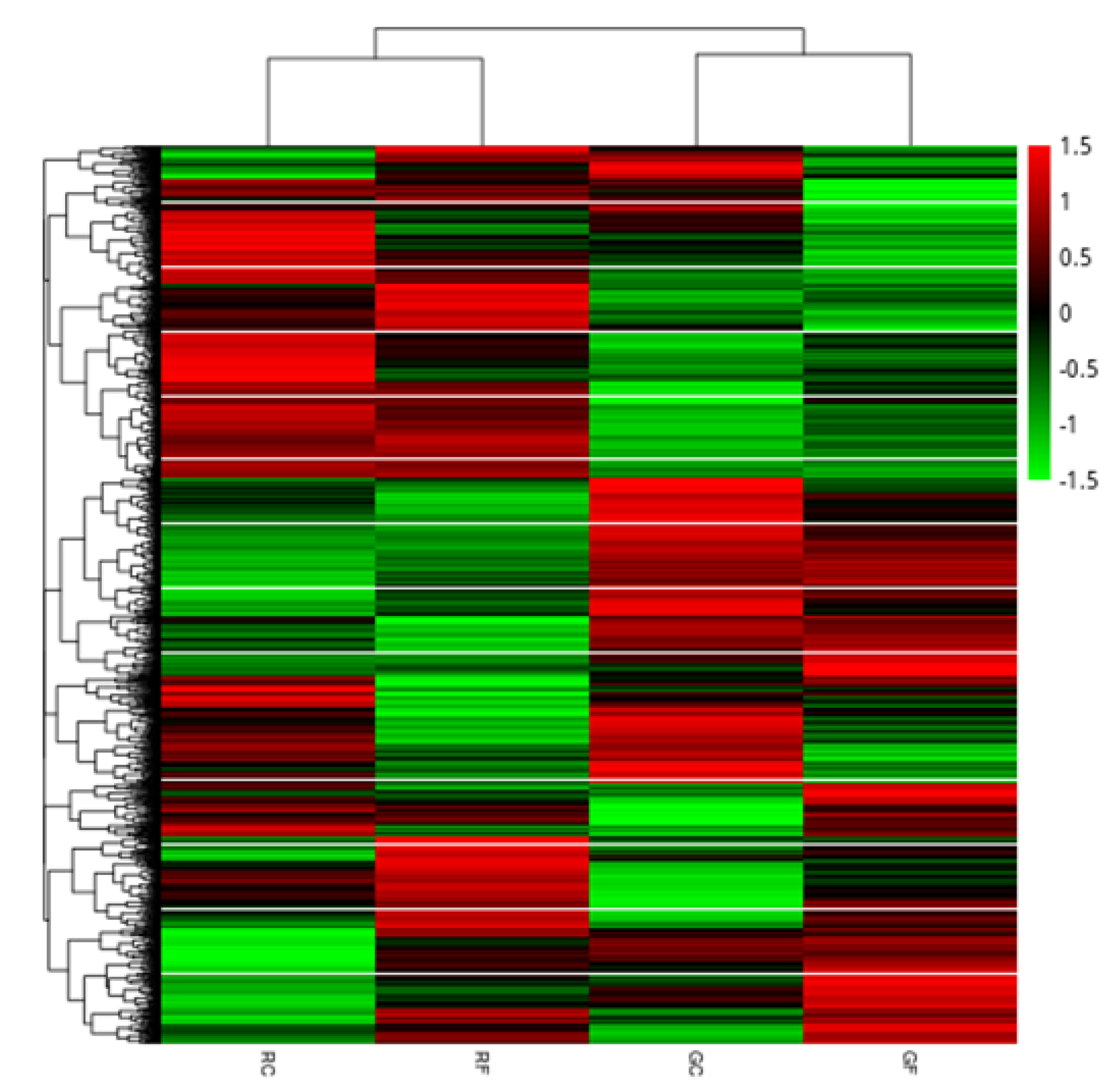
2.5. Analysis of Differentially Expressed Genes
2.5.1. Summary of DEGs under Different Conditions
2.5.2. GO and KEGG Enrichment Analysis of Green Stage DEGs
2.5.3. KEGG Enrichment Analysis of Red Stage DEGs
2.6. Effects of Fluridone on Specific Metabolic Pathways of Haematococcus pluvialis
2.6.1. Effects on Carotenoid Biosynthesis
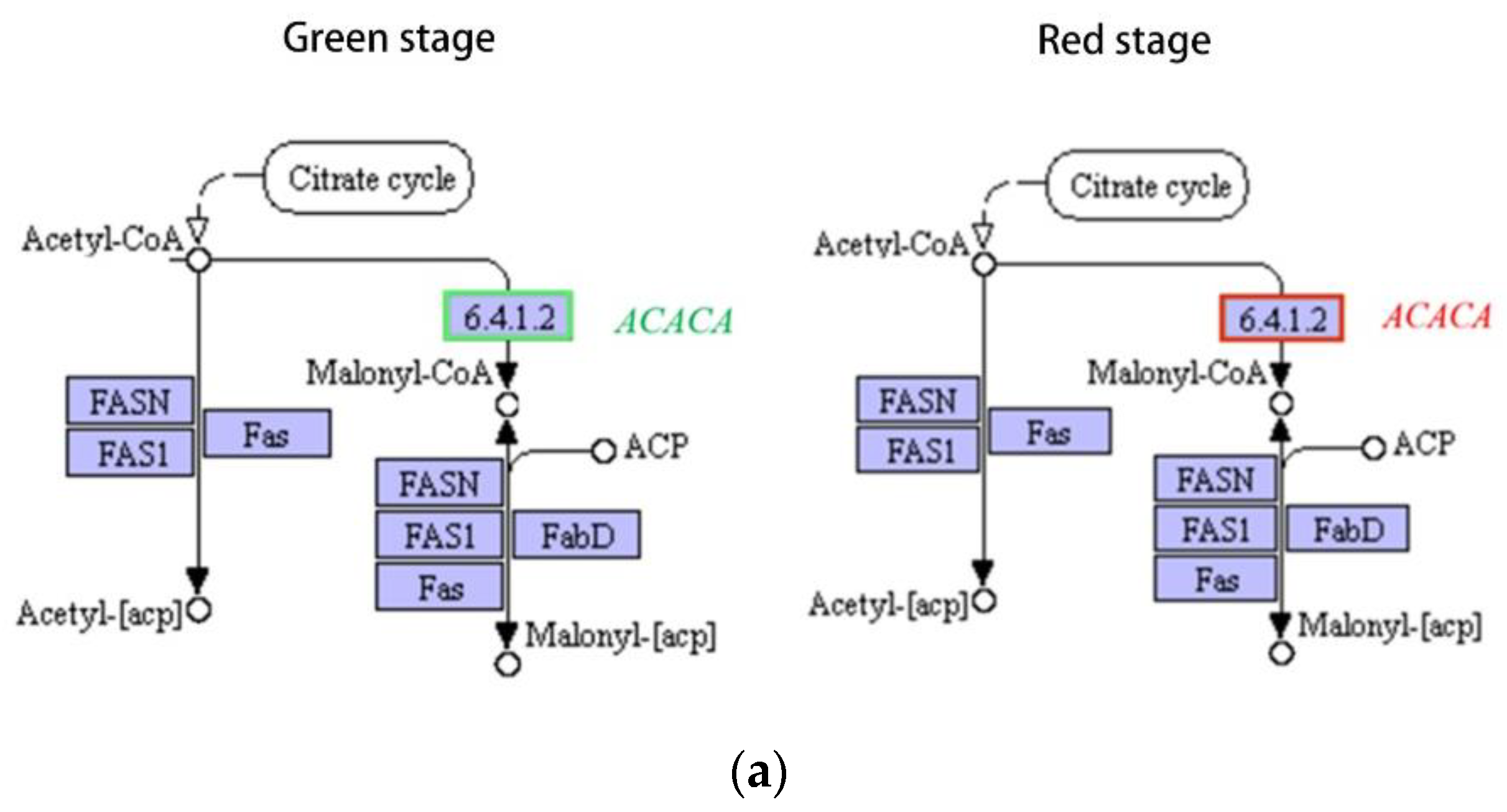
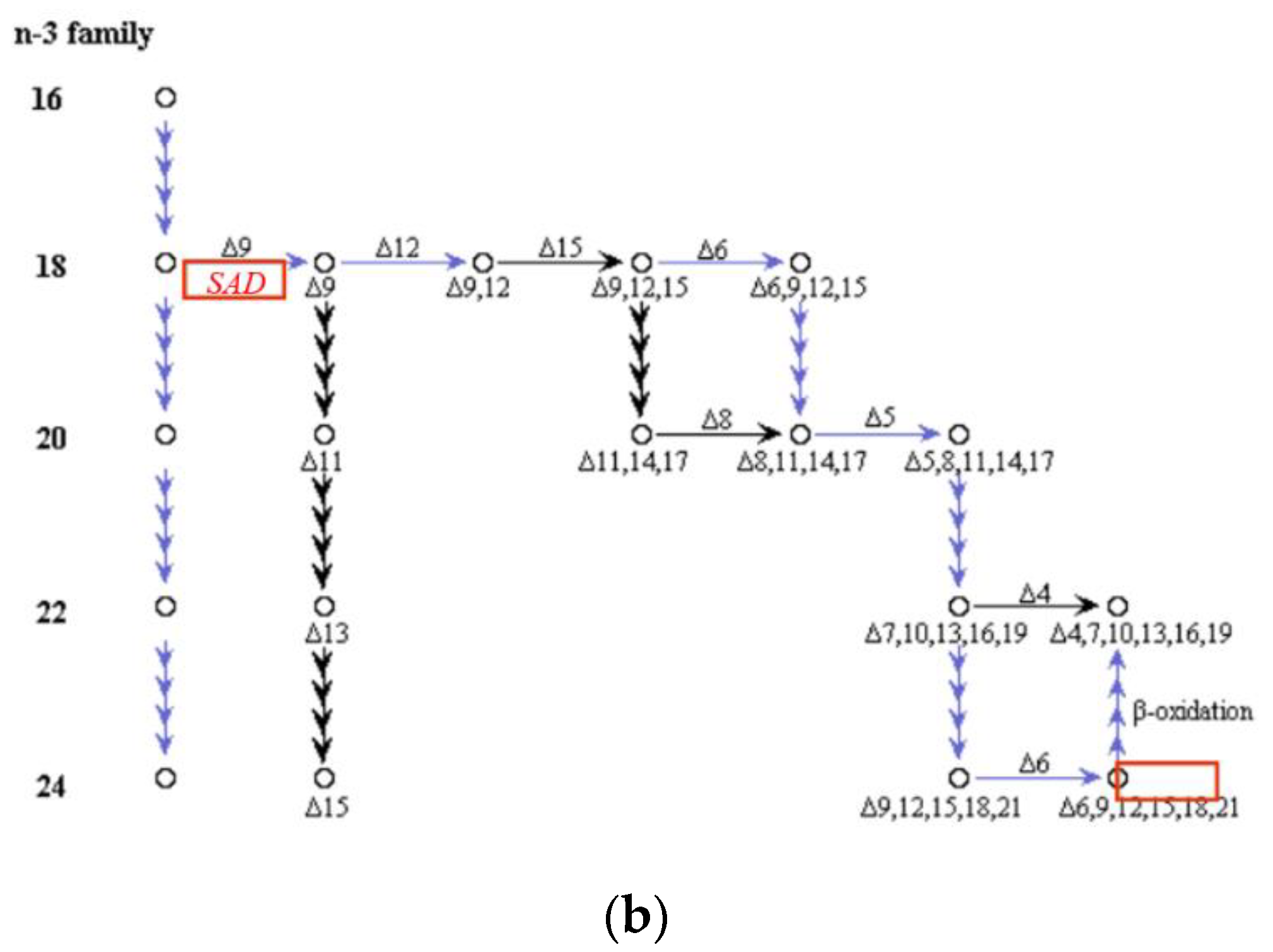
2.6.2. Effects on Fatty Acid Metabolism
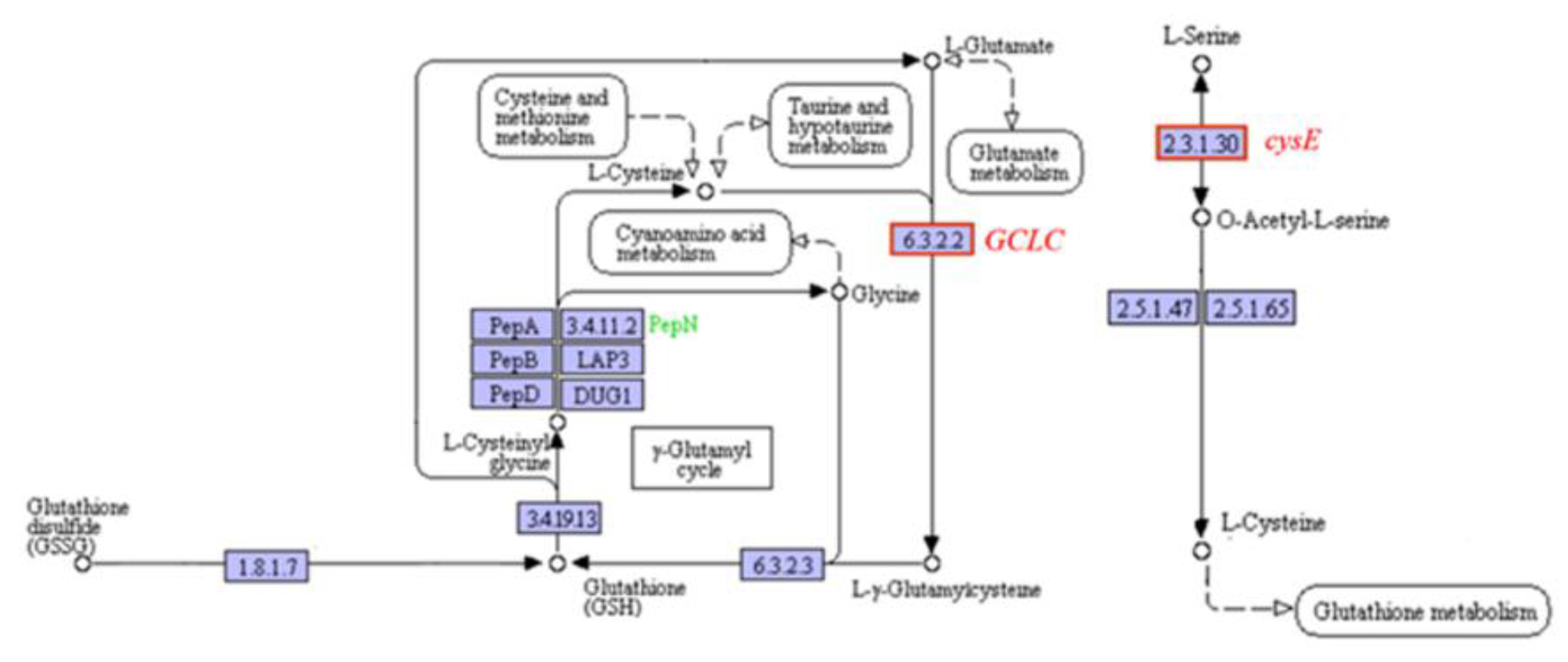
2.6.3. Effects on Cellular Antioxidant Capacity
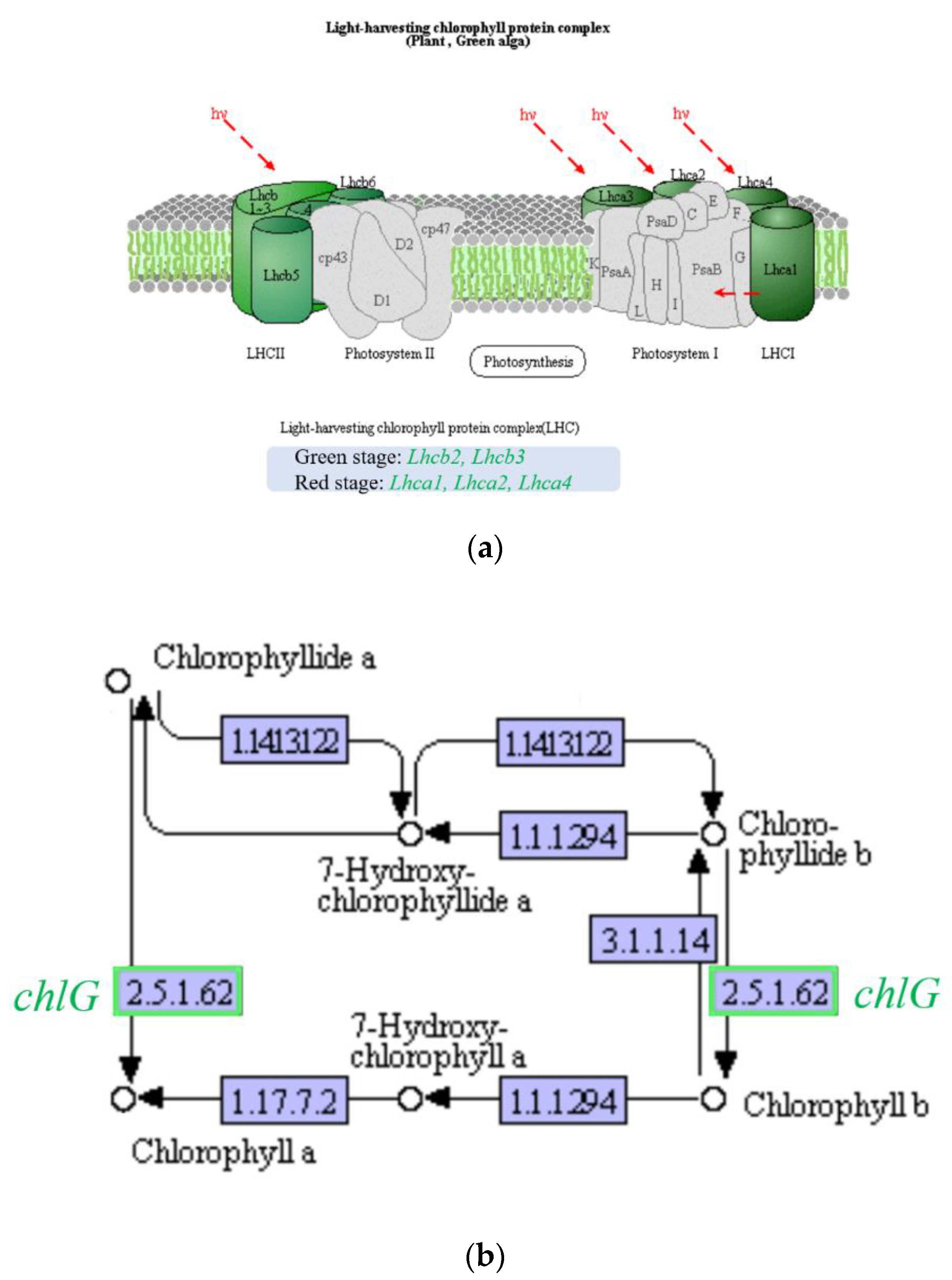
2.6.4. Effects on Photosynthesis
2.7. Real-Time PCR Verification
3. Discussion
4. Materials and Methods
4.1. Cultivation of Algae
4.2. Biomass Determination and Pigment Analysis
4.3. Sample Preparation for Transcriptome Sequencing
4.4. Construction of cDNA Library and Transcriptome Sequencing
4.5. Transcript Splicing and Gene Function Annotation
4.6. Differentially Expressed Gene Analysis
4.7. Quantitative PCR Validation of Gene Expression
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Shah, M.M.; Liang, Y.; Cheng, J.J.; Daroch, M. Astaxanthin-Producing Green Microalga Haematococcus pluvialis: From Single Cellto High Value Commercia Products. Front. Plant Sci. 2016, 