Abstract
Chronic viral hepatitis is a main cause of liver disease and hepatocellular carcinoma. There are striking similarities in the pathological impact of hepatitis B, C, and D, although these diseases are caused by very different viruses. Paired with the conventional study of protein–host interactions, the rapid technological development of -omics and bioinformatics has allowed highlighting the important role of signaling networks in viral pathogenesis. In this review, we provide an integrated look on the three major viruses associated with chronic viral hepatitis in patients, summarizing similarities and differences in virus-induced cellular signaling relevant to the viral life cycles and liver disease progression.
1. Introduction
Viral hepatitis predominantly affects and damages the liver by commonly causing the progression from chronic inflammation to fibrosis, cirrhosis, and ultimately cancer. It is estimated that approximatively 350 million people worldwide are chronically infected with hepatitis viruses [1]. During a chronic infection of the liver, hepatic viruses persistently tweak and attenuate the host antiviral defenses and modulate cellular pathways that impact liver homeostasis and disease progression. Viral hepatitis is a major risk factor for liver fibrosis, cirrhosis, and hepatocellular carcinoma (HCC), which is the second leading and fastest rising cause of cancer death worldwide [2,3]. Although caused by very different viruses, virus-induced liver disease displays similar features, suggesting common molecular drivers. Moreover, chronic viral hepatitis may serve also as a model to understand the mechanism of non-viral etiologies like non-alcoholic fatty liver disease (NAFLD) and non-alcoholic steato-hepatitis (NASH).
Although replication strategies of hepatotropic viruses are very diverse, we can highlight common molecular mechanisms that occur during chronic viral hepatitis: (1) induction of intrahepatic oxidative stress damage by viral proteins, (2) dysregulation of cellular metabolic pathways, (3) persistence of liver inflammation, (4) activation of pro-fibrotic, pro-oncogenic processes that can lead to the accumulation of genetic alterations and genomic instability. Therefore, the common denominator of these events comprises a virus-induced dysregulation of signaling events that holds the potential for the identification of novel host-targeting and chemo-preventive strategies targeting the viral life cycle and/or liver disease progression. In this review, we summarize similarities and differences in virus-induced cellular signaling associated with the three major viruses that cause chronic viral hepatitis in patients.
2. Hepatitis B, C, D Viruses
The three hepatotropic viruses causing chronic liver infection are Hepatitis B virus (HBV), Hepatitis C virus (HCV), and Hepatitis Delta virus (HDV). HBV is a DNA virus of the Hepadnaviridae family whose partially double-stranded genome is translocated into the host nucleus. Here, a covalently closed circular DNA (cccDNA) is formed and serves as a template for transcription. cccDNA is highly persistent and epigenetically regulated as a host chromosome, a critical feature that makes it difficult to achieve a complete cure for HBV infection [4,5]. HBV is the only hepatotropic virus that causes integration of the viral DNA into the host genome. It thus contributes directly to an elevated liver cancer risk even in non-cirrhotic patients by cis-mediated insertional mutagenesis, chromosomal instability, and expression of aberrant viral proteins [6]. HDV is a single-stranded RNA virus and the only virus of the genus Deltavirus. It is a satellite virus of HBV, which requires the HBV surface antigen (HBsAg) for its lifecycle [7,8]. Importantly, HBV/HDV coinfection causes the most severe form of chronic viral hepatitis, with accelerated liver disease progression to cirrhosis and HCC and increased liver-related and overall mortality [9]. However, little is known about the HBV/HDV–host interactions driving these complications. HCV, a member of the Flaviviridae family, is a positive-sensed single-stranded RNA virus that depends on and interacts with hepatocyte lipid metabolism during its lifecycle [10]. HCV triggers phenotypic changes closely resembling metabolic liver disease, including hepatic steatosis and insulin resistance [11], and profoundly influences the proteogenomic landscape of the host cell [12].
Challenges in the treatment of these viruses differ very much. While an efficient HBV vaccine is available protecting from HBV and HDV, the development of an HCV vaccine is hampered by its lipoviral composition and its highly variable quasispecies that contribute to its shielding and escape from neutralizing antibodies. However, over the last decade, novel and highly efficient antivirals have been developed to cure HCV infection [13]. In contrast, chronic HBV infection can only be controlled by long-term antiviral strategies due to the persistence of cccDNA pools in patients’ liver [14,15]. For HDV, interferon-based therapies had only limited success; however, novel antivirals such as entry inhibitors have shown encouraging results in clinical practice to control HDV infection and to improve liver function [16]. Nevertheless, even if a viral infection is controlled or cured, the risk of developing HCC may not be fully reversed, depending on the duration of chronic infection, liver disease stage, and type of virus [17,18]. Moreover, evidence points towards an epigenetic imprinting by hepatic viruses (HCV, HBV) and underlying liver fibrosis in the host genome which maintains a persistent transcriptomic environment in cured livers that acts in a pro-oncogenic manner [17,19,20].
3. Virus-Induced Oxidative Stress Signaling
In healthy cells, reactive oxygen species (ROS) are predominantly produced through mitochondria oxidative phosphorylation, protein folding in the endoplasmic reticulum (ER), and the catabolism of lipids and amino acids [21,22,23]. ROS are considered to be harmful for the cell, exerting damage-promoting, detrimental effects. However, ROS are also an essential signaling trigger regulating apoptosis and immune response against pathogens [24]. ROS are neutralized by the enzymatic and non-enzymatic cellular antioxidant system. The enzymatic antioxidant system includes various types of ROS-scavenging phase II enzymes such as glutathione peroxidase (GPx), superoxide dismutase (SOD), and catalase (CAT) catalyzing free radicals’ neutralization. In contrast, the non-enzymatic antioxidants system is composed of low-molecular-weight compounds such as glutathione and vitamin C, scavenging ROS with a slow kinetics. Both systems are regulated by the expression of genes comprising antioxidant response elements (ARE), which are controlled by the transcription factor Nrf2 [25,26]. A persistent imbalance of ROS is an important driver of chronic liver disease, and the associated redox imbalance has been suggested to be highly relevant to NAFLD pathogenesis [27]. Moreover, oxidative stress has been associated with oncogenic transformation in patients with chronic viral hepatitis [28,29].
The mechanism of virus-induced oxidative stress by HBV, HCV, HDV can be summarized in four main categories: (1) alteration of mitochondrial function mediated by Ca2+ uptake; (2) triggering of ER stress and unfolded protein response (UPR); (3) virus-induced expression of ROS-producing enzymes; (4) dysregulation of antioxidative pathways (Figure 1). In the case of HCV, viral core proteins [30], E1/E2 [31], and NS4B [32,33] induce oxidative stress via calcium efflux through the induction of ER stress and UPR, which is a component of the ER adaptative system. In addition, the HCV core at the mitochondrial outer membrane [34] interacts with heat shock protein Hsp60 [35], triggering the release of Ca2+ from the ER and its accumulation in the mitochondria. Moreover, the HCV core stimulates the expression oxidoreductin 1α (ERO1α) in the ER. This promotes the formation of mitochondria-associated membranes and induces Ca2+ translocation from the ER to the mitochondria. Mitochondrial Ca2+ accumulation alters the respiratory chain and promotes ROS production [36,37]. For HBV, HBx protein expression reduces the activity of several respiratory chain complexes, causing the loss of mitochondrial membrane potential and therefore enhancing the production of ROS [38]. Moreover, HBx dysregulates mitochondrial functioning by interacting with two partners: voltage-dependent anion channel 3 (VDAC3), involved in calcium transport across the mitochondrial outer membrane [39], and cytochrome c oxidase subunit III (COX3) [40,41]. Other HBV proteins may also be involved in the induction of oxidative stress. HBsAg is generally secreted during the HBV lifecycle; however, secretion-deficient mutants can appear during infection and accumulate in the ER. This also occurs with the HBV core antigen (HBcAg). Both proteins induce ER stress and UPR signaling, leading to calcium release from the ER and subsequent ROS production [42,43]. Viral components also enhance oxidative stress by inducing the expression of ROS-producing enzymes. The HCV core proteins and NS5A enhance the expression of cytochrome P450 2E1 (CYP2E1) and NADPH oxidase 1 and 4 (NOX1 and 4), leading to elevated levels of ROS, including superoxide and hydrogen peroxide [37,44]. Similarly, the large HDV surface antigen (L-HDAg) induces oxidative stress by promoting NOX4 expression [45].
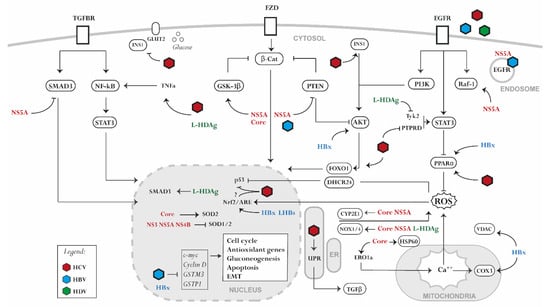
Figure 1.
Signaling pathways perturbed by hepatotropic viral proteins. HCV, HBV, and HDV alter liver homeostasis by disrupting several signaling processes associated with (1) the generation of oxidative stress and the dysregulation of the antioxidant system, (2) the alteration of a pro-inflammatory signaling, (3) the hijacking of glucose and lipid metabolism, (4) the dysregulation of host genome expression. NS3, NS4B, NS5A, (non-structural protein 3/4B/5A); HBx, (hepatitis b X antigen); LHBs, (large HBV surface antigen); L-HDAg, (large HDV antigen).
A virus-induced, but not always consistent, dysregulation of the antioxidant system is observed during HCV and HBV infection. For HCV, the expression of nonstructural proteins downregulates SOD1 and SOD2 and induce catalase, whereas HCV core alone enhances the expression of SOD2 [46]. The expression of full-length HCV or the nonstructural proteins and core leads to impaired Nrf2/ARE activity [47], whereas a full HCV infection activates the Nrf2/ARE axis [48,49,50]. Overexpression studies in Huh7 cells point towards Nrf2-activating phosphorylation during HCV infection by protein kinase C in response to ROS or by casein kinase 2 and phosphoinositide 3-kinase (PI3K) in a ROS-independent manner [48]. Moreover, in an HCV infection model, Nrf2/ARE activation was promoted by the inhibitory phosphorylation of glycogen synthase kinase 3β (GSK3β) [49]. While in non-transformed hepatocytes several Nrf2-dependent genes are induced by HCV [51], downregulation of a wide spectrum of antioxidant defense proteins can be observed in hepatoma cell line-based models [52,53]. Several theories have been proposed to explain this discrepancy, one of which is the bi-phasic nature of oxidative stress, which at low and moderate ROS levels activates antioxidants, whereas at high ROS level induces damage and inhibits the expression of antioxidant genes. The used study model and the readout have thus a significant impact on the results. HCV-induced gene expression is often not translated into a protein response to blunt the antiviral response of the host cells [12]. For HBV, the Nrf2/ARE pathway is activated by the virus in both infected cells and liver tissues of chronic HBV carriers in a genotype-dependent manner via HBx and the large surface antigen (LHBs) [54]. HBx sequesters the Nrf2 partner protein, Keap1, forming a HBx–p62–Keap1 triple complex in a ROS-independent manner [55]. However, the activation of the antioxidant system by HBV is challenged by several studies in HBV-infected cells and in HBV patients [56,57,58]. Indeed, some Nrf2-dependent genes such as GSTM3 [59] and GSTP1 [60] are epigenetically silenced by HBx expression or HBV infection. HBx also alters type II enzyme expression by interfering with the expression of other regulatory elements/factors of the Nrf2/ARE signaling pathway [61]. Furthermore, HBV suppresses the expression of proteins indirectly implicated in the antioxidant system such as selenoprotein P (SeP) and selenium-binding protein 2 (Selenbp2) [62,63]. Beside the observed effects of HBV on ROS, also HDV promotes oxidative stress in the ER through the interaction between L-HDAg and NOX4. The activation of the NOX4 pathway induces the release of ROS from the ER, activating the signal transducer and activator of transcription-3 (STAT3) and nuclear factor kappa B (NF-κB) signaling [45]. Moreover, the small hepatitis delta antigen (S-HDAg) can directly bind to glutathione S-transferase P1 mRNA causing the downregulation of its expression, therefore increasing ROS and promoting apoptosis [64].
