Development and Regeneration of Muscle, Tendon, and Myotendinous Junctions in Striated Skeletal Muscle
Abstract
1. Introduction
2. Part 1: Maturation
2.1. Muscle and Myotendinous Junctions
- (1)
- Developmental mechanisms of muscles and myotendinous junctions revealed by scRNA-seq
- (2)
- Morphogenesis of the tongue dependent on Dlx5 and Dlx6 expression in mice
2.2. Tendon
3. Part 2: Regeneration
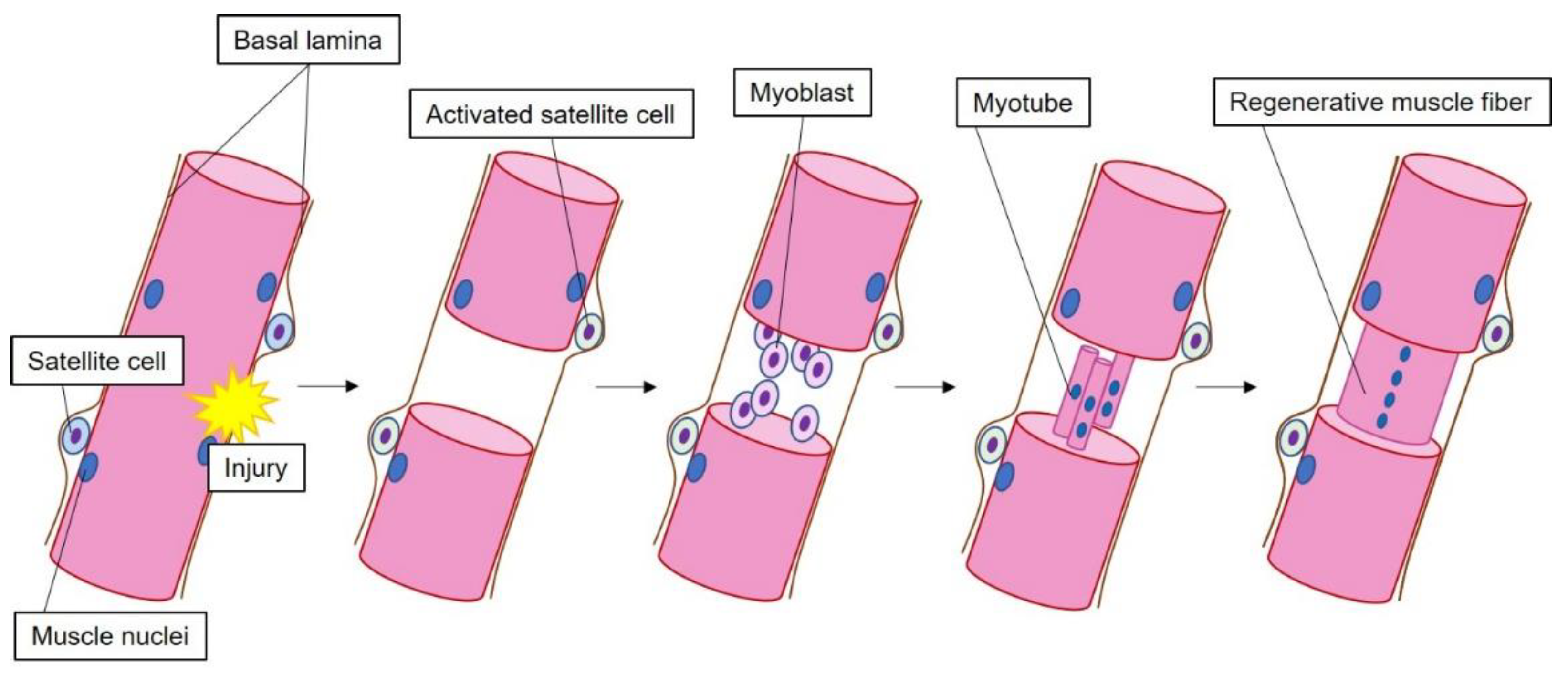
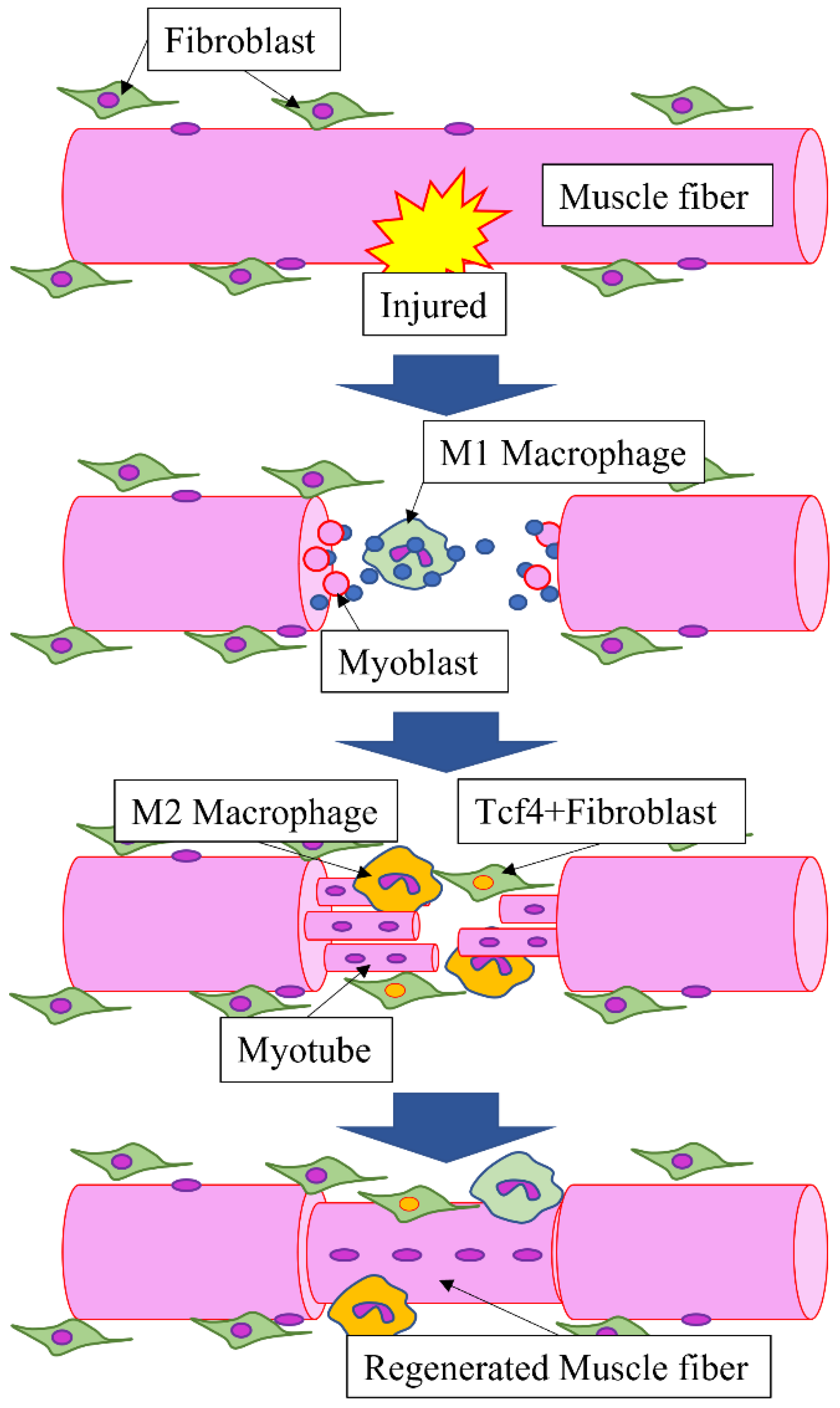
3.1. Role of HMGB1 during Muscle Regeneration
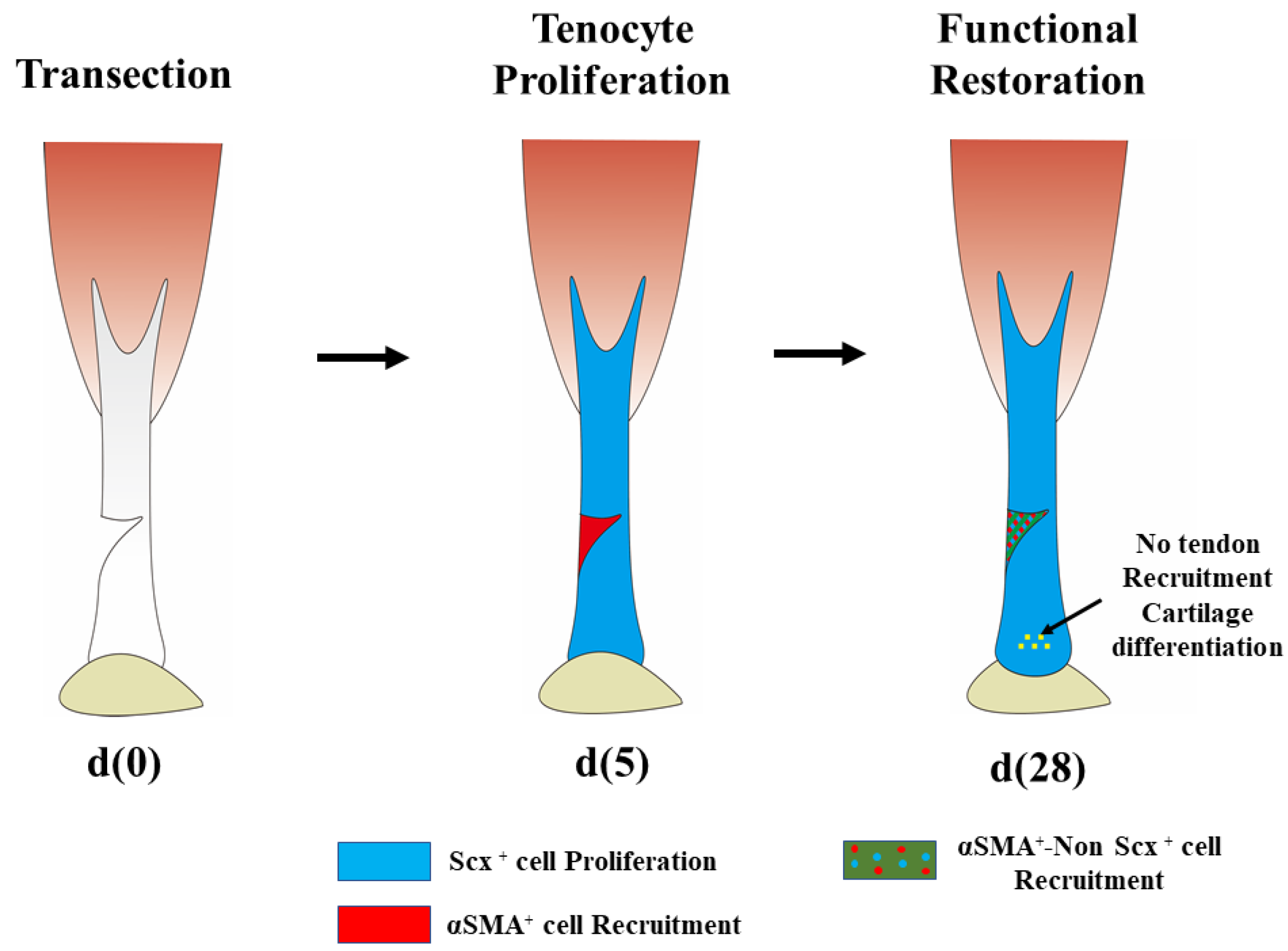
3.2. Tendon Regeneration in Myotendinous Junctions
- (1)
- Tendons in neonates and regeneration
- (2)
- Tendon regeneration in adult animals
3.3. Effects of Mechanical Load on the Growth of Muscle Attachment Sites
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Uezumi, A.; Fukada, S.; Yamamoto, N.; Takeda, S.; Tsuchida, K. Mesenchymal progenitors distinct from satellite cells contribute to ectopic fat cell formation in skeletal muscle. Nat. Cell Biol. 2010, 12, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Joe, A.W.B.; Yi, L.; Natarajan, A.; Grand, F.L.; So, L.; Wang, J.; Rudnicki, M.A.; Rossi, F.M.V. Muscle injury activates resident fibro/adipogenic progenitors that facilitate myogenesis. Nat. Cell Biol. 2010, 12, 153–163. [Google Scholar] [CrossRef] [PubMed]
- Lees-Shepard, J.B.; Yamamoto, M.; Biswas, A.A.; Stoessel, S.J.; Nicholas, S.E.; Congswell, C.A.; Devarakonda, P.M.; Schneider, M.J.; Cummins, S.M.; Legendre, N.P.; et al. Activin-dependent signaling in fibro/adipogenic progenitors causes fibrodysplasia ossificans progressive. Nat. Commun. 2018, 9, 471. [Google Scholar] [CrossRef] [PubMed]
- Eisner, C.; Cummings, M.; Johnston, G.; Tung, L.W.; Groppa, E.; Chang, C.; Rossi, F.M.V. Murine Tissue-Resident PDGFRα+ Fibro-Adipogenic Progenitors Spontaneously Acquire Osteogenic Phenotype in an Altered Inflammatory Environment. J. Bone Miner. Metab. 2020, 35, 1525–1534. [Google Scholar] [CrossRef] [PubMed]
- Neto, J.P.; Rocha, L.C.; Barbosa, G.K.; Jacob, C.S.; Neto, W.K.; Watanabe, I.; Ciena, A.P. Myotendinous junction adaptations to ladder-based resistance training: Identification of a new telocyte niche. Sci. Rep. 2020, 10, 14124. [Google Scholar] [CrossRef]
- Cretoiu, S.M.; Popescu, L.M. Telocytes revised. Biomol. Concepts 2014, 5, 353–369. [Google Scholar] [CrossRef]
- Mathew, S.J.; Hansen, J.M.; Merrell, A.J.; Murphy, M.M.; Lawson, J.A.; Hutcheson, D.A.; Hansen, M.S.; Angus-Hill, M.; Kardon, G. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 2011, 138, 371–384. [Google Scholar] [CrossRef]
- Murphy, M.M.; Lawson, J.A.; Mathew, S.J.; Hutcheson, D.A.; Kardon, G. Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 2011, 138, 3625–3637. [Google Scholar] [CrossRef]