7, 531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davinelli, S.; Nielsen, M.E.; Scapagnini, G. Astaxanthin in Skin Health, Repair, and Disease: A Comprehensive Review. Nutrients 2018, 10, 522. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khoo, K.S.; Lee, S.Y.; Ooi, C.W.; Fu, X.; Miao, X.; Ling, T.C.; Show, P.L. Recent advances in biorefinery of astaxanthin from Haematococcus pluvialis. Bioresour. Technol. 2019, 288, 121606. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.C.; Yusoff, F.M.; Shariff, M.; Kamarudin, M.S. Astaxanthin as feed supplement in aquatic animals. Rev. Aquac. 2018, 10, 738–773. [Google Scholar] [CrossRef]
- Yuan, J.-P.; Peng, J.; Yin, K.; Wang, J.-H. Potential health-promoting effects of astaxanthin: A high-value carotenoid mostly from microalgae. Mol. Nutr. Food Res. 2011, 55, 150–165. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, U.; Venkateshwaran, G.; Sarada, R.; Ravishankar, G. Studies on Haematococcus pluvialis for improved production of astaxanthin by mutagenesis. World J. Microbiol. Biotechnol. 2001, 17, 143–148. [Google Scholar] [CrossRef]
- Tjahjono, A.E.; Kakizono, T.; Hayama, Y.; Nishio, N.; Nagai, S. Isolation of resistant mutants against carotenoid biosynthesis inhibitors for a green-alga Haematococcus pluvialis, and their hybrid formation by protoplast fusion for breeding of higher astaxanthin producers. J. Ferment. Bioeng. 1994, 77, 352–357. [Google Scholar]