Oxidative stress, ER stress, and UPR trigger a cascade of signaling events that may protect but also damage the liver, depending on the duration of the insult. During HCV infection, elevated ROS levels induce the production of pro-inflammatory cytokines such as tumor necrosis factor alpha (TNF-α) and IL-8 [65]. The underlying NF-κB pathway regulating their production is very sensitive to oxidative stimuli. HCV core proteins, NS4B, and NS5A activate NF-κB as well as STAT3 expression through the elevation of ROS and the disruption of calcium homeostasis [66,67]. Interestingly, the induction of oxidative stress during HCV infection is positively associated with the progression of liver fibrosis, which is characterized by the overproduction of extracellular matrix. A key mechanism of fibrosis is the activation of hepatic stellate cells (HSCs) by pro-inflammatory cytokines, leading to collagen deposition [68]. HCV core proteins, NS3A/4A, NS4B, and NS5A activate transforming growth factor beta 1 (TGF-β1) secretion through ROS and calcium-dependent mechanisms [69]. Viruses also benefit from manipulating ROS levels. Studies revealed that HCV activates the Nrf2/ARE axis, promoting ROS scavenging and preventing ROS accumulation to levels with antiviral and/or lethal effects in the host cell [48,70]. Moreover, under conditions of oxidative stress, viruses promote cell survival and proliferation via associated signaling. HCV activates β-catenin that induces c-Myc and cyclin D1 expression, thus promoting cell cycle progression [71]. Moreover, ROS disrupt p53 binding to Mdm2 via the upregulation of DHCR24 expression, thus attenuating apoptosis [72]. HCV further prevents apoptosis by activating the peroxisome proliferator-activated receptor alpha (PPARα) and suppressing the voltage-gated K+ channel Kv2.1 through NS5A and ROS [73,74]. ROS also suppress the expression of p14, which is implicated in the induction of the pro-apoptotic p53/Mdm2 pathway [75]. ROS upregulates p21, a cyclin-dependent kinase inhibitor activating Nrf2 [76]. While in low stress conditions p21 induces cell cycle arrest, in the presence of high oxidative stress levels, it induces apoptosis [77,78]. HCV core proteins and NS5A inhibit p21 and therefore render Nrf2 less sensitive to ROS. This hampers the induction of apoptosis and stimulates the proliferation of damaged hepatocytes [79,80].
4. Virus-Induced Pro-Inflammatory Signaling
A common consequence of chronic viral hepatis is the induction of liver inflammation (hepatitis). Upon viral sensing, infected hepatocytes trigger the activation of innate immune receptors and sensors that are referred to as the inflammasome and are large protein complexes. The inflammasome serves as a signaling hub triggering type I interferons and the processing and release of proinflammatory cytokines. It is activated by pattern recognition receptors (PRRs) triggered by pathogen-related molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs) (reviewed in more detail in [81]). The host inflammasome represents an important line of defense and is a decision maker with regard to fight (antiviral response) and containment (apoptosis). It is therefore not surprising that hepatic viruses evade the host innate immune response and twist pro-inflammatory signaling to their own benefit to persist and prevent apoptosis of the infected cells. The induction of the inflammasome by viral hepatitis was already evident in the previous section highlighting the common mechanism of virus-induced ROS and ER stress activating STAT3 and NF-κB signaling. Presumably, ROS-independent or -related mechanisms comprise the induction of NF-κB signaling by HDV. Indeed, L-HDAg renders NF-κB signaling more susceptible to TNF-α [82] and induces STAT3 [83]. NF-κB-independent induction of inflammation has been observed for HCV, which involves NS5A induction of cyclooxygenase-2 (COX-2) and, consequently, second messenger signaling and prostaglandin production [84]. The JAK/STAT signaling pathway is an important mediator of the host innate immune response as well as of cellular apoptosis and survival. Viral sensing triggers type I and II interferon responses via STAT1 and STAT2, promoting the expression of antiviral interferon response genes and apoptosis if the pathogen is not cleared [85]. Viruses causing chronic hepatitis have developed elaborated strategies to evade the innate response (reviewed in [86,87,88,89]). An important aspect in this evasion strategy is the pro-viral role of STAT3 signaling. In the liver, STAT3 signaling is a mediator of liver regeneration and balances the pro-apoptotic role of STAT1 by heterodimerization in response to IL-6 and proliferative signaling by HGF and EGF [90]. Attenuation of STAT3 signaling in functional studies impaired the replication of HCV and HBV, suggesting a pro-viral role of STAT3 signaling [90,91,92]. Proliferative signaling via the epidermal growth factor receptor (EGFR) is required for HBV, HCV, and HDV infection [93,94]. Interestingly, EGFR further promotes STAT3 activity by repressing a negative regulator of STAT3, i.e., suppressor of cytokine signaling 3 (SOCS3). Consequently, this tempers the pro-apoptotic, antiviral effect of type I interferon signaling by promoting STAT1/STAT3 heterodimerization over STAT1 homodimerization [95]. STAT3 signaling is further maintained by HCV-induced downregulation of another negative regulator of STAT3, protein tyrosine phosphatase delta (PTPRD) [96]. A potential similar effect may be triggered by HDV infection, which attenuates STAT1/STAT2 signaling via the suppression of Tyk2 [89], which is another negative regulator of STAT3 transcriptional activity [97]. Whether this may promote the pro-viral effect of STAT3 signaling in HBV/HDV-infected hepatocytes remains to be demonstrated.
5. Deregulation of Cellular Metabolism Pathways
The liver is an essential hub for metabolic processes and energy storage. Chronic viral hepatitis has an important impact on metabolic processes in the liver with distinct virus-specific manifestations. While chronic HCV infection strongly resembles clinical manifestations caused by NAFLD and NASH [98], the role of HBV in metabolic disease is controversial, and it has been suggested that HBV may potentially exhibit protective effects towards NAFLD development [99,100]. However, metabolic disease and obesity in HBV- or HCV-infected patients are considered important co-morbidities promoting liver disease progression and increasing cancer risk [101]. The accumulation of free fatty acids induces mitochondrial and ER oxidative stress. Moreover, the accumulation of ROS stimulates lipid peroxidation and inflammatory cascades such as those associated with TNF-α and IL-6, leading to the development of hepatic steatosis and insulin resistance [102].
HCV lifecycle is tightly linked to human lipid metabolism also because HCV requires lipid droplets to replicate and circulates as lipoviral particles to evade the host immune response [103]. HCV proteins directly interact with or regulate the expression of key effector molecules of the lipid metabolism, including apolipoproteins and diacylglycerol acyltransferase-1 [104,105,106]. HCV also activates IKK, which induces the expression of lipogenic genes and promotes lipid droplet formation [107]. Indeed, recent proteogenomic analysis revealed a massive suppression of pathways required for fatty acid metabolism, perturbing the capacity of infected hepatocytes to use fat as an energy source. This coincided with a shift towards a highly glutamine-/glucose-dependent metabolism, which promoted HCV replication [12,108] and resembled the high energy dependence of tumor cells. HCV also tweaks host signal transduction, promoting a favorable metabolic environment for its replication and persistence.
A central role in metabolic liver disease is a chronically dysregulated STAT3 and NF-κB signaling [109,110]. As reviewed above, both pathways are induced by chronic viral hepatitis and associated oxidative stress and are associated with liver disease progression and increased HCC risk [12,90,111,112,113,114]. However, while during HCV infection STAT3 signaling contributes to the accumulation of free fatty acids by suppressing peroxisomal beta-oxidation via inhibiting PPARα [12], HBV-induced STAT3 signaling does not produce the same phenotype. In contrast, HBV replication requires PPARα [115,116] and therefore prevents PPARα inhibition by inducing STAT3 activity. Indeed, HBV infection of primary human hepatocytes induces peroxisomal function, which may be a consequence of a direct rescue of PPARα activity by an HBV protein [12], as observed for PPARγ activation by the protein HBx [117]. HBx maintains fatty acid oxidation [118] and has been observed to bind PPARα in cell culture [119]. This matches observations that PPARα stimulation improves NAFLD in pre-clinical models [120,121]. However, evidence points also to a pro-steatotic impact of HBV infection, even though this is more rarely observed in patients compared to patients infected with HCV [122]. HBV infection does promote the biosynthesis of fatty acids in HBV transgenic (HBV-Tg) mice by the upregulation of fatty acid-binding protein 5 (FABBP5) and acyl-CoA-binding protein (ACBP) [123]. Moreover, HBV-Tg mice display the upregulation of lipid biosynthetic pathways such as those involving retinol-binding protein 1 (RBP1), sterol regulatory element-binding protein 2 (SREBP2), ATP citrate lyase, and fatty acid synthase (FAS) [124]. Additionally, other factors participating in fatty acid transport and biogenesis are dysregulated by HCV, including fatty acid-binding protein 1 (FABP1), responsible for the uptake and transport of long-chain fatty acids (LFA) [125,126], SREBP1, and PPARγ, which induces the expression of hepatic lipogenic and adipogenic genes, accompanied by the accumulation of lipid droplets [127,128,129,130]. HDV infection also impacts metabolic pathways, since HDV decreases the availability of triosephosphate isomerase and pyruvate carboxylase, leading to an abnormal retention of lipids. This effect may also be responsible for microvesicular steatosis during HDV infection [131].
A particular feature of HCV infection is its association with insulin resistance (IR) in patients, which is less frequently observed in HBV- or HBV/HDV-infected patients [132], although a recent genetic screen highlighted the importance of metabolic pathways in HDV lifecycle, including insulin resistance-related genes [133]. This may be due to the different dependency of the hepatic viruses on intracellular glucose levels. Insulin is a central regulator of glucose levels in the blood and of gluconeogenesis in the liver. It therefore also impacts the glucose levels within the hepatocyte. During IR, insulin fails to suppress gluconeogenesis in the hepatocytes [134]. While HCV is a highly glucose-dependent virus [12], HBV replication favors low glucose levels, and thus glucose-induced mTOR signaling hampers HBV replication [135]. HCV proteins directly promote IR by interacting with insulin pathways’ components, including insulin receptor substrate 1 (IRS-1) [136,137,138]. It also alters Akt-induced forkhead box O1 (FOXO1) phosphorylation and its nuclear exclusion, which is required for the transcription of the gluconeogenic gene phosphoenolpyruvate carboxykinase 1 (PCK1) in hepatocytes [139]. Moreover, HCV suppresses glucose transporter 2 (GLUT2) and IRS-2, contributing to higher endogenous glucose levels. Indirect mechanisms of IR involve HCV-induced oxidative stress, steatosis, and pro-inflammatory cytokines, e.g., TNF-α. These indirect effects induce the expression of gluconeogenic genes, such as glucose 6 phosphatase (G6P) and PCK2 [140,141]. Moreover, HCV mediates oxidative stress, leading to hypoxia. This activates H1Fα via c-Myc and Nrf2, controlling the expression of key enzymes in glycolysis [108]. One may speculate that HBV and HDV infection rather indirectly contribute to insulin resistance in patients via virus-induced inflammation and oxidative stress, which is further promoted by comorbidities such as overweight. Direct effects are observed, however, in HBx-Tg mice, which develop hyperglycemia and impaired glucose tolerance [142], and in the HBV-expressing cell line HepG2.2.15, with stimulated TCA cycle and glycolysis [143].