- Uezumi, A.; Fukada, S.; Yamamoto, N.; Ikemoto-Uezumi, M.; Nakatani, M.; Moita, M.; Yamaguhi, A.; Yamada, H.; Nishino, I.; Hamada, Y.; et al. Identification and characterization of PDGFRα+ mesenchymal progenitors in human skeletal muscle. Cell Death Dis. 2014, 5, e1186. [Google Scholar] [CrossRef]
- Ogawa, Y.; Yamamoto, M.; Sato, M.; Odaka, K.; Kasahara, M.; Hinata, N.; Sakiyama, K.; Abe, S. Localization of T-cell factor 4 positive fibroblasts and CD206-positive macrophages during skeletal muscle regeneration in mice. Ann. Anat. 2021, 235, 151694. [Google Scholar] [CrossRef]
- Kim, M.; Franke, V.; Brandt, B.; Lowenstein, E.D.; Schöwel, V.; Spuler, S.; Akalin, A.; Birchmeier, C. Single-nucleus transcriptomics reveals functional compartmentalization in syncytial skeletal muscle cells. Nat. Commun. 2020, 11, 6375. [Google Scholar] [CrossRef] [PubMed]
- Petrany, M.J.; Swoboda, C.O.; Sun, C.; Chetal, K.; Chen, X.; Weirauch, M.T.; Salomonis, N.; Millay, D.P. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 2020, 11, 6374. [Google Scholar] [CrossRef] [PubMed]
- Yaseen, W.; Kraft-Sheleg, O.; Zaffryar-Eilot, S.; Melamed, S.; Sun, C.; Millay, D.P.; Hasson, P. Fibroblast fusion to the muscle fiber regulates myotendinous junction formation. Nat. Commun. 2021, 12, 3852. [Google Scholar] [CrossRef] [PubMed]
- Lima, J.E.; Blavet, C.; Bonnin, M.A.; Hirsinger, E.; Comai, G.; Yvernogeau, L.; Delfini, M.C.; Bellenger, L.; Mella, S.; Nassari, S.; et al. Unexpected contribution of fibroblasts to muscle lineage as a mechanism for limb muscle patterning. Nat. Commun. 2021, 12, 3851. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, A.; Kishi, A.; Yamamoto, M.; Yamane, S.; Umezawa, T.; Ide, Y.; Abe, S. Expression of intermediate filaments in the development of genioglossus muscle. J. Hard Tissue Biol. 2012, 21, 421–425. [Google Scholar] [CrossRef][Green Version]
- Kishi, A.; Yamamoto, M.; Kikuchi, A.; Iwanuma, O.; Watanabe, Y.; Ide, Y.; Abe, S. Gene and protein expressions of vimentin and desmin during embryonic development of the mylohyoid muscle. Anat. Sci. Int. 2012, 87, 126–131. [Google Scholar] [CrossRef]
- Noden, D.M.; Francis-West, P. The differentiation and morphogenesis of craniofacial muscles. Dev. Dyn. 2006, 235, 1194–1218. [Google Scholar] [CrossRef]
- Parada, C.; Han, D.; Chai, Y. Molecular and cellular regulatory mechanisms of tongue myogenesis. J. Dent. Res. 2012, 91, 528–535. [Google Scholar] [CrossRef]
- Heude, E.; Bouhali, K.; Kurihara, Y.; Kurihara, H.; Couly, G.; Janvier, P.; Levi, G. Jaw muscularization requires Dlx expression by cranial neural crest cells. Proc. Natl. Acad. Sci. USA 2010, 107, 11441–11446. [Google Scholar] [CrossRef]
- Anakwe, K.; Robson, L.; Hadley, J.; Buxton, P.; Church, V.; Allen, S.; Hartmann, C.; Harfe, B.; Nohno, T.; Brown, A.M.; et al. Wnt signalling regulates myogenic differentiation in the developing avian wing. Development 2003, 130, 3503–3514. [Google Scholar] [CrossRef]