- Bartels, P.G.; Watson, C.W. Inhibition of Carotenoid Synthesis by Fluridone and Norflurazon. Weed Sci. 1978, 26, 198–203. [Google Scholar] [CrossRef]
- Chalifour, A.; Arts, M.T.; Kainz, M.J.; Juneau, P. Combined effect of temperature and bleaching herbicides on photosynthesis, pigment and fatty acid composition of Chlamydomonas reinhardtii. Eur. J. Phycol. 2014, 49, 508–515. [Google Scholar] [CrossRef] [Green Version]
- Laje, K.; Seger, M.; Dungan, B.; Cooke, P.; Polle, J.; Holguin, F.O. Phytoene Accumulation in the Novel Microalga Chlorococcum sp. Using the Pigment Synthesis Inhibitor Fluridone. Mar. Drugs 2019, 17, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, Y.; Liu, Y.; Rong, W. A new technology for transcriptome research: RNA-Seq and its application. Genetics 2011, 33, 1191–1202. [Google Scholar]
- Li, Q.; Zhang, L.; Liu, J. Comparative transcriptome analysis at seven time points during Haematococcus pluvialis motile cell growth and astaxanthin accumulation. Aquaculture 2019, 503, 304–311. [Google Scholar] [CrossRef]
- Gao, Z.; Li, Y.; Wu, G.; Li, G.; Sun, H.; Deng, S.; Shen, Y.; Chen, G.; Zhang, R.; Meng, C.; et al. Transcriptome Analysis in Haematococcus pluvialis: Astaxanthin Induction by Salicylic Acid (SA) and Jasmonic Acid (JA). PLoS ONE 2015, 10, e0140609. [Google Scholar] [CrossRef]
- Li, K.; Cheng, J.; Lu, H.; Yang, W.; Zhou, J.; Cen, K. Transcriptome-based analysis on carbon metabolism of Haematococcus pluvialis mutant under 15% CO2. Bioresour. Technol. 2017, 233, 313–321. [Google Scholar] [CrossRef] [PubMed]
- He, B.; Hou, L.; Dong, M.; Shi, J.; Huang, X.; Ding, Y.; Cong, X.; Zhang, F.; Zhang, X.; Zang, X. Transcriptome Analysis in Haematococcus pluvialis: Astaxanthin Induction by High Light with Acetate and Fe2+. Int. J. Mol. Sci. 2018, 19, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhekisheva, M.; Boussiba, S.; Khozin-Goldberg, I.; Zarka, A.; Cohen, Z. Accumulation of oleic acid in Haematococcus pluvialis (Chlorophyceae) under nitrogen starvation or high light is correlated with that of astaxanthin esters. J. Phycol. 2002, 38, 325–331. [Google Scholar] [CrossRef] [Green Version]
- Ren, Y.; Deng, J.; Huang, J.; Wu, Z.; Yi, L.; Bi, Y.; Chen, F. Using green alga Haematococcus pluvialis for astaxanthin and lipid co-production: Advances and outlook. Bioresour. Technol. 2021, 340, 125736. [Google Scholar] [CrossRef]