6. Virus-Induced Pro-Fibrotic/Pro-Oncogenic Signaling
Many of the above-mentioned dysregulated signaling pathways promote viral replication and persistence mostly by diverting the host antiviral response to prevent apoptosis and ensure the survival of the infected cell. Strikingly, many of these survival signals are also involved in regenerative processes during liver injury and orchestrate a delicate balance between pro-inflammatory and proliferative signals [144]. As mentioned earlier in this review, all three hepatis viruses chronically infecting the liver engage EGFR signaling to maintain their life cycle [93,94]. EGFR orchestrates the entry of HBV [93] and HCV [94]. EGFR signaling is active during ligand-induced receptor dimerization and internalization and is regulated by phosphatases and endosomal recycling/degradation [145]. HCV has developed strategies to maintain EGFR signaling to its own benefit. HCV infection induces EGFR signaling [146,147] and prolongs EGFR signaling by retaining EGFR in the early endosome via NS5A. This prevents EGFR degradation and leads to EGFR accumulation in infected cells [148,149]. HCV also alters the expression of other ErbB receptors in favor of EGFR [150]. In contrast to HCV, HBV internalization requires EGFR transport to the late endosome, which is critical for efficient HBV infection [151]. Consistently, the inhibition of EGFR degradation abrogated the internalization of HBV via its receptor sodium/taurocholate cotransporter (NTCP) and prevented viral infection [151]. Downstream of EGFR signaling, several viral proteins interact with the MAPK signaling pathways and stimulate cell proliferation [152,153,154,155]. For HCV, NS5A associates with Raf-1 kinase, promoting HCV replication [152]. Consistently, inhibiting Raf kinases with sorafenib blocked the infection, while a further downstream inhibition of MEK1/2 and Erk1/2 showed only marginal effects. This suggests a direct virus–host dependency independent of pathway-associated transcriptional changes. The same holds true for HBV, for which the inhibition of EGFR-associated MAPK or PI3K signaling during infection seems only to have marginal effects [151]. Although no studies have so far demonstrated a role of EGFR during HDV infection, it can be assumed that EGFR may also be required for HDV internalization, since this virus uses the HBV envelope to enter the cell and shares the same entry pathway through HSPG and NTCP.
PI3K/Akt signaling regulates glucose metabolism, cell growth, and survival [156] and it is tightly regulated by phosphatase and tensin homolog deleted on chromosome 10 (PTEN) [157,158]. Independently of its role in insulin signaling, HCV NS5A downregulates PTEN expression through a cooperation of ROS-dependent and -independent pathways that subsequently drives a PTEN–PI3K/Akt feedback loop supporting cell survival [159]. For HBV, the role of PI3K/Akt signaling is more diverse. HBx activates Akt in hepatocytes thereby self-limiting HBV replication [160]. This is consistent with a decreased HBV replication upon PI3K/Akt pathway inhibition using a small-molecule inhibitor in cell culture [161]. However, despite the self-limiting effect on HBV replication, HBx inhibits hepatocyte apoptosis via Akt stimulation and potentially facilitates the persistent, noncytopathic HBV replication [160]. As observed for HCV, HBV impairs PTEN expression, promoting β-catenin/c-Myc signaling and PD-L1 expression [162]. The authors found that PTEN rescue in hepatocytes inhibited β-catenin/PD-L1 signaling and promoted HBV clearance.
Wnt/ß-catenin signaling is essentially involved in the regulation of cell fate during embryogenesis and hepatobiliary development, as well as in liver homeostasis, epithelial–mesenchymal transition (EMT), and tissue regeneration during adulthood. If dysregulated, it promotes liver disease and cancer [163]. The Wnt/β-catenin signaling pathway is activated by hepatic viruses via direct engagement of viral proteins. β-catenin signaling is stimulated by HCV infection via NS3 and NS5A [164] or the phospho-inactivation of GSK-3β by NS5A and core proteins [165,166]. Strikingly, despite a highly genetic heterogeneity, a relative higher frequency of mutations in the β-catenin gene CTNNB1 can be observed in HCC associated with HCV than with HBV [164]. Wnt/ß-catenin signaling is involved in EMT, which is a hallmark of wound healing and liver fibrosis [167]. During chronic infection, dysregulated wound healing processes cause an excessive deposition of extracellular matrix in the liver, leading to liver fibrosis and cirrhosis, which involve not only hepatocytes but also non-parenchymal cells like HSCs and liver macrophages [167]. The Wnt/ß-catenin cascade has a central role in regulating profibrotic pathways in hepatocytes, which involve oxidative stress signaling and transforming growth factor beta (TGF-β)/SMAD signaling [167]. HCV induces TGF-β signaling indirectly via UPR [168]. Interestingly, TGF-β signaling seems to limit HCV infection in hepatocytes [169]. The virus counteracts this activation by an NS5A-mediated inhibition of the phosphorylation and transcriptional activity of SMAD2 and SMAD3/4 heterodimers [170]. HBV infection is also restricted by TGF-β [171], while HDV seems to stimulate TGF-β in luciferase reporter gene assays [172]. This is consistent with a reported activation of TGF-β expression by HDV via an L-HDAg-mediated activation of the Twist promoter through binding to SMAD3 on Smad-binding elements (SBEs) [173]. This is an interesting finding that may help to understand the aggravation of HBV liver disease and the rapid fibrosis progression in HDV/HBV-infected patients [174].
7. Discussion
Chronic liver disease and associated complications including cancer constitute an important burden for public health, with a long-lasting impact on affected individuals even after viral infection cure. The comparison of virus-induced signaling during chronic infection with HBV, HCV, and HDV outlined common pathogenic mechanisms that predominantly result in the failure of the antiviral response to clear infection and in a diversion of the final antiviral safeguard apoptosis towards cell survival. It is evident that the involved signaling pathways that are thereto manipulated largely overlap for different viruses (Table 1), although the detailed strategies differ (Figure 2). Also hepatitis E virus (HEV) seem to dysregulate similar pro-oncogenic signaling pathways linked to oxidative stress, inflammation, apoptosis, and cell proliferation, as reviewed elsewhere [175]. However, chronic infections are relatively rare, and only a fraction of patients progress to fibrosis and HCC [176]. The majority of studies reviewed here were based on cell culture models and performed a limited analysis of canonical pathways. Given these limitations, the currently available literature for some viruses is biased by functional studies of individual proteins (e.g., HBx for HBV) and does not consider protein dynamics or synergic effects of the virus interactome. However, the fast-moving technological development in the recent years and the diffusion of -omics studies in the scientific routine are allowing a more profound study of virus-induced signaling. This should be combined with the use of better infection models representing the three-dimensional architecture of the liver, the heterogeneity of its cell populations, and the contribution of immune cells. Signaling pathways are established targets in cancer therapy [177] and have previously drawn attention as targets for cancer prevention attenuating liver disease progression [178,179]. Host signaling-targeting approaches to battle chronic infection have been discussed [133,180,181] as they hold the potential to lower the genetic barrier of resistance to direct-acting antivirals. However, currently, only interferons are in clinical use targeting chronic viral hepatitis. Thus, a better understanding of virus-induced signaling could promote the development of common therapeutic strategies to help not only patients with chronic infection but also patients suffering from non-viral disease etiologies that display a similar course of liver disease and fibrosis-associated carcinogenesis.

Table 1.
Virus-perturbed signaling pathways during chronic viral hepatitis.
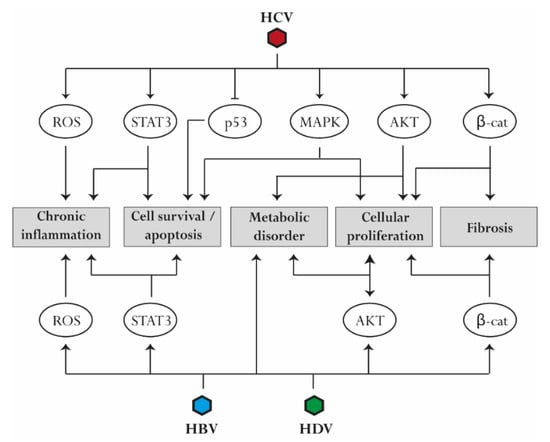
Figure 2.
Common pathways associated with virus-induced liver disease progression. Several perturbations are mediated by HBV, HCV, and HDV infection. Reactive oxygen species (ROS) production and activation of STAT3 contribute to the establishment of chronic liver inflammation. Upregulation of mitogen-activated protein kinase (MAPK) and STAT3 signaling as well as downregulation of p53 reduce apoptosis and promote cell survival. Similarly, activation of AKT, MAPK, and β-catenin induces cell proliferation. AKT upregulation contributes to the development of metabolic disorders, while β-catenin is involved in the progression of liver fibrosis.
Author Contributions
Conceptualization, Z.B., A.V., J.L.; writing—original draft preparation, Z.B. and J.L.; writing—review and editing, Z.B., A.V., T.F.B., E.R.V., J.L.; visualization, Z.B. and A.V.; funding acquisition, T.F.B., E.R.V., J.L. All authors have read and agreed to the published version of the manuscript.