- Liu, H.X.; Grosse, A.S.; Iwatsuki, K.; Mishina, Y.; Gumucio, D.L.; Mistretta, C.M. Separate and distinctive roles for Wnt5a in tongue, lingual tissue and taste papilla development. Dev. Biol. 2012, 361, 39–56. [Google Scholar] [CrossRef] [PubMed]
- Cong, W.; Liu, B.; Liu, S.; Sun, M.; Liu, H.; Yang, Y.; Wang, R.; Xiao, J. Implications of the Wnt5a/CaMKII pathway in retinoic acid-induced myogenic tongue abnormalities of developing mice. Sci. Rep. 2014, 4, 6082. [Google Scholar] [CrossRef] [PubMed]
- Kardon, G. Muscle and tendon morphogenesis in the avian hind limb. Development 1998, 125, 4019–4032. [Google Scholar] [CrossRef]
- Ros, M.A.; Rivero, F.B.; Hinchliffe, J.R.; Hurle, J.M. Immunohistological and ultrastructural study of the developing tendons of the avian foot. Anat. Embryol. 1995, 192, 483–496. [Google Scholar] [CrossRef]
- Tucker, R.P.; Spring, J.; Baumgartner, S.; Martin, D.; Hagios, C.; Poss, P.M.; Chiquet-Ehrismann, R. Novel tenascin variants with a distinctive pattern of expression in the avian embryo. Development 1994, 120, 637–647. [Google Scholar] [CrossRef] [PubMed]
- Docheva, D.; Hunziker, E.B.; Fässler, R.; Brandau, O. Tenomodulin is necessary for tenocyte proliferation and tendon maturation. Mol. Cell Biol. 2005, 25, 699–705. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, R.; Chyung, J.H.; Murtaugh, L.C.; Brent, A.E.; Rosen, V.; Olson, E.N.; Lassar, A.; Tabin, C.J. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 2001, 128, 3855–3866. [Google Scholar] [CrossRef]
- Brent, A.E.; Braun, T.; Tabin, C.J. Genetic analysis of interactions between the somitic muscle, cartilage and tendon cell lineages during mouse development. Development 2005, 132, 515–528. [Google Scholar] [CrossRef]
- Shukunami, C.; Takimoto, A.; Oro, M.; Hiraki, Y. Scleraxis positively regulates the expression of tenomodulin, a differentiation marker of tenocytes. Dev. Biol. 2006, 298, 234–247. [Google Scholar] [CrossRef]
- Ito, Y.; Toriuchi, N.; Yoshitaka, T.; Ueno-Kudoh, H.; Sato, T.; Yokoyama, S.; Nishida, K.; Akimoto, T.; Takahashi, M.; Miyaki, S.; et al. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc. Natl. Acad. Sci. USA 2010, 107, 10538–10542. [Google Scholar] [CrossRef]
- Yoshimoto, Y.; Takimoto, A.; Watanabe, H.; Hiraki, Y.; Kondoh, G.; Shukunami, C. Scleraxis is required for maturation of tissue domains for proper integration of the musculoskeletal system. Sci. Rep. 2017, 7, 45010. [Google Scholar] [CrossRef] [PubMed]
- Nagakura, R.; Yamamoto, M.; Jeong, J.; Hinata, N.; Katori, Y.; Chang, W.J.; Abe, S. Switching of Sox9 expression during musculoskeletal system development. Sci. Rep. 2020, 21, 8425. [Google Scholar] [CrossRef] [PubMed]
- Huang, A.H.; Lu, H.H.; Schweitzer, R. Molecular regulation of tendon cell fate during development. J. Orthop. Res. 2015, 33, 800–812. [Google Scholar] [CrossRef] [PubMed]