- Grünewald, K.; Hirschberg, J.; Hagen, C. Ketocarotenoid Biosynthesis Outside of Plastids in the Unicellular Green Alga Haematococcus pluvialis. J. Biol. Chem. 2001, 276, 6023–6029. [Google Scholar] [CrossRef] [Green Version]
- Damiani, M.C.; Popovich, C.A.; Constenla, D.; Leonardi, P.I. Lipid analysis in Haematococcus pluvialis to assess its potential use as a biodiesel feedstock. Bioresour. Technol. 2010, 101, 3801–3807. [Google Scholar] [CrossRef] [PubMed]
- Zhekisheva, M.; Zarka, A.; Khozin-Goldberg, I.; Cohen, Z.; Boussiba, S. Inhibition of astaxanthin synthesis under high irradiance does not abolish triacylglycerol accumulation in the green alga Haematococcus pluvialis (Chlorophyceae). J. Phycol. 2005, 41, 819–826. [Google Scholar] [CrossRef]
- Kim, J.; DellaPenna, D. Defining the primary route for lutein synthesis in plants: The role of Arabidopsis carotenoid beta-ring hydroxylase CYP97A3. Proc. Natl. Acad. Sci. USA 2006, 103, 3474–3479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boussiba, S.; Fan, L.; Vonshak, A. Enhancement and Determination of Astaxanthin Accumulation in Green Alga Haematococcus pluvialis. Methods Enzymol. 1992, 213, 386–391. [Google Scholar]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, N.M.; Oshlack, A. Corset: Enabling differential gene expression analysis for de novo assembled transcriptomes. Genome Biol. 2014, 15, 410–424. [Google