Funding
The authors acknowledge research support by the European Union (EU H2020-HEPCAR #667273 to J.L. and T.F.B., ERC AdG ERC-AdG-2014-HEPCIR #671231, and ERC PoC-HEPCAN #862551 to T.F.B.), the French Cancer Agency (TheraHCC2.0 IHU201901299), the Agence Nationale de Re-cherche sur le Sida et les hépatites virales (ANRS ECTZ103701, ECTZ131760, ECTZ160436, ECTZ171594 to J.L., ANRS ECTZ104017 and ECTZ75178 to T.F.B.), The French National Research Agency (ANR-21-CE15-0035-01 DELTArget to E.R.V.), the University of Strasbourg (IdEx AAP2021 DeltaSig to E.R.V), the Fondation de l’Université de Strasbourg (HEPKIN) (TBA-DON-0002) and the Inserm Plan Cancer 2019–2023, the US National Institute of Health (R01CA233794), and the French state funds managed within the “Plan Investissements d’Avenir” and by the ANR (references ANR-10-IAHU-02 and ANR-10-LABX-0028). This work of the Interdisciplinary Thematic Institute IMCBio, as part of the ITI 2021-2028 program of the University of Strasbourg, CNRS and Inserm, was supported by IdEx Unistra (ANR-10-IDEX-0002), and by SFRI-STRAT’US project (ANR 20-SFRI-0012) and EUR IMCBio (ANR-17-EURE-0023) under the framework of the French Investments for the Future Program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- WHO Team Global HIV, H. a. S. T. I. P. Global Progress Report on HIV, Viral Hepatitis and Sexually Transmitted Infections, 2021; World Health Organization: Geneva, Switzerland, 2021; p. 108. [Google Scholar]
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, E.M.; Sherman, R.L.; Henley, S.J.; Jemal, A.; Siegel, D.A.; Feuer, E.J.; Firth, A.U.; Kohler, B.A.; Scott, S.; Ma, J.; et al. Annual Report to the Nation on the Status of Cancer, Featuring Cancer in Men and Women Age 20–49 Years. J. Natl. Cancer Inst. 2019, 111, 1279–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuen, M.F.; Chen, D.S.; Dusheiko, G.M.; Janssen, H.L.A.; Lau, D.T.Y.; Locarnini, S.A.; Peters, M.G.; Lai, C.L. Hepatitis B virus infection. Nat. Rev. Dis. Primers 2018, 4, 18035. [Google Scholar] [CrossRef] [PubMed]
- Lucifora, J.; Protzer, U. Attacking hepatitis B virus cccDNA—The holy grail to hepatitis B cure. J. Hepatol 2016, 64, S41–S48. [Google Scholar] [CrossRef] [PubMed]
- Tu, T.; Budzinska, M.A.; Shackel, N.A.; Urban, S. HBV DNA Integration: Molecular Mechanisms and Clinical Implications. Viruses 2017, 9, 75. [Google Scholar] [CrossRef]
- Mentha, N.; Clement, S.; Negro, F.; Alfaiate, D. A review on hepatitis D: From virology to new therapies. J. Adv. Res. 2019, 17, 3–15. [Google Scholar] [CrossRef]
- Turon-Lagot, V.; Saviano, A.; Schuster, C.; Baumert, T.F.; Verrier, E.R. Targeting the Host for New Therapeutic Perspectives in Hepatitis D. J. Clin. Med. 2020, 9, 222. [Google Scholar] [CrossRef] [Green Version]
- Wedemeyer, H.; Negro, F. Devil hepatitis D: An orphan disease or largely underdiagnosed? Gut 2019, 68, 381–382. [Google Scholar] [CrossRef]
- Dubuisson, J.; Cosset, F.L. Virology and cell biology of the hepatitis C virus life cycle: An update. J. Hepatol. 2014, 61, S3–S13. [Google Scholar] [CrossRef] [Green Version]
- Comparcola, D.; Alisi, A.; Nobili, V. Hepatitis C virus and nonalcoholic Fatty liver disease: Similar risk factors for necroinflammation, fibrosis, and cirrhosis. Clin. Gastroenterol. Hepatol. 2010, 8, 97. [Google Scholar] [CrossRef]
- Lupberger, J.; Croonenborghs, T.; Roca Suarez, A.A.; Van Renne, N.; Juhling, F.; Oudot, M.A.; Virzi, A.; Bandiera, S.; Jamey, C.; Meszaros, G.; et al. Combined Analysis of Metabolomes, Proteomes, and Transcriptomes of Hepatitis C Virus-Infected Cells and Liver to Identify Pathways Associated with Disease Development. Gastroenterology 2019, 157, 537–551.e9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pawlotsky, J.M.; Negro, F.; Aghemo, A.; Berenguer, M.; Dalgard, O.; Dusheiko, G.; Marra, F.; Puoti, M.; Wedemeyer, H.; European Association for the Study of the Liver. EASL recommendations on treatment of hepatitis C: Final update of the series. J. Hepatol. 2020, 73, 1170–1218. [Google Scholar] [CrossRef] [PubMed]
- Martinez, M.G.; Boyd, A.; Combe, E.; Testoni, B.; Zoulim, F. Covalently closed circular DNA: The ultimate therapeutic target for curing HBV infections. J. Hepatol. 2021, 75, 706–717. [Google Scholar] [CrossRef]
- Roca Suarez, A.A.; Testoni, B.; Zoulim, F. HBV 2021: New therapeutic strategies against an old foe. Liver Int. 2021, 41 (Suppl. S1), 15–23. [Google Scholar] [CrossRef]
- Asselah, T.; Loureiro, D.; Le Gal, F.; Narguet, S.; Brichler, S.; Bouton, V.; Abazid, M.; Boyer, N.; Giuly, N.; Gerber, A.; et al. Early virological response in six patients with hepatitis D virus infection and compensated cirrhosis treated with Bulevirtide in real-life. Liver Int. 2021, 41, 1509–1517. [Google Scholar] [CrossRef]
- Hamdane, N.; Juhling, F.; Crouchet, E.; El Saghire, H.; Thumann, C.; Oudot, M.A.; Bandiera, S.; Saviano, A.; Ponsolles, C.; Roca Suarez, A.A.; et al. HCV-Induced Epigenetic Changes Associated with Liver Cancer Risk Persist after Sustained Virologic Response. Gastroenterology 2019, 156, 2313–2329.e7. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.U.; Seo, Y.S.; Lee, H.A.; Kim, M.N.; Lee, E.J.; Shin, H.J.; Lee, Y.R.; Lee, H.W.; Park, J.Y.; Kim, D.Y.; et al. Hepatocellular Carcinoma Risk Steadily Persists over Time Despite Long-Term Antiviral Therapy for Hepatitis B: A Multicenter Study. Cancer Epidemiol. Biomark. Prev. 2020, 29, 832–837. [Google Scholar] [CrossRef] [Green Version]
- Juhling, F.; Hamdane, N.; Crouchet, E.; Li, S.; El Saghire, H.; Mukherji, A.; Fujiwara, N.; Oudot, M.A.; Thumann, C.; Saviano, A.; et al. Targeting clinical epigenetic reprogramming for chemoprevention of metabolic and viral hepatocellular carcinoma. Gut 2021, 70, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Andrisani, O. Epigenetic mechanisms in hepatitis B virus-associated hepatocellular carcinoma. Hepatoma Res. 2021, 2021, 7. [Google Scholar] [CrossRef] [PubMed]
- Phaniendra, A.; Jestadi, D.B.; Periyasamy, L. Free radicals: Properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 2015, 30, 11–26. [Google Scholar] [CrossRef] [Green Version]
- Santos, C.X.; Tanaka, L.Y.; Wosniak, J.; Laurindo, F.R. Mechanisms and implications of reactive oxygen species generation during the unfolded protein response: Roles of endoplasmic reticulum oxidoreductases, mitochondrial electron transport, and NADPH oxidase. Antioxid. Redox Signal. 2009, 11, 2409–2427. [Google Scholar] [CrossRef] [PubMed]
- Scialo, F.; Fernandez-Ayala, D.J.; Sanz, A. Role of Mitochondrial Reverse Electron Transport in ROS Signaling: Potential Roles in Health and Disease. Front. Physiol. 2017, 8, 428. [Google Scholar] [CrossRef]
- Chen, Y.; Zhou, Z.; Min, W. Mitochondria, Oxidative Stress and Innate Immunity. Front. Physiol. 2018, 9, 1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Xu, M.; Jeong, S.; Qian, Y.; Wu, H.; Xia, Q.; Kong, X. The Role of Nrf2 in Liver Disease: Novel Molecular Mechanisms and Therapeutic Approaches. Front. Pharmacol. 2018, 9, 1428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aleksunes, L.M.; Manautou, J.E. Emerging role of Nrf2 in protecting against hepatic and gastrointestinal disease. Toxicol. Pathol. 2007, 35, 459–473. [Google Scholar] [CrossRef] [PubMed]
- Spahis, S.; Delvin, E.; Borys, J.M.; Levy, E. Oxidative Stress as a Critical Factor in Nonalcoholic Fatty Liver Disease Pathogenesis. Antioxid. Redox Signal. 2017, 26, 519–541. [Google Scholar] [CrossRef] [PubMed]
- Ivanov, A.V.; Valuev-Elliston, V.T.; Tyurina, D.A.; Ivanova, O.N.; Kochetkov, S.N.; Bartosch, B.; Isaguliants, M.G. Oxidative stress, a trigger of hepatitis C and B virus-induced liver carcinogenesis. Oncotarget 2017, 8, 3895–3932. [Google Scholar] [CrossRef] [Green Version]
- Block, T.M.; Mehta, A.S.; Fimmel, C.J.; Jordan, R. Molecular viral oncology of hepatocellular carcinoma. Oncogene 2003, 22, 5093–5107. [Google Scholar] [CrossRef] [Green Version]
- Benali-Furet, N.L.; Chami, M.; Houel, L.; De Giorgi, F.; Vernejoul, F.; Lagorce, D.; Buscail, L.; Bartenschlager, R.; Ichas, F.; Rizzuto, R.; et al. Hepatitis C virus core triggers apoptosis in liver cells by inducing ER stress and ER calcium depletion. Oncogene 2005, 24, 4921–4933. [Google Scholar] [CrossRef] [Green Version]
- Chan, S.W.; Egan, P.A. Hepatitis C virus envelope proteins regulate CHOP via induction of the unfolded protein response. FASEB J. 2005, 19, 1510–1512. [Google Scholar] [CrossRef] [Green Version]
- Li, S.; Ye, L.; Yu, X.; Xu, B.; Li, K.; Zhu, X.; Liu, H.; Wu, X.; Kong, L. Hepatitis C virus NS4B induces unfolded protein response and endoplasmic reticulum overload response-dependent NF-kappaB activation. Virology 2009, 391, 257–264. [Google Scholar] [CrossRef] [Green Version]
- Zheng, Y.; Gao, B.; Ye, L.; Kong, L.; Jing, W.; Yang, X.; Wu, Z.; Ye, L. Hepatitis C virus non-structural protein NS4B can modulate an unfolded protein response. J. Microbiol. 2005, 43, 529–536. [Google Scholar]
- Wang, T.; Campbell, R.V.; Yi, M.K.; Lemon, S.M.; Weinman, S.A. Role of Hepatitis C virus core protein in viral-induced mitochondrial dysfunction. J. Viral Hepat. 2010, 17, 784–793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kang, S.M.; Kim, S.J.; Kim, J.H.; Lee, W.; Kim, G.W.; Lee, K.H.; Choi, K.Y.; Oh, J.W. Interaction of hepatitis C virus core protein with Hsp60 triggers the production of reactive oxygen species and enhances TNF-alpha-mediated apoptosis. Cancer Lett. 2009, 279, 230–237. [Google Scholar] [CrossRef] [PubMed]