- Schweitzer, R.; Zelzer, E.; Volk, T. Connecting muscles to tendons: Tendons and musculoskeletal development in flies and vertebrates. Development 2010, 137, 2807–2817. [Google Scholar] [CrossRef]
- Lejard, V.; Blais, F.; Guerquin, M.J.; Bonnet, A.; Bonnin, M.A.; Havis, E.; Malbouyres, M.; Bidaud, C.B.; Maro, G.; Gilardi-Hebenstreit, P.; et al. EGR1 and EGR2 involvement in vertebrate tendon differentiation. J. Biol. Chem. 2011, 286, 5855–5867. [Google Scholar] [CrossRef]
- Frommer, G.; Vorbrüggen, G.; Pasca, G.; Jäckle, H.; Volk, T. Epidermal egr-like zinc finger protein of Drosophila participates in myotube guidance. EMBO J. 1996, 15, 1642–1649. [Google Scholar] [CrossRef]
- Goodwin, G.H.; Sander, C.; Johns, E.W. A new group of chromatin-associated proteins with a high content of acidic and basic amino acids. Eur. J. Biochem. 1973, 38, 14–19. [Google Scholar] [CrossRef]
- Ulloa, L.; Messmer, D. High-mobility group box 1 (HMGB1) protein: Friend and foe. Cytokine Growth Factor Rev. 2006, 17, 189–201. [Google Scholar] [CrossRef]
- Takizawa, S.; Sakiyama, K.; Bando, Y.; Inoue, K.; Sakashita, H.; Ogasawara, Y.; Amano, O.; Sakashita, H. Influence of high mobility group box 1 (HMGB1) derived from SCC7 cells on mouse normal tongue muscle fibers. J. Oral Maxillofac. Surg. Med. Pathol. 2018, 30, 466–474. [Google Scholar] [CrossRef]
- Tang, D.; Kang, R.; Zeh, H.J.; Lotze, M.T. High-mobility group box 1 and cancer. Biochim. Biophys. Acta 2009, 1799, 131–140. [Google Scholar] [CrossRef]
- Nestl, A.; Stein, D.V.; Zatloukal, K.; Thies, W.G.; Herrlich, P.; Hofmann, M.; Sleeman, J.P. Gene expression patterns associated with the metastatic phenotype in rodent and human tumors. Cancer Res. 2001, 61, 1569–1577. [Google Scholar] [PubMed]
- Schierbeck, H.; Pullerits, R.; Pruunsild, C.; Fischer, M.; Holzinger, D.; Leastadius, Å.; Sundberg, E.; Harris, H.E. HMGB1 levels are increased in patients with juvenile idiopathic arthritis, correlate with early onset of disease, and are independent of disease duration. J. Rheumatol. 2013, 40, 1604–1613. [Google Scholar] [CrossRef] [PubMed]
- Sakiyama, K.; Takizawa, S.; Bando, Y.; Inoue, K.; Sasaki, A.; Kurokawa, K.; Shimoo, Y.; Suzuki, M.; Abe, S.; Amano, O. Characteristics and Effects of Muscle Fibers surrounding Lingual Carcinoma. J. Hard Tissue Biol. 2013, 22, 215–220. [Google Scholar] [CrossRef]
- Wake, H.; Mori, S.; Liu, K.; Takahashi, H.K.; Nishibori, M. Histidine-rich glycoprotein inhibited high mobility group box 1 in complex with heparin-induced angiogenesis in Matrigel plug assay. Eur. J. Pharmacol. 2009, 623, 89–95. [Google Scholar] [CrossRef]
- Aikawa, E.; Fujita, R.; Kikuchi, Y.; Kaneda, Y.; Tamai, K. Systemic high-mobility group box 1 administration suppresses skin inflammation by inducing an accumulation of PDGFRα(+) mesenchymal cells from bone marrow. Sci. Rep. 2015, 5, 11008. [Google Scholar] [CrossRef]
