An Efficient Aequorea victoria Green Fluorescent Protein for Stimulated Emission Depletion Super-Resolution Microscopy
Abstract
1. Introduction
2. Results
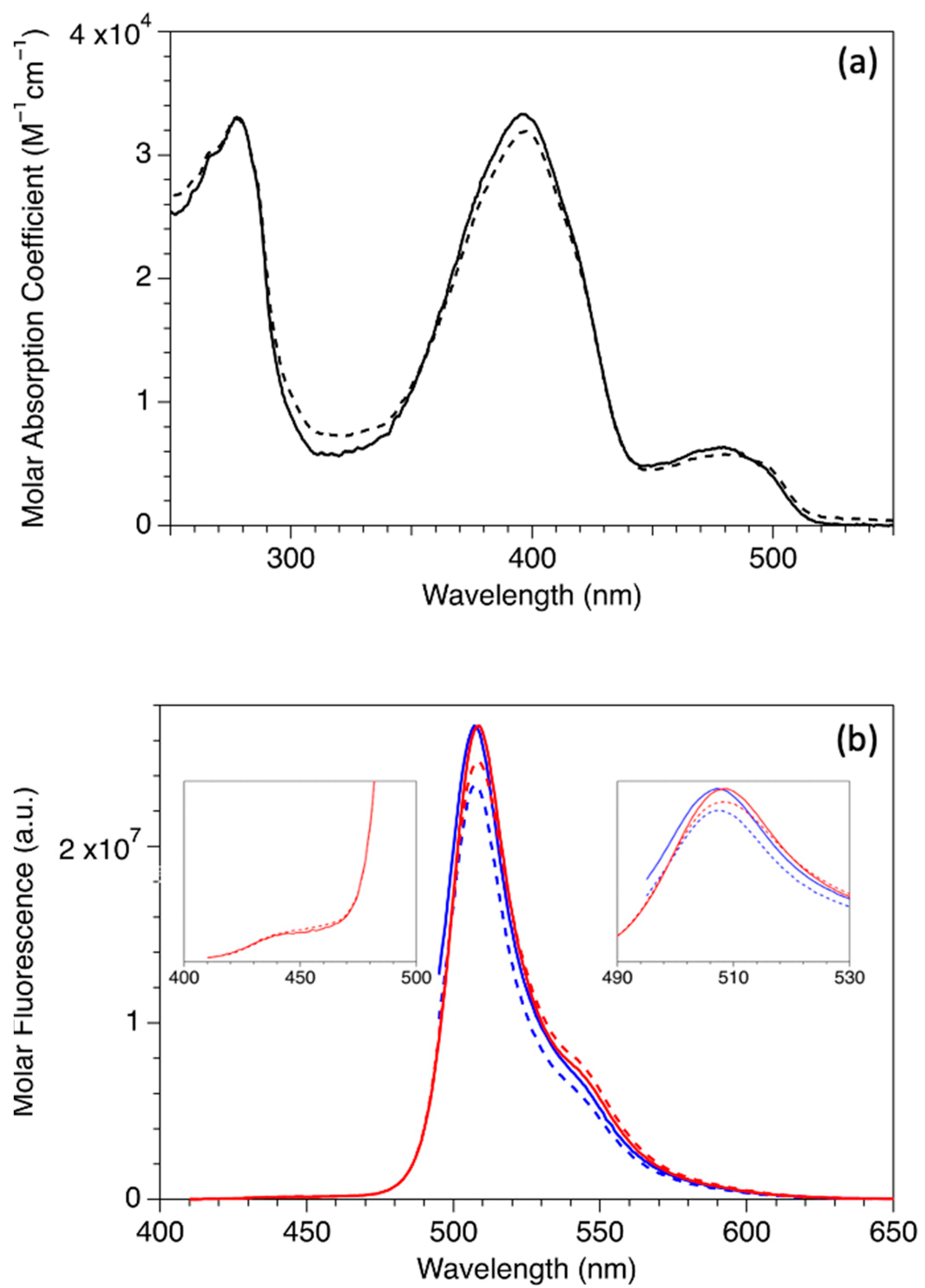