- Anelli, T.; Bergamelli, L.; Margittai, E.; Rimessi, A.; Fagioli, C.; Malgaroli, A.; Pinton, P.; Ripamonti, M.; Rizzuto, R.; Sitia, R. Ero1alpha regulates Ca(2+) fluxes at the endoplasmic reticulum-mitochondria interface (MAM). Antioxid. Redox Signal. 2012, 16, 1077–1087. [Google Scholar] [CrossRef]
- Ivanov, A.V.; Smirnova, O.A.; Petrushanko, I.Y.; Ivanova, O.N.; Karpenko, I.L.; Alekseeva, E.; Sominskaya, I.; Makarov, A.A.; Bartosch, B.; Kochetkov, S.N.; et al. HCV core protein uses multiple mechanisms to induce oxidative stress in human hepatoma Huh7 cells. Viruses 2015, 7, 2745–2770. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.I.; Hwang, J.M.; Im, J.H.; Lee, Y.I.; Kim, N.S.; Kim, D.G.; Yu, D.Y.; Moon, H.B.; Park, S.K. Human hepatitis B virus-X protein alters mitochondrial function and physiology in human liver cells. J. Biol. Chem. 2004, 279, 15460–15471. [Google Scholar] [CrossRef] [Green Version]
- Rahmani, Z.; Huh, K.W.; Lasher, R.; Siddiqui, A. Hepatitis B virus X protein colocalizes to mitochondria with a human voltage-dependent anion channel, HVDAC3, and alters its transmembrane potential. J. Virol. 2000, 74, 2840–2846. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, L.Y.; Zheng, B.Y.; Fang, X.F.; Li, D.; Huang, Y.H.; Chen, Z.X.; Zhou, L.Y.; Wang, X.Z. HBx co-localizes with COXIII in HL-7702 cells to upregulate mitochondrial function and ROS generation. Oncol. Rep. 2015, 33, 2461–2467. [Google Scholar] [CrossRef] [Green Version]
- Li, D.; Ding, J.; Chen, Z.; Chen, Y.; Lin, N.; Chen, F.; Wang, X. Accurately mapping the location of the binding site for the interaction between hepatitis B virus X protein and cytochrome c oxidase III. Int. J. Mol. Med. 2015, 35, 319–324. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Kim, H.; Lee, S.A.; Won, Y.S.; Kim, H.I.; Inn, K.S.; Kim, B.J. Upregulation of endoplasmic reticulum stress and reactive oxygen species by naturally occurring mutations in hepatitis B virus core antigen. J. Gen. Virol. 2015, 96, 1850–1854. [Google Scholar] [CrossRef]
- Lee, I.K.; Lee, S.A.; Kim, H.; Won, Y.S.; Kim, B.J. Induction of endoplasmic reticulum-derived oxidative stress by an occult infection related S surface antigen variant. World J. Gastroenterol. 2015, 21, 6872–6883. [Google Scholar] [CrossRef] [PubMed]
- Smirnova, O.A.; Ivanova, O.N.; Bartosch, B.; Valuev-Elliston, V.T.; Mukhtarov, F.; Kochetkov, S.N.; Ivanov, A.V. Hepatitis C Virus NS5A Protein Triggers Oxidative Stress by Inducing NADPH Oxidases 1 and 4 and Cytochrome P450 2E1. Oxid. Med. Cell. Longev. 2016, 2016, 8341937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, V.; Brichler, S.; Khan, E.; Chami, M.; Deny, P.; Kremsdorf, D.; Gordien, E. Large hepatitis delta antigen activates STAT-3 and NF-kappaB via oxidative stress. J. Viral Hepat. 2012, 19, 744–753. [Google Scholar] [CrossRef] [PubMed]
- Abdalla, M.Y.; Ahmad, I.M.; Spitz, D.R.; Schmidt, W.N.; Britigan, B.E. Hepatitis C virus-core and non structural proteins lead to different effects on cellular antioxidant defenses. J. Med. Virol. 2005, 76, 489–497. [Google Scholar] [CrossRef]
- Carvajal-Yepes, M.; Himmelsbach, K.; Schaedler, S.; Ploen, D.; Krause, J.; Ludwig, L.; Weiss, T.; Klingel, K.; Hildt, E. Hepatitis C virus impairs the induction of cytoprotective Nrf2 target genes by delocalization of small Maf proteins. J. Biol. Chem. 2011, 286, 8941–8951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ivanov, A.V.; Smirnova, O.A.; Ivanova, O.N.; Masalova, O.V.; Kochetkov, S.N.; Isaguliants, M.G. Hepatitis C virus proteins activate NRF2/ARE pathway by distinct ROS-dependent and independent mechanisms in HUH7 cells. PLoS ONE 2011, 6, e24957. [Google Scholar] [CrossRef] [Green Version]
- Jiang, Y.; Bao, H.; Ge, Y.; Tang, W.; Cheng, D.; Luo, K.; Gong, G.; Gong, R. Therapeutic targeting of GSK3beta enhances the Nrf2 antioxidant response and confers hepatic cytoprotection in hepatitis C. Gut 2015, 64, 168–179. [Google Scholar] [CrossRef] [Green Version]
- Smirnova, O.A.; Ivanova, O.N.; Mukhtarov, F.S.; Tunitskaya, V.L.; Jansons, J.; Isaguliants, M.G.; Kochetkov, S.N.; Ivanov, A.V. Analysis of the Domains of Hepatitis C Virus Core and NS5A Proteins that Activate the Nrf2/ARE Cascade. Acta Nat. 2016, 8, 123–127. [Google Scholar] [CrossRef]
- Tang, W.; Lazaro, C.A.; Campbell, J.S.; Parks, W.T.; Katze, M.G.; Fausto, N. Responses of nontransformed human hepatocytes to conditional expression of full-length hepatitis C virus open reading frame. Am. J. Pathol. 2007, 171, 1831–1846. [Google Scholar] [CrossRef] [Green Version]
- Blackham, S.; Baillie, A.; Al-Hababi, F.; Remlinger, K.; You, S.; Hamatake, R.; McGarvey, M.J. Gene expression profiling indicates the roles of host oxidative stress, apoptosis, lipid metabolism, and intracellular transport genes in the replication of hepatitis C virus. J. Virol. 2010, 84, 5404–5414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, K.A.; Syder, A.J.; Lederer, S.L.; Diamond, D.L.; Paeper, B.; Rice, C.M.; Katze, M.G. Genomic analysis reveals a potential role for cell cycle perturbation in HCV-mediated apoptosis of cultured hepatocytes. PLoS Pathog. 2009, 5, e1000269. [Google Scholar] [CrossRef] [PubMed]
- Schaedler, S.; Krause, J.; Himmelsbach, K.; Carvajal-Yepes, M.; Lieder, F.; Klingel, K.; Nassal, M.; Weiss, T.S.; Werner, S.; Hildt, E. Hepatitis B virus induces expression of antioxidant response element-regulated genes by activation of Nrf2. J. Biol. Chem. 2010, 285, 41074–41086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, B.; Fang, M.; He, Z.; Cui, D.; Jia, S.; Lin, X.; Xu, X.; Zhou, T.; Liu, W. Hepatitis B virus stimulates G6PD expression through HBx-mediated Nrf2 activation. Cell Death Dis. 2015, 6, e1980. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, Y.; Lin, Z.; Han, M.; Cheng, H. Altered serum copper homeostasis suggests higher oxidative stress and lower antioxidant capability in patients with chronic hepatitis B. Medicine 2018, 97, e11137. [Google Scholar] [CrossRef]
- Nakashima, T.; Sumida, Y.; Yoh, T.; Kakisaka, Y.; Nakajima, Y.; Ishikawa, H.; Mitsuyoshi, H.; Kashima, K.; Nakamura, H.; Yodoi, J. Thioredoxin levels in the sera of untreated viral hepatitis patients and those treated with glycyrrhizin or ursodeoxycholic acid. Antioxid. Redox Signal. 2000, 2, 687–694. [Google Scholar] [CrossRef]
- Wang, Q.; Na, B.; Ou, J.H.; Pulliam, L.; Yen, T.S. Hepatitis B virus alters the antioxidant system in transgenic mice and sensitizes hepatocytes to Fas signaling. PLoS ONE 2012, 7, e36818. [Google Scholar] [CrossRef] [Green Version]
- Qi, L.; Zou, Z.Q.; Wang, L.Y.; Gao, S.; Fan, Y.C.; Long, B.; Guo, Y.M.; Xu, A.L.; Han, J.; Li, T.; et al. Methylation of the glutathione-S-transferase M3 gene promoter is associated with oxidative stress in acute-on-chronic hepatitis B liver failure. Tohoku J. Exp. Med. 2012, 228, 43–51. [Google Scholar] [CrossRef] [Green Version]
- Niu, D.; Zhang, J.; Ren, Y.; Feng, H.; Chen, W.N. HBx genotype D represses GSTP1 expression and increases the oxidative level and apoptosis in HepG2 cells. Mol. Oncol. 2009, 3, 67–76. [Google Scholar] [CrossRef]
- Cho, I.J.; Ki, S.H.; Brooks, C., 3rd; Kim, S.G. Role of hepatitis B virus X repression of C/EBPbeta activity in the down-regulation of glutathione S-transferase A2 gene: Implications in other phase II detoxifying enzyme expression. Xenobiotica 2009, 39, 182–192. [Google Scholar] [CrossRef]
- Ding, C.; Wei, H.; Sun, R.; Zhang, J.; Tian, Z. Hepatocytes proteomic alteration and seroproteome analysis of HBV-transgenic mice. Proteomics 2009, 9, 87–105. [Google Scholar] [CrossRef] [PubMed]
- Yi, Y.S.; Park, S.G.; Byeon, S.M.; Kwon, Y.G.; Jung, G. Hepatitis B virus X protein induces TNF-alpha expression via down-regulation of selenoprotein P in human hepatoma cell line, HepG2. Biochim. Biophys. Acta 2003, 1638, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Chen, M.; Du, D.; Zheng, W.; Liao, M.; Zhang, L.; Liang, G.; Gong, M. Small hepatitis delta antigen selectively binds to target mRNA in hepatic cells: A potential mechanism by which hepatitis D virus downregulates glutathione S-transferase P1 and induces liver injury and hepatocarcinogenesis. Biochem. Cell Biol. 2019, 97, 130–139. [Google Scholar] [CrossRef]
- Jaeschke, H. Reactive oxygen and mechanisms of inflammatory liver injury. J. Gastroenterol. Hepatol. 2000, 15, 718–724. [Google Scholar] [CrossRef]
- Gong, G.; Waris, G.; Tanveer, R.; Siddiqui, A. Human hepatitis C virus NS5A protein alters intracellular calcium levels, induces oxidative stress, and activates STAT-3 and NF-kappa B. Proc. Natl. Acad. Sci. USA 2001, 98, 9599–9604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, W.; Tsai, W.L.; Shao, R.X.; Wu, G.; Peng, L.F.; Barlow, L.L.; Chung, W.J.; Zhang, L.; Zhao, H.; Jang, J.Y.; et al. Hepatitis C virus regulates transforming growth factor beta1 production through the generation of reactive oxygen species in a nuclear factor kappaB-dependent manner. Gastroenterology 2010, 138, 2509–2518.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bataller, R.; Lemon, S.M. Fueling fibrosis in chronic hepatitis C. Proc. Natl. Acad. Sci. USA 2012, 109, 14293–14294. [Google Scholar] [CrossRef] [Green Version]