- Zhou, Z.; Han, J.Y.; Xia, C.X.; Xie, J.X.; Feng, X.; Wang, C.Y.; Mei, L.; Xiong, W.C. HMGB1 regulates RANKL-induced osteoclastogenesis in a manner dependent on RAGE. J. Bone Miner. Res. 2008, 23, 1084–1096. [Google Scholar] [CrossRef]
- Zhou, Z.; Xiong, W.C. RAGE and its ligands in bone metabolism. Front. Biosci. 2011, 3, 768–776. [Google Scholar]
- Jing, M.; Li, F.; Xu, J.; Liu, Y.D.; Hu, T.; Chen, J.T. rhHMGB1 drives osteoblast migration in a TLR2/TLR4-and NF-κB-dependent manner. Biosci. Rep. 2016, 36, 300. [Google Scholar]
- Murchison, N.D.; Price, B.A.; Conner, D.A.; Keene, D.R.; Olson, E.N.; Tabin, C.J.; Schweitzer, R. Regulation of tendon differentiation by scleraxis distinguishes force-transmitting tendons from muscle-anchoring tendons. Development 2007, 134, 2697–2708. [Google Scholar] [CrossRef]
- Liu, W.; Watson, S.S.; Lan, Y.; Keene, D.R.; Ovitt, C.E.; Liu, H.; Schweitzer, R.; Jiang, R. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol. Cell 2010, 30, 4797–4807. [Google Scholar] [CrossRef]
- Howell, K.; Chien, C.; Bell, R.; Laudier, D.; Tufa, S.F.; Keene, D.R.; Andarawis-Puri, N.; Huang, A.H. Novel model of tendon regeneration reveals distinct cell mechanisms underlying regenerative and fibrotic tendon healing. Sci. Rep. 2017, 7, 45238. [Google Scholar] [CrossRef] [PubMed]
- Sakabe, T.; Sakai, K.; Maeda, T.; Sunaga, A.; Furuta, N.; Schweitzer, R.; Sasaki, T.; Sakai, T. Transcription factor scleraxis vitally contributes to progenitor lineage direction in wound healing of adult tendon in mice. J. Biol. Chem. 2018, 293, 5766–5780. [Google Scholar] [CrossRef] [PubMed]
- Dyment, N.A.; Hagiwara, Y.; Matthews, B.G.; Li, Y.; Kalajzic, I.; Rowe, D.W. Lineage tracing of resident tendon progenitor cells during growth and natural healing. PLoS ONE 2014, 9, e96113. [Google Scholar] [CrossRef] [PubMed]
- Dyment, N.A.; Liu, C.F.; Kazemi, N.; Aschbacher-Smith, L.E.; Kenter, K.; Breidenbach, A.P.; Sheam, J.T.; Whlie, C.; Rowe, D.; Butler, D.L. The Paratenon contributes to scleraxis-expressing cells during patellar tendon healing. PLoS ONE 2013, 8, e59944. [Google Scholar] [CrossRef]
- Gumucio, J.P.; Phan, A.C.; Ruehlmann, D.G.; Noah, A.C.; Mendias, C.L. Synergist ablation induces rapid tendon growth through the synthesis of a neotendon matrix. J. Appl. Physiol. 2014, 117, 1287–1291. [Google Scholar] [CrossRef]
- Mendias, C.L.; Gumucio, J.P.; Bakhurin, K.I.; Lynch, E.B.; Brooks, S.V. Physiological loading of tendons induces scleraxis expression in epitenon fibroblasts. J. Orthop. Res. 2012, 30, 606–612. [Google Scholar] [CrossRef]
- Mienaltowski, M.J.; Adams, S.M.; Brik, D.E. Regional differences in stem cell/progenitor cell populations from the mouse achilles tendon. Tissue Eng. Part A 2013, 19, 199–210. [Google Scholar] [CrossRef]
- Kaji, D.A.; Howell, K.L.; Balic, Z.; Hubmacher, D.; Huang, A.H. Tgfb signaling is required for tenocyte recruitment and functional neonatal tendon regeneration. eLife 2020, 9, e51779. [Google Scholar] [CrossRef]