Scholar] [CrossRef]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; Van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Zang, X.; Wu, F.; Jin, Y.; Wang, H.; Liu, C.; Ding, Y.; He, B.; Xiao, D.; Song, X.; et al. Transcriptome Sequencing of Gracilariopsis lemaneiformisto Analyze the Genes Related to Optically Active Phycoerythrin Synthesis. PLoS ONE 2017, 12, e0170855. [Google Scholar]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787. [Google Scholar] [CrossRef] [PubMed]




| Database | Number of Unigenes | Percentage |
|---|---|---|
| Annotated in NR | 12,375 | 26.45 |
| Annotated in NT | 4656 | 9.95 |
| Annotated in KO | 5256 | 11.23 |
| Annotated in SwissProt | 7541 | 16.11 |
| Annotated in PFAM | 16,653 | 35.59 |
| Annotated in GO | 16,653 | 35.59 |
| Annotated in KOG | 3866 | 8.26 |
| Annotated in all Databases | 1177 | 2.51 |
| Annotated in at least one Database | 21,704 | 46.39 |
| Total Unigenes | 46,785 | 100 |
| Gene Name | Primers |
|---|---|
| Rbcl | F: ACGAATGTTTACGCGGTGGTCT |
| R: GGTACACCCAACTCCTTAGCA | |
| cysE | F: TGCCGCCCAGGGACACT |
| R: CCCATGCCATCCAGAGCAAG | |
| LUT5 | F: GATGTCGAGGTCGAGGAGTTGT |
| R: CCAGGGATGATGAAGTTAGGG | |
| ACACA | F: AAGTAGTAGCAGCCCTCCTCCA |
| R: CGCCACCAGAAGATCATAGAA | |
| lcyB | F: TGAGCAGGCAACACGAACAGAC |
| R: ACCCGCTCTGCTACAGCTATG | |