- Presser, L.D.; Haskett, A.; Waris, G. Hepatitis C virus-induced furin and thrombospondin-1 activate TGF-beta1: Role of TGF-beta1 in HCV replication. Virology 2011, 412, 284–296. [Google Scholar] [CrossRef] [Green Version]
- Ramos-Tovar, E.; Muriel, P. Free radicals, antioxidants, nuclear factor-E2-related factor-2 and liver damage. J. Appl. Toxicol. 2020, 40, 151–168. [Google Scholar] [CrossRef]
- Higgs, M.R.; Lerat, H.; Pawlotsky, J.M. Hepatitis C virus-induced activation of beta-catenin promotes c-Myc expression and a cascade of pro-carcinogenetic events. Oncogene 2013, 32, 4683–4693. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, T.; Kohara, M.; Izumi, K.; Kasama, Y.; Hirata, Y.; Huang, Y.; Shuda, M.; Mukaidani, C.; Takano, T.; Tokunaga, Y.; et al. Hepatitis C virus impairs p53 via persistent overexpression of 3beta-hydroxysterol Delta24-reductase. J. Biol. Chem. 2009, 284, 36442–36452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mankouri, J.; Dallas, M.L.; Hughes, M.E.; Griffin, S.D.; Macdonald, A.; Peers, C.; Harris, M. Suppression of a pro-apoptotic K+ channel as a mechanism for hepatitis C virus persistence. Proc. Natl. Acad. Sci. USA 2009, 106, 15903–15908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tanaka, N.; Moriya, K.; Kiyosawa, K.; Koike, K.; Gonzalez, F.J.; Aoyama, T. PPARalpha activation is essential for HCV core protein-induced hepatic steatosis and hepatocellular carcinoma in mice. J. Clin. Investig. 2008, 118, 683–694. [Google Scholar] [CrossRef] [PubMed]
- Seo, Y.L.; Heo, S.; Jang, K.L. Hepatitis C virus core protein overcomes H2O2-induced apoptosis by downregulating p14 expression via DNA methylation. J. Gen. Virol. 2015, 96, 822–832. [Google Scholar] [CrossRef]
- O’Reilly, M.A. Redox activation of p21Cip1/WAF1/Sdi1: A multifunctional regulator of cell survival and death. Antioxid. Redox Signal. 2005, 7, 108–118. [Google Scholar] [CrossRef]
- Inoue, T.; Kato, K.; Kato, H.; Asanoma, K.; Kuboyama, A.; Ueoka, Y.; Yamaguchi, S.; Ohgami, T.; Wake, N. Level of reactive oxygen species induced by p21Waf1/CIP1 is critical for the determination of cell fate. Cancer Sci. 2009, 100, 1275–1283. [Google Scholar] [CrossRef]
- Villeneuve, N.F.; Sun, Z.; Chen, W.; Zhang, D.D. Nrf2 and p21 regulate the fine balance between life and death by controlling ROS levels. Cell Cycle 2009, 8, 3255–3256. [Google Scholar] [CrossRef] [Green Version]
- Majumder, M.; Ghosh, A.K.; Steele, R.; Ray, R.; Ray, R.B. Hepatitis C virus NS5A physically associates with p53 and regulates p21/waf1 gene expression in a p53-dependent manner. J. Virol. 2001, 75, 1401–1407. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Yoshida, I.; Takamatsu, M.; Ishido, S.; Fujita, T.; Oka, K.; Hotta, H. Complex formation between hepatitis C virus core protein and p21Waf1/Cip1/Sdi1. Biochem. Biophys. Res. Commun. 2000, 273, 479–484. [Google Scholar] [CrossRef]
- Elpek, G.O. Molecular pathways in viral hepatitis-associated liver carcinogenesis: An update. World J. Clin. Cases 2021, 9, 4890–4917. [Google Scholar] [CrossRef]
- Park, C.Y.; Oh, S.H.; Kang, S.M.; Lim, Y.S.; Hwang, S.B. Hepatitis delta virus large antigen sensitizes to TNF-alpha-induced NF-kappaB signaling. Mol. Cells 2009, 28, 49–55. [Google Scholar] [CrossRef]
- Williams, V.; Brichler, S.; Radjef, N.; Lebon, P.; Goffard, A.; Hober, D.; Fagard, R.; Kremsdorf, D.; Deny, P.; Gordien, E. Hepatitis delta virus proteins repress hepatitis B virus enhancers and activate the alpha/beta interferon-inducible MxA gene. J. Gen. Virol. 2009, 90, 2759–2767. [Google Scholar] [CrossRef]
- Zampino, R.; Marrone, A.; Restivo, L.; Guerrera, B.; Sellitto, A.; Rinaldi, L.; Romano, C.; Adinolfi, L.E. Chronic HCV infection and inflammation: Clinical impact on hepatic and extra-hepatic manifestations. World J. Hepatol. 2013, 5, 528–540. [Google Scholar] [CrossRef]
- McNab, F.; Mayer-Barber, K.; Sher, A.; Wack, A.; O’Garra, A. Type I interferons in infectious disease. Nat. Rev. Immunol. 2015, 15, 87–103. [Google Scholar] [CrossRef] [PubMed]
- Brezgin, S.; Kostyusheva, A.; Bayurova, E.; Volchkova, E.; Gegechkori, V.; Gordeychuk, I.; Glebe, D.; Kostyushev, D.; Chulanov, V. Immunity and Viral Infections: Modulating Antiviral Response via CRISPR-Cas Systems. Viruses 2021, 13, 1373. [Google Scholar] [CrossRef] [PubMed]
- Heim, M.H.; Thimme, R. Innate and adaptive immune responses in HCV infections. J. Hepatol. 2014, 61, S14–S25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, S.; Altstetter, S.M.; Protzer, U. Innate immune recognition and modulation in hepatitis D virus infection. World J. Gastroenterol. 2020, 26, 2781–2791. [Google Scholar] [CrossRef] [PubMed]
- Pugnale, P.; Pazienza, V.; Guilloux, K.; Negro, F. Hepatitis delta virus inhibits alpha interferon signaling. Hepatology 2009, 49, 398–406. [Google Scholar] [CrossRef]
- Lupberger, J.; Duong, F.H.; Fofana, I.; Zona, L.; Xiao, F.; Thumann, C.; Durand, S.C.; Pessaux, P.; Zeisel, M.B.; Heim, M.H.; et al. Epidermal growth factor receptor signaling impairs the antiviral activity of interferon-alpha. Hepatology 2013, 58, 1225–1235. [Google Scholar] [CrossRef]
- Hosel, M.; Quasdorff, M.; Ringelhan, M.; Kashkar, H.; Debey-Pascher, S.; Sprinzl, M.F.; Bockmann, J.H.; Arzberger, S.; Webb, D.; von Olshausen, G.; et al. Hepatitis B Virus Activates Signal Transducer and Activator of Transcription 3 Supporting Hepatocyte Survival and Virus Replication. Cell. Mol. Gastroenterol. Hepatol. 2017, 4, 339–363. [Google Scholar] [CrossRef]
- Yang, Y.; Zheng, B.; Han, Q.; Zhang, C.; Tian, Z.; Zhang, J. Targeting blockage of STAT3 inhibits hepatitis B virus-related hepatocellular carcinoma. Cancer Biol. Ther. 2016, 17, 449–456. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Saso, W.; Sugiyama, R.; Ishii, K.; Ohki, M.; Nagamori, S.; Suzuki, R.; Aizaki, H.; Ryo, A.; Yun, J.H.; et al. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc. Natl. Acad. Sci. USA 2019, 116, 8487–8492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lupberger, J.; Zeisel, M.B.; Xiao, F.; Thumann, C.; Fofana, I.; Zona, L.; Davis, C.; Mee, C.J.; Turek, M.; Gorke, S.; et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011, 17, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zona, L.; Lupberger, J.; Sidahmed-Adrar, N.; Thumann, C.; Harris, H.J.; Barnes, A.; Florentin, J.; Tawar, R.G.; Xiao, F.; Turek, M.; et al. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe 2013, 13, 302–313. [Google Scholar] [CrossRef] [Green Version]
- Van Renne, N.; Roca Suarez, A.A.; Duong, F.H.T.; Gondeau, C.; Calabrese, D.; Fontaine, N.; Ababsa, A.; Bandiera, S.; Croonenborghs, T.; Pochet, N.; et al. miR-135a-5p-mediated downregulation of protein tyrosine phosphatase receptor delta is a candidate driver of HCV-associated hepatocarcinogenesis. Gut 2018, 67, 953–962. [Google Scholar] [CrossRef]
- Mori, R.; Wauman, J.; Icardi, L.; Van der Heyden, J.; De Cauwer, L.; Peelman, F.; De Bosscher, K.; Tavernier, J. TYK2-induced phosphorylation of Y640 suppresses STAT3 transcriptional activity. Sci. Rep. 2017, 7, 15919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.C.; Cheng, P.N.; Kao, J.H. Systematic review: Chronic viral hepatitis and metabolic derangement. Aliment. Pharmacol. Ther. 2020, 51, 216–230. [Google Scholar] [CrossRef]
- Hui, R.W.H.; Seto, W.K.; Cheung, K.S.; Mak, L.Y.; Liu, K.S.H.; Fung, J.; Wong, D.K.; Lai, C.L.; Yuen, M.F. Inverse relationship between hepatic steatosis and hepatitis B viremia: Results of a large case-control study. J. Viral Hepat. 2018, 25, 97–104. [Google Scholar] [CrossRef]
- Joo, E.J.; Chang, Y.; Yeom, J.S.; Ryu, S. Hepatitis B virus infection and decreased risk of nonalcoholic fatty liver disease: A cohort study. Hepatology 2017, 65, 828–835. [Google Scholar] [CrossRef] [Green Version]
- Seto, W.K. Chronic hepatitis B and metabolic risk factors: A call for rigorous longitudinal studies. World J. Gastroenterol. 2019, 25, 282–286. [Google Scholar] [CrossRef]
- Patel, A.; Harrison, S.A. Hepatitis C virus infection and nonalcoholic steatohepatitis. Gastroenterol. Hepatol. 2012, 8, 305–312. [Google Scholar]
- Wrensch, F.; Crouchet, E.; Ligat, G.; Zeisel, M.B.; Keck, Z.Y.; Foung, S.K.H.; Schuster, C.; Baumert, T.F. Hepatitis C Virus (HCV)-Apolipoprotein Interactions and Immune Evasion and Their Impact on HCV Vaccine Design. Front. Immunol. 2018, 9, 1436. [Google Scholar] [CrossRef]
- Benga, W.J.; Krieger, S.E.; Dimitrova, M.; Zeisel, M.B.; Parnot, M.; Lupberger, J.; Hildt, E.; Luo, G.; McLauchlan, J.; Baumert, T.F.; et al. Apolipoprotein E interacts with hepatitis C virus nonstructural protein 5A and determines assembly of infectious particles. Hepatology 2010, 51, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Chang, K.S.; Jiang, J.; Cai, Z.; Luo, G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 2007, 81, 13783–13793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herker, E.; Harris, C.; Hernandez, C.; Carpentier, A.; Kaehlcke, K.; Rosenberg, A.R.; Farese, R.V., Jr.; Ott, M. Efficient hepatitis C virus particle formation requires diacylglycerol acyltransferase-1. Nat. Med. 2010, 16, 1295–1298. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zhang, Y.Y.; Chiu, S.; Hu, Z.; Lan, K.H.; Cha, H.; Sodroski, C.; Zhang, F.; Hsu, C.S.; Thomas, E.; et al. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog. 2014, 10, e1004163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy, P.L.; Duponchel, S.; Eischeid, H.; Molle, J.; Michelet, M.; Diserens, G.; Vermathen, M.; Vermathen, P.; Dufour, J.F.; Dienes, H.P.; et al. Hepatitis C virus infection triggers a tumor-like glutamine metabolism. Hepatology 2017, 65, 789–803. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Karin, M. NF-kappaB and STAT3—Key players in liver inflammation and cancer. Cell Res. 2011, 21, 159–168. [Google Scholar] [CrossRef] [Green Version]