- Agarwal, S.; Loder, S.J.; Cholok, D.; Peterson, J.; Li, J.; Breuler, C.; Brownley, R.C.; Sung, H.H.; Chung, M.T.; Kamiya, N.; et al. Scleraxis-lineage cells contribute to ectopic bone formation in muscle and tendon. Stem. Cells 2017, 35, 705–710. [Google Scholar] [CrossRef]
- Dey, D.; Bagarova, J.; Hatsell, S.J.; Armstrong, K.A.; Huang, L.; Ermann, J.; Vonner, A.J.; Shen, Y.; Mohedas, A.H.; Lee, A.; et al. Two tissue-resident progenitor lineages drive distinct phenotypes of heterotopic ossification. Sci. Transl. Med. 2016, 8, 366ra163. [Google Scholar] [CrossRef]
- Blitz, E.; Sharir, A.; Akiyama, H.; Zelzer, E. Tendon-bone attachment unit is formed modularly by a distinct pool of Scx- and Sox9-positive progenitors. Development 2013, 140, 2680–2690. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, Y.; Takimoto, A.; Akiyama, H.; Kist, R.; Scherer, G.; Nakamura, T.; Hiraki, Y.; Schukunami, C. Scx+/Sox9+ progenitors contribute to the establishment of the junction between cartilage and tendon/ligament. Development 2013, 140, 2280–2288. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, H.; Ito, Y.; Shinohara, M.; Yamashita, S.; Ichinose, S.; Kishida, A.; Oyaizu, T.; Kayama, T.; Nakamichi, R.; Kada, N.; et al. Gene targeting of the transcription factor Mohawk in rats causes heterotopic ossification of Achilles tendon via failed tenogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, 7840–7845. [Google Scholar] [CrossRef]
- Maeda, T.; Sakabe, T.; Sunaga, A.; Sakai, K.; Rivera, A.L.; Keene, D.R.; Sasaki, T.; Stavnezer, E.; Iannotti, J.; Schweitzer, R.; et al. Conversion of mechanical force into TGF-medi ated biochemical signals. Curr. Biol. 2011, 21, 933–941. [Google Scholar] [CrossRef]
- Davies, M.R.; Liu, X.; Lee, L.; Laron, D.; Ning, A.Y.; Kim, H.T.; Feeley, B.T. TGF-β small molecule inhibitor SB431542 reduces rotator cuff muscle fibrosis and fatty infiltration by promoting fibro/Adipogenic progenitor apoptosis. PLoS ONE 2016, 11, e0155486. [Google Scholar] [CrossRef]
- Katzel, E.B.; Wolenski, M.; Loiselle, A.E.; Basile, P.; Flick, L.M.; Langstein, H.N.; Hilton, M.J.; Awad, H.A.; Hammert, W.C.; O’Keefe, R.J. Impact of Smad3 loss of function on scarring and adhesion formation during tendon healing. J. Orthop. Res. 2011, 29, 684–693. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Jin, S.; Luo, D.; He, D.; Shi, C.; Zhu, L.; Zixin, L.; Zhang, T.; Zhou, Y.; Wang, C.Y.; et al. Functional regeneration and repair of tendons using biomimetic scaffolds loaded with recombinant periostin. Nat. Commun. 2021, 12, 1293. [Google Scholar] [CrossRef] [PubMed]
- Curzi, D.; Salucci, S.; Marini, M.; Esposito, F.; Aqnello, L.; Veicsteinas, A.; Burattini, S.; Falcieri, E. How physical exercise changes rat myotendinous junctions: An ultrastructural study. Eur. J. Histochem. 2012, 56, e19. [Google Scholar] [CrossRef]