2.1. Steady-State Spectrophotometric Measurements
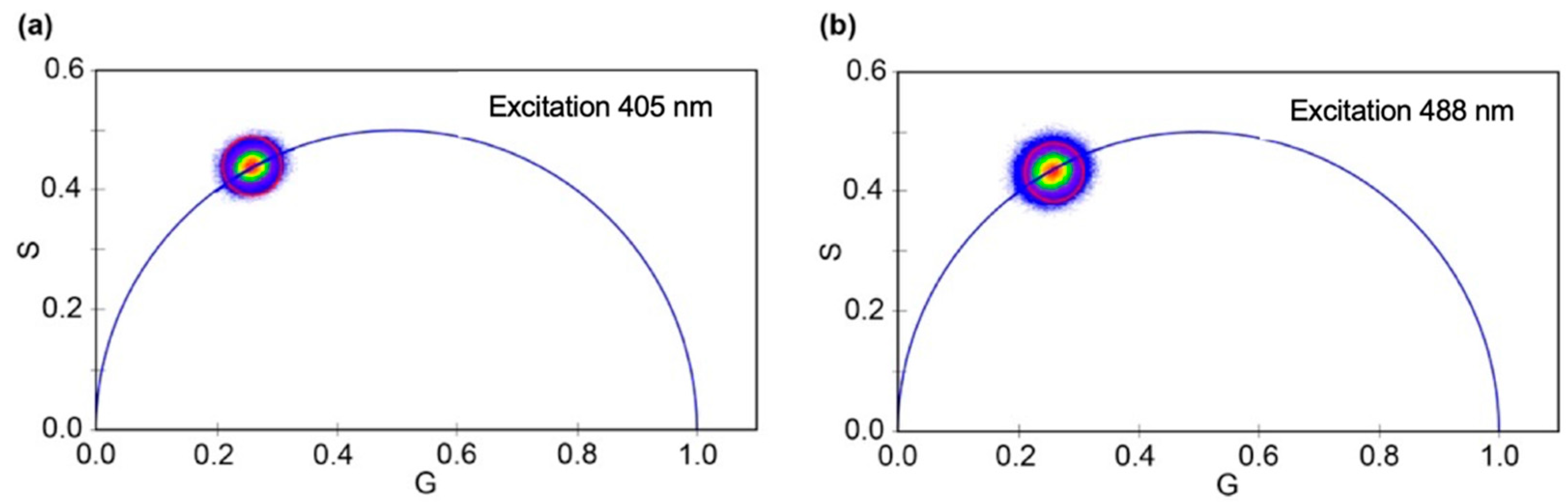
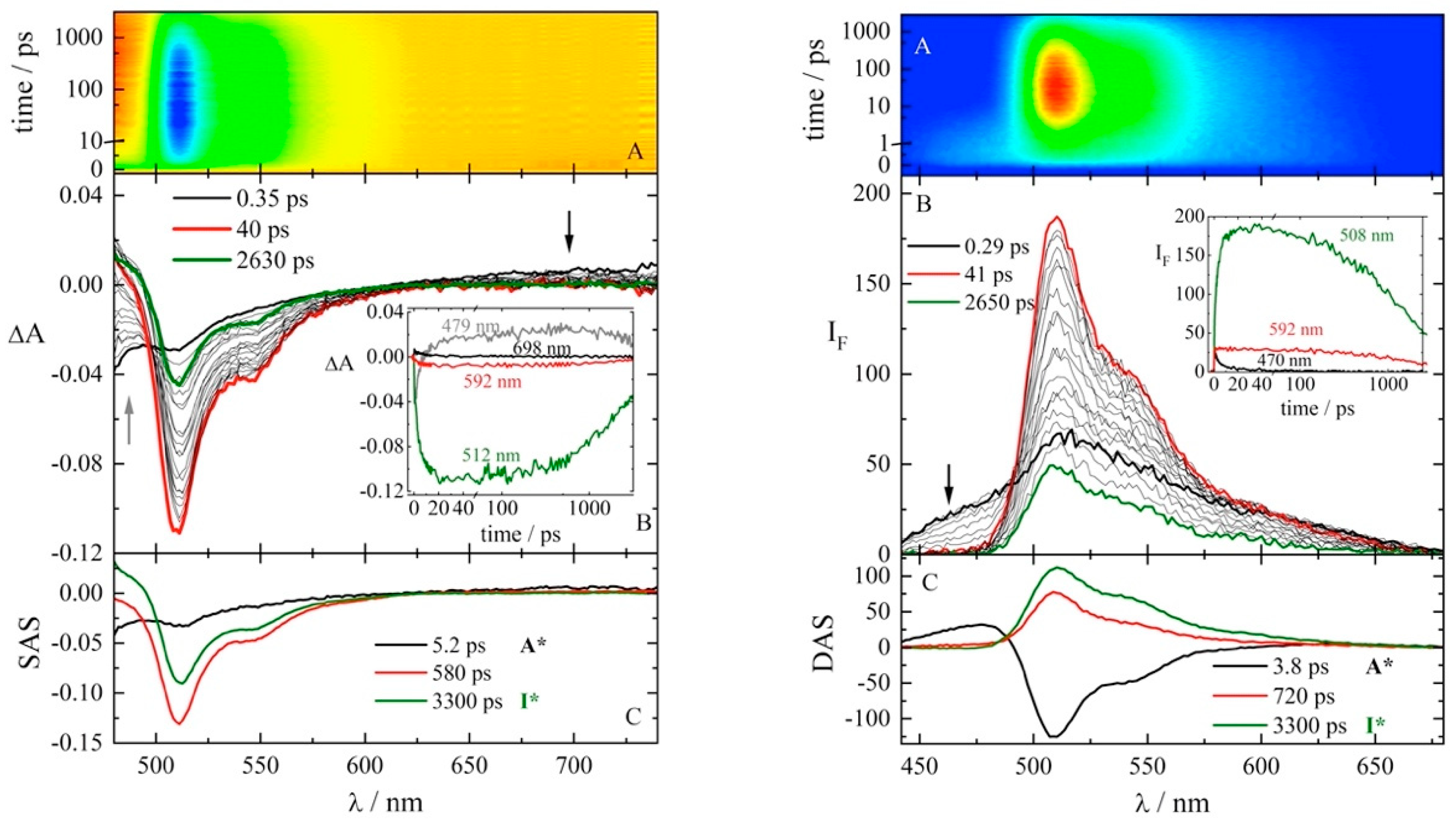
2.2. Time-Resolved Spectrophotometric Measurements