- Park, E.J.; Lee, J.H.; Yu, G.Y.; He, G.; Ali, S.R.; Holzer, R.G.; Osterreicher, C.H.; Takahashi, H.; Karin, M. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell 2010, 140, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Chang, Z.; Wang, Y.; Zhou, X.; Long, J.E. STAT3 roles in viral infection: Antiviral or proviral? Future Virol. 2018, 13, 557–574. [Google Scholar] [CrossRef]
- Hiscott, J.; Kwon, H.; Genin, P. Hostile takeovers: Viral appropriation of the NF-kappaB pathway. J. Clin. Investig. 2001, 107, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Waris, G.; Huh, K.W.; Siddiqui, A. Mitochondrially associated hepatitis B virus X protein constitutively activates transcription factors STAT-3 and NF-kappa B via oxidative stress. Mol. Cell. Biol. 2001, 21, 7721–7730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, T.; Hanada, T.; Tokuhisa, T.; Kosai, K.; Sata, M.; Kohara, M.; Yoshimura, A. Activation of STAT3 by the hepatitis C virus core protein leads to cellular transformation. J. Exp. Med. 2002, 196, 641–653. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Ma, Y.; Liu, M.; Yan, L.; Tang, H. Peroxisome Proliferators Activated Receptor (PPAR) agonists activate hepatitis B virus replication in vivo. Virol J. 2017, 14, 96. [Google Scholar] [CrossRef] [Green Version]
- Hu, W.; Wang, X.; Ding, X.; Li, Y.; Zhang, X.; Xie, P.; Yang, J.; Wang, S. MicroRNA-141 represses HBV replication by targeting PPARA. PLoS ONE 2012, 7, e34165. [Google Scholar] [CrossRef] [Green Version]
- Dubuquoy, L.; Louvet, A.; Hollebecque, A.; Mathurin, P.; Dharancy, S. Peroxisome proliferator-activated receptors in HBV-related infection. PPAR Res. 2009, 2009, 145124. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.D.; Wu, H.; Huang, S.; Zhang, H.L.; Qin, C.J.; Zhao, L.H.; Fu, G.B.; Zhou, X.; Wang, X.M.; Tang, L.; et al. HBx regulates fatty acid oxidation to promote hepatocellular carcinoma survival during metabolic stress. Oncotarget 2016, 7, 6711–6726. [Google Scholar] [CrossRef]
- Wu, Y.L.; Peng, X.E.; Zhu, Y.B.; Yan, X.L.; Chen, W.N.; Lin, X. Hepatitis B Virus X Protein Induces Hepatic Steatosis by Enhancing the Expression of Liver Fatty Acid Binding Protein. J. Virol. 2016, 90, 1729–1740. [Google Scholar] [CrossRef] [Green Version]
- Pawlak, M.; Lefebvre, P.; Staels, B. Molecular mechanism of PPARalpha action and its impact on lipid metabolism, inflammation and fibrosis in non-alcoholic fatty liver disease. J. Hepatol. 2015, 62, 720–733. [Google Scholar] [CrossRef] [Green Version]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD development and therapeutic strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Haga, Y.; Kanda, T.; Sasaki, R.; Nakamura, M.; Nakamoto, S.; Yokosuka, O. Nonalcoholic fatty liver disease and hepatic cirrhosis: Comparison with viral hepatitis-associated steatosis. World J. Gastroenterol. 2015, 21, 12989–12995. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Yan, S.; He, Y.; Wang, F.; Song, S.; Guo, Y.; Zhou, Q.; Wang, Y.; Lin, Z.; Yang, Y.; et al. Expression of hepatitis B virus proteins in transgenic mice alters lipid metabolism and induces oxidative stress in the liver. J. Hepatol. 2008, 48, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Hajjou, M.; Norel, R.; Carver, R.; Marion, P.; Cullen, J.; Rogler, L.E.; Rogler, C.E. cDNA microarray analysis of HBV transgenic mouse liver identifies genes in lipid biosynthetic and growth control pathways affected by HBV. J. Med. Virol. 2005, 77, 57–65. [Google Scholar] [CrossRef]
- Hertzel, A.V.; Bernlohr, D.A. The mammalian fatty acid-binding protein multigene family: Molecular and genetic insights into function. Trends Endocrinol. Metab. 2000, 11, 175–180. [Google Scholar] [CrossRef]
- Liu, G.; Liu, G.; Cui, X.; Xu, Y. Transcriptomic Data Analyses Reveal a Reprogramed Lipid Metabolism in HCV-Derived Hepatocellular Cancer. Front. Cell Dev. Biol. 2020, 8, 581863. [Google Scholar] [CrossRef]
- Shieh, Y.S.; Chang, Y.S.; Hong, J.R.; Chen, L.J.; Jou, L.K.; Hsu, C.C.; Her, G.M. Increase of hepatic fat accumulation by liver specific expression of Hepatitis B virus X protein in zebrafish. Biochim. Biophys. Acta 2010, 1801, 721–730. [Google Scholar] [CrossRef]
- Na, T.Y.; Shin, Y.K.; Roh, K.J.; Kang, S.A.; Hong, I.; Oh, S.J.; Seong, J.K.; Park, C.K.; Choi, Y.L.; Lee, M.O. Liver X receptor mediates hepatitis B virus X protein-induced lipogenesis in hepatitis B virus-associated hepatocellular carcinoma. Hepatology 2009, 49, 1122–1131. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Kim, K.H.; Kim, H.H.; Cheong, J. Hepatitis B virus X protein induces lipogenic transcription factor SREBP1 and fatty acid synthase through the activation of nuclear receptor LXRalpha. Biochem. J. 2008, 416, 219–230. [Google Scholar] [CrossRef]
- Kim, K.H.; Shin, H.J.; Kim, K.; Choi, H.M.; Rhee, S.H.; Moon, H.B.; Kim, H.H.; Yang, U.S.; Yu, D.Y.; Cheong, J. Hepatitis B virus X protein induces hepatic steatosis via transcriptional activation of SREBP1 and PPARgamma. Gastroenterology 2007, 132, 1955–1967. [Google Scholar] [CrossRef]
- Mota, S.; Mendes, M.; Freitas, N.; Penque, D.; Coelho, A.V.; Cunha, C. Proteome analysis of a human liver carcinoma cell line stably expressing hepatitis delta virus ribonucleoproteins. J. Proteomics 2009, 72, 616–627. [Google Scholar] [CrossRef]
- Petta, S.; Camma, C.; Di Marco, V.; Macaluso, F.S.; Maida, M.; Pizzolanti, G.; Belmonte, B.; Cabibi, D.; Di Stefano, R.; Ferraro, D.; et al. Hepatic steatosis and insulin resistance are associated with severe fibrosis in patients with chronic hepatitis caused by HBV or HCV infection. Liver Int. 2011, 31, 507–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verrier, E.R.; Weiss, A.; Bach, C.; Heydmann, L.; Turon-Lagot, V.; Kopp, A.; El Saghire, H.; Crouchet, E.; Pessaux, P.; Garcia, T.; et al. Combined small molecule and loss-of-function screen uncovers estrogen receptor alpha and CAD as host factors for HDV infection and antiviral targets. Gut 2020, 69, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Hatting, M.; Tavares, C.D.J.; Sharabi, K.; Rines, A.K.; Puigserver, P. Insulin regulation of gluconeogenesis. Ann. N. Y. Acad. Sci. 2018, 1411, 21–35. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Lin, Y.; Kemper, T.; Chen, J.; Yuan, Z.; Liu, S.; Zhu, Y.; Broering, R.; Lu, M. AMPK and Akt/mTOR signalling pathways participate in glucose-mediated regulation of hepatitis B virus replication and cellular autophagy. Cell. Microbiol. 2020, 22, e13131. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Saito, K.; Ait-Goughoulte, M.; Meyer, K.; Ray, R.B.; Ray, R. Hepatitis C virus core protein upregulates serine phosphorylation of insulin receptor substrate-1 and impairs the downstream akt/protein kinase B signaling pathway for insulin resistance. J. Virol. 2008, 82, 2606–2612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, T.T.; Qin, Z.L.; Ren, H.; Zhao, P.; Qi, Z.T. Inhibition of IRS-1 by hepatitis C virus infection leads to insulin resistance in a PTEN-dependent manner. Virol. J. 2015, 12, 12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parvaiz, F.; Manzoor, S.; Iqbal, J.; Sarkar-Dutta, M.; Imran, M.; Waris, G. Hepatitis C virus NS5A promotes insulin resistance through IRS-1 serine phosphorylation and increased gluconeogenesis. World J. Gastroenterol. 2015, 21, 12361–12369. [Google Scholar] [CrossRef]
- Arai, T.; Kano, F.; Murata, M. Translocation of forkhead box O1 to the nuclear periphery induces histone modifications that regulate transcriptional repression of PCK1 in HepG2 cells. Genes Cells 2015, 20, 340–357. [Google Scholar] [CrossRef] [Green Version]
- Bose, S.K.; Shrivastava, S.; Meyer, K.; Ray, R.B.; Ray, R. Hepatitis C virus activates the mTOR/S6K1 signaling pathway in inhibiting IRS-1 function for insulin resistance. J. Virol. 2012, 86, 6315–6322. [Google Scholar] [CrossRef] [Green Version]
- Deng, L.; Shoji, I.; Ogawa, W.; Kaneda, S.; Soga, T.; Jiang, D.P.; Ide, Y.H.; Hotta, H. Hepatitis C virus infection promotes hepatic gluconeogenesis through an NS5A-mediated, FoxO1-dependent pathway. J. Virol. 2011, 85, 8556–8568. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.J.; Park, Y.H.; Kim, S.U.; Moon, H.B.; Park, D.S.; Han, Y.H.; Lee, C.H.; Lee, D.S.; Song, I.S.; Lee, D.H.; et al. Hepatitis B virus X protein regulates hepatic glucose homeostasis via activation of inducible nitric oxide synthase. J. Biol. Chem. 2011, 286, 29872–29881. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Zhu, W.; Zhang, L.; Lei, H.; Wu, X.; Guo, L.; Chen, X.; Wang, Y.; Tang, H. The metabolic responses to hepatitis B virus infection shed new light on pathogenesis and targets for treatment. Sci. Rep. 2015, 5, 8421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michalopoulos, G.K.; Bhushan, B. Liver regeneration: Biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 40–55. [Google Scholar] [CrossRef] [PubMed]
- Tomas, A.; Futter, C.E.; Eden, E.R. EGF receptor trafficking: Consequences for signaling and cancer. Trends Cell Biol. 2014, 24, 26–34. [Google Scholar] [CrossRef] [Green Version]