- Yamamoto, M.; Takada, H.; Ishizuka, S.; Kitamura, K.; Jeong, J.; Sato, M.; Hinata, N.; Abe, S. Morphological association between the muscles and bones in the craniofacial region. PLoS ONE 2020, 15, e0227301. [Google Scholar] [CrossRef]
- Zelzer, E.; Blitz1, E.; Killian, M.; Thomopoulos, S. Tendon-to-bone attachment: From development to maturity. Birth Defects Res. C Embryo Today 2014, 102, 101–112. [Google Scholar] [CrossRef]
- Schwartz, G.; Pasteris, D.; Genin, M.; Daulton, T.; Thomopoulos, S. Mineral distributions at the developing tendon enthesis. PLoS ONE 2012, 7, e48630. [Google Scholar] [CrossRef] [PubMed]
- Orfei, C.; Viganò, M.; Pearson, J.; Colombini, A.; Luca, P.; Ragni, E.; Santos-Ruiz, L.; Girolamo, L. In Vitro Induction of Tendon-Specific Markers in Tendon Cells, Adipose- and Bone Marrow-Derived Stem Cells is Dependent on TGFβ3, BMP-12 and Ascorbic Acid Stimulation. Int. J. Mol. Sci. 2019, 20, 149. [Google Scholar] [CrossRef] [PubMed]
- Furumatsu, T.; Shukunami, C.; Amemiya-Kudo, M.; Shimano, H.; Ozaki, T. Scleraxis and E47 cooperatively regulate the Sox9-dependent transcription. Int. J. Biochem. Cell Biol. 2010, 42, 148–156. [Google Scholar] [CrossRef] [PubMed]
- Blitz, E.; Viukov, S.; Sharir, A.; Shwartz, Y.; Galloway, L.; Pryce, A.; Johnson, L.; Tabin, J.; Schweitzer, R.; Zelzer, E. Bone ridge patterning during musculoskeletal assembly is mediated through SCX regulation of Bmp4 at the tendon-skeleton junction. Dev. Cell 2009, 17, 861–873. [Google Scholar] [CrossRef]
- Abe, S.; Suzuki, M.; Cho, M.K.; Murakami, G.; Cho, B.H.; Ide, Y. CD34-positive developing vessels and other structures in human fetuses: An immunohistochemical study. Surg. Radiol. Anat. 2011, 33, 919–927. [Google Scholar] [CrossRef]
- Hiroki, E.; Abe, S.; Iwanuma, O.; Sakiyama, K.; Yanagisawa, N.; Shiozaki, K.; Ide, Y. A comparative study of myostatin, follistatin and decorin expression in muscle of different origin. Anat. Sci. Int. 2011, 86, 151–159. [Google Scholar] [CrossRef]
- Honda, H.; Abe, S.; Ishida, R.; Watanabe, Y.; Iwanuma, O.; Sakiyama, K.; Ide, Y. Expression of HGF and IGF-1 during regeneration of masseter muscle in mdx mice. J. Muscle Res. Cell Mtil. 2010, 31, 71–77. [Google Scholar] [CrossRef]
- Honda, A.; Abe, S.; Hiroki, E.; Honda, H.; Iwanuma, O.; Yanagisawa, N.; Ide, Y. Activation of caspase 3, 9, 12, and Bax in masseter muscle of mdx mice during necrosis. J. Muscle Res. Cell Mtil. 2007, 28, 243–247. [Google Scholar] [CrossRef][Green Version]
- Abe, S.; Maejima, M.; Watanabe, H.; Shibahara, T.; Agematsu, H.; Doi, T.; Sakiyama, K.; Usami, A.; Gojyo, K.; Hashimoto, M.; et al. Muscle—fiber characteristics in adult mouse-tongue muscles. Anat. Sci. Int. 2002, 77, 145–148. [Google Scholar] [CrossRef]
- Suzuki, K.; Abe, S.; Kim, H.J.; Usami, A.; Iwanuma, O.; Okubo, H.; Ide, Y. Changes in the muscle fibre properties of the mouse temporal muscle after weaning. Anat. Histol. Embryol. 2007, 36, 103–106. [Google Scholar] [CrossRef]