| Lhca2 | F: TCGCCCTGCCCAACCAC |
| R: TGGTGGGCGCAAAGTACTGGAA | |
| GCLC | F: GGGATAATTCCCGCATCAGTTA |
| R: GACCTGGTACAAGTGGCTATTC | |
| chlG | F: CTCACAGGTTACACCCAGACAA |
| R: GATGAAGGAGCCAAACAAGG |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Zan, J.; Zang, X. Research of Fluridone’s Effects on Growth and Pigment Accumulation of Haematococcus pluvialis Based on Transcriptome Sequencing. Int. J. Mol. Sci. 2022, 23, 3122. https://doi.org/10.3390/ijms23063122
Sun J, Zan J, Zang X. Research of Fluridone’s Effects on Growth and Pigment Accumulation of Haematococcus pluvialis Based on Transcriptome Sequencing. International Journal of Molecular Sciences. 2022; 23(6):3122. https://doi.org/10.3390/ijms23063122
Chicago/Turabian StyleSun, Jinpeng, Jiawei Zan, and Xiaonan Zang. 2022. "Research of Fluridone’s Effects on Growth and Pigment Accumulation of Haematococcus pluvialis Based on Transcriptome Sequencing" International Journal of Molecular Sciences 23, no. 6: 3122. https://doi.org/10.3390/ijms23063122
APA StyleSun, J., Zan, J., & Zang, X. (2022). Research of Fluridone’s Effects on Growth and Pigment Accumulation of Haematococcus pluvialis Based on Transcriptome Sequencing. International Journal of Molecular Sciences, 23(6), 3122. https://doi.org/10.3390/ijms23063122