- Mailly, L.; Xiao, F.; Lupberger, J.; Wilson, G.K.; Aubert, P.; Duong, F.H.T.; Calabrese, D.; Leboeuf, C.; Fofana, I.; Thumann, C.; et al. Clearance of persistent hepatitis C virus infection in humanized mice using a claudin-1-targeting monoclonal antibody. Nat. Biotechnol. 2015, 33, 549–554. [Google Scholar] [CrossRef] [Green Version]
- Diao, J.; Pantua, H.; Ngu, H.; Komuves, L.; Diehl, L.; Schaefer, G.; Kapadia, S.B. Hepatitis C virus induces epidermal growth factor receptor activation via CD81 binding for viral internalization and entry. J. Virol. 2012, 86, 10935–10949. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mankouri, J.; Griffin, S.; Harris, M. The hepatitis C virus non-structural protein NS5A alters the trafficking profile of the epidermal growth factor receptor. Traffic 2008, 9, 1497–1509. [Google Scholar] [CrossRef] [PubMed]
- Igloi, Z.; Kazlauskas, A.; Saksela, K.; Macdonald, A.; Mankouri, J.; Harris, M. Hepatitis C virus NS5A protein blocks epidermal growth factor receptor degradation via a proline motif- dependent interaction. J. Gen. Virol. 2015, 96, 2133–2144. [Google Scholar] [CrossRef]
- Stindt, S.; Cebula, P.; Albrecht, U.; Keitel, V.; Schulte am Esch, J.; Knoefel, W.T.; Bartenschlager, R.; Haussinger, D.; Bode, J.G. Hepatitis C Virus Activates a Neuregulin-Driven Circuit to Modify Surface Expression of Growth Factor Receptors of the ErbB Family. PLoS ONE 2016, 11, e0148711. [Google Scholar] [CrossRef] [Green Version]
- Iwamoto, M.; Saso, W.; Nishioka, K.; Ohashi, H.; Sugiyama, R.; Ryo, A.; Ohki, M.; Yun, J.H.; Park, S.Y.; Ohshima, T.; et al. The machinery for endocytosis of epidermal growth factor receptor coordinates the transport of incoming hepatitis B virus to the endosomal network. J. Biol. Chem. 2020, 295, 800–807. [Google Scholar] [CrossRef]
- Burckstummer, T.; Kriegs, M.; Lupberger, J.; Pauli, E.K.; Schmittel, S.; Hildt, E. Raf-1 kinase associates with Hepatitis C virus NS5A and regulates viral replication. FEBS Lett. 2006, 580, 575–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, D.Y.; Sun, Y.; Cheng, R.X.; Ouyang, X.M.; Zheng, H. Effect of hepatitis C virus nonstructural protein NS3 on proliferation and MAPK phosphorylation of normal hepatocyte line. World J. Gastroenterol. 2005, 11, 2157–2161. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, J.; Aoki, H.; Kajino, K.; Moriyama, M.; Arakawa, Y.; Hino, O. Hepatitis C virus core protein activates the MAPK/ERK cascade synergistically with tumor promoter TPA, but not with epidermal growth factor or transforming growth factor alpha. Hepatology 2000, 32, 958–961. [Google Scholar] [CrossRef]
- Zhao, L.J.; Wang, L.; Ren, H.; Cao, J.; Li, L.; Ke, J.S.; Qi, Z.T. Hepatitis C virus E2 protein promotes human hepatoma cell proliferation through the MAPK/ERK signaling pathway via cellular receptors. Exp. Cell Res. 2005, 305, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Hoxhaj, G.; Manning, B.D. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat. Rev. Cancer 2020, 20, 74–88. [Google Scholar] [CrossRef]
- Chen, C.Y.; Chen, J.; He, L.; Stiles, B.L. PTEN: Tumor Suppressor and Metabolic Regulator. Front. Endocrinol. 2018, 9, 338. [Google Scholar] [CrossRef] [Green Version]
- Hemmings, B.A.; Restuccia, D.F. PI3K-PKB/Akt pathway. Cold Spring Harb. Perspect. Biol. 2012, 4, a011189. [Google Scholar] [CrossRef] [Green Version]
- Cheng, D.; Zhang, L.; Yang, G.; Zhao, L.; Peng, F.; Tian, Y.; Xiao, X.; Chung, R.T.; Gong, G. Hepatitis C virus NS5A drives a PTEN-PI3K/Akt feedback loop to support cell survival. Liver Int. 2015, 35, 1682–1691. [Google Scholar] [CrossRef]
- Rawat, S.; Bouchard, M.J. The hepatitis B virus (HBV) HBx protein activates AKT to simultaneously regulate HBV replication and hepatocyte survival. J. Virol. 2015, 89, 999–1012. [Google Scholar] [CrossRef] [Green Version]
- Xiang, K.; Wang, B. Role of the PI3KAKTmTOR pathway in hepatitis B virus infection and replication. Mol. Med. Rep. 2018, 17, 4713–4719. [Google Scholar] [CrossRef]
- Sun, Y.; Yu, M.; Qu, M.; Ma, Y.; Zheng, D.; Yue, Y.; Guo, S.; Tang, L.; Li, G.; Zheng, W.; et al. Hepatitis B virus-triggered PTEN/beta-catenin/c-Myc signaling enhances PD-L1 expression to promote immune evasion. Am. J. Physiol. Gastrointest Liver Physiol. 2020, 318, G162–G173. [Google Scholar] [CrossRef] [PubMed]
- Perugorria, M.J.; Olaizola, P.; Labiano, I.; Esparza-Baquer, A.; Marzioni, M.; Marin, J.J.G.; Bujanda, L.; Banales, J.M. Wnt-beta-catenin signalling in liver development, health and disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 121–136. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Pan, Q.; Fuhler, G.M.; Smits, R.; Peppelenbosch, M.P. Action and function of Wnt/beta-catenin signaling in the progression from chronic hepatitis C to hepatocellular carcinoma. J. Gastroenterol. 2017, 52, 419–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Ding, X.; Tang, J.; Cao, Y.; Hu, P.; Zhou, F.; Shan, X.; Cai, X.; Chen, Q.; Ling, N.; et al. Enhancement of canonical Wnt/beta-catenin signaling activity by HCV core protein promotes cell growth of hepatocellular carcinoma cells. PLoS ONE 2011, 6, e27496. [Google Scholar] [CrossRef]
- Street, A.; Macdonald, A.; McCormick, C.; Harris, M. Hepatitis C virus NS5A-mediated activation of phosphoinositide 3-kinase results in stabilization of cellular beta-catenin and stimulation of beta-catenin-responsive transcription. J. Virol. 2005, 79, 5006–5016. [Google Scholar] [CrossRef] [Green Version]
- Roehlen, N.; Crouchet, E.; Baumert, T.F. Liver Fibrosis: Mechanistic Concepts and Therapeutic Perspectives. Cells 2020, 9, 875. [Google Scholar] [CrossRef] [Green Version]
- Chusri, P.; Kumthip, K.; Hong, J.; Zhu, C.; Duan, X.; Jilg, N.; Fusco, D.N.; Brisac, C.; Schaefer, E.A.; Cai, D.; et al. HCV induces transforming growth factor beta1 through activation of endoplasmic reticulum stress and the unfolded protein response. Sci. Rep. 2016, 6, 22487. [Google Scholar] [CrossRef]
- Zou, L.L.; Li, J.R.; Li, H.; Tan, J.L.; Wang, M.X.; Liu, N.N.; Gao, R.M.; Yan, H.Y.; Wang, X.K.; Dong, B.; et al. TGF-beta isoforms inhibit hepatitis C virus propagation in transforming growth factor beta/SMAD protein signalling pathway dependent and independent manners. J. Cell. Mol. Med. 2021, 25, 3498–3510. [Google Scholar] [CrossRef]
- Choi, S.H.; Hwang, S.B. Modulation of the transforming growth factor-beta signal transduction pathway by hepatitis C virus nonstructural 5A protein. J. Biol. Chem. 2006, 281, 7468–7478. [Google Scholar] [CrossRef] [Green Version]
- Liang, G.; Liu, G.; Kitamura, K.; Wang, Z.; Chowdhury, S.; Monjurul, A.M.; Wakae, K.; Koura, M.; Shimadu, M.; Kinoshita, K.; et al. TGF-beta suppression of HBV RNA through AID-dependent recruitment of an RNA exosome complex. PLoS Pathog. 2015, 11, e1004780. [Google Scholar] [CrossRef]
- Choi, S.H.; Jeong, S.H.; Hwang, S.B. Large hepatitis delta antigen modulates transforming growth factor-beta signaling cascades: Implication of hepatitis delta virus-induced liver fibrosis. Gastroenterology 2007, 132, 343–357. [Google Scholar] [CrossRef]
- Liang, Y.J.; Sun, C.P.; Hsu, Y.C.; Chen, Y.W.; Wang, I.A.; Su, C.W.; Tao, M.H.; Wu, J.C. Statin inhibits large hepatitis delta antigen-Smad3 -twist-mediated epithelial-to-mesenchymal transition and hepatitis D virus secretion. J. Biomed. Sci. 2020, 27, 65. [Google Scholar] [CrossRef] [PubMed]
- Wedemeyer, H.; Manns, M.P. Epidemiology, pathogenesis and management of hepatitis D: Update and challenges ahead. Nat. Rev. Gastroenterol. Hepatol. 2010, 7, 31–40. [Google Scholar] [CrossRef]
- Klohn, M.; Schrader, J.A.; Bruggemann, Y.; Todt, D.; Steinmann, E. Beyond the Usual Suspects: Hepatitis E Virus and Its Implications in Hepatocellular Carcinoma. Cancers 2021, 13, 5867. [Google Scholar] [CrossRef] [PubMed]
- Khuroo, M.S.; Khuroo, M.S.; Khuroo, N.S. Hepatitis E: Discovery, global impact, control and cure. World J. Gastroenterol. 2016, 22, 7030–7045. [Google Scholar] [CrossRef] [PubMed]
- Pottier, C.; Fresnais, M.; Gilon, M.; Jerusalem, G.; Longuespee, R.; Sounni, N.E. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers 2020, 12, 731. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, B.C.; Hoshida, Y.; Fujii, T.; Wei, L.; Yamada, S.; Lauwers, G.Y.; McGinn, C.M.; DePeralta, D.K.; Chen, X.; Kuroda, T.; et al. Epidermal growth factor receptor inhibition attenuates liver fibrosis and development of hepatocellular carcinoma. Hepatology 2014, 59, 1577–1590. [Google Scholar] [CrossRef] [Green Version]
- El-Far, Y.M.; Khodir, A.E.; Emarah, Z.A.; Ebrahim, M.A.; Al-Gayyar, M.M.H. Chemopreventive and hepatoprotective effects of genistein via inhibition of oxidative stress and the versican/PDGF/PKC signaling pathway in experimentally induced hepatocellular carcinoma in rats by thioacetamide. Redox Rep. 2022, 27, 9–20. [Google Scholar] [CrossRef]
- Faure-Dupuy, S.; Riedl, T.; Rolland, M.; Hizir, Z.; Reisinger, F.; Neuhaus, K.; Schuehle, S.; Remouchamps, C.; Gillet, N.; Schonung, M.; et al. Control of APOBEC3B induction and cccDNA decay by NF-kappaB and miR-138-5p. JHEP Rep. 2021, 3, 100354. [Google Scholar] [CrossRef]
- Zeisel, M.B.; Lupberger, J.; Fofana, I.; Baumert, T.F. Host-targeting agents for prevention and treatment of chronic hepatitis C—Perspectives and challenges. J. Hepatol. 2013, 58, 375–384. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).