- Okubo, K.; Abe, S.; Usami, A.; Agematsu, H.; Nakamura, H.; Hashimoto, M.; Ide, Y. Changes in muscle-fiber properties of the murine digastric muscle before and after weaning. Zool Sci. 2006, 23, 1079–1084. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abe, S.; Hirose, D.; Kado, S.; Iwanuma, O.; Saka, H.; Yanagisawa, N.; Ide, Y. Increased expression of decorin during the regeneration stage of mdx mouse. Anat. Sci. Int. 2009, 84, 305–311. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.H.; Abe, S.; Kim, H.J.; Usami, A.; Honda, A.; Sakiyama, K.; Ide, Y. Characteristics of muscle fibers reconstituted in the regeneration process of masseter muscle in an mdx mouse model of muscular dystrophy. J. Muscle Res. Cell Mtil. 2006, 27, 235–240. [Google Scholar] [CrossRef]
- Doi, T.; Abe, S.; Ide, Y. Masticatory function and properties of masseter muscle fibers in microphthalmic (mi/mi) mice during postnatal development. Ann. Anat. 2003, 185, 435–440. [Google Scholar] [CrossRef]
- Takagi, T.; Yamamoto, M.; Sugano, A.; Kanehira, C.; Kitamura, K.; Katayama, M.; Sakai, K.; Sato, M.; Abe, S. Alteration of oral and perioral soft tissue in mice following incisor tooth extraction. Int. J. Mol. Sci. 2022, in press. [CrossRef]
- Abe, S.; Yamamoto, M. Factors involved in morphogenesis in the muscle-tendon-bone complex. Int. J. Mol. Sci. 2021, 22, 6365. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yamamoto, M.; Sakiyama, K.; Kitamura, K.; Yamamoto, Y.; Takagi, T.; Sekiya, S.; Watanabe, G.; Taniguchi, S.; Ogawa, Y.; Ishizuka, S.; et al. Development and Regeneration of Muscle, Tendon, and Myotendinous Junctions in Striated Skeletal Muscle. Int. J. Mol. Sci. 2022, 23, 3006. https://doi.org/10.3390/ijms23063006
Yamamoto M, Sakiyama K, Kitamura K, Yamamoto Y, Takagi T, Sekiya S, Watanabe G, Taniguchi S, Ogawa Y, Ishizuka S, et al. Development and Regeneration of Muscle, Tendon, and Myotendinous Junctions in Striated Skeletal Muscle. International Journal of Molecular Sciences. 2022; 23(6):3006. https://doi.org/10.3390/ijms23063006
Chicago/Turabian StyleYamamoto, Masahito, Koji Sakiyama, Kei Kitamura, Yutaro Yamamoto, Takahiro Takagi, Sayo Sekiya, Genji Watanabe, Shuichiro Taniguchi, Yudai Ogawa, Satoshi Ishizuka, and et al. 2022. "Development and Regeneration of Muscle, Tendon, and Myotendinous Junctions in Striated Skeletal Muscle" International Journal of Molecular Sciences 23, no. 6: 3006. https://doi.org/10.3390/ijms23063006
APA StyleYamamoto, M., Sakiyama, K., Kitamura, K., Yamamoto, Y., Takagi, T., Sekiya, S., Watanabe, G., Taniguchi, S., Ogawa, Y., Ishizuka, S., Sugiyama, Y., Takayama, T., Hayashi, K., Chang, W.-J., & Abe, S. (2022). Development and Regeneration of Muscle, Tendon, and Myotendinous Junctions in Striated Skeletal Muscle. International Journal of Molecular Sciences, 23(6), 3006. https://doi.org/10.3390/ijms23063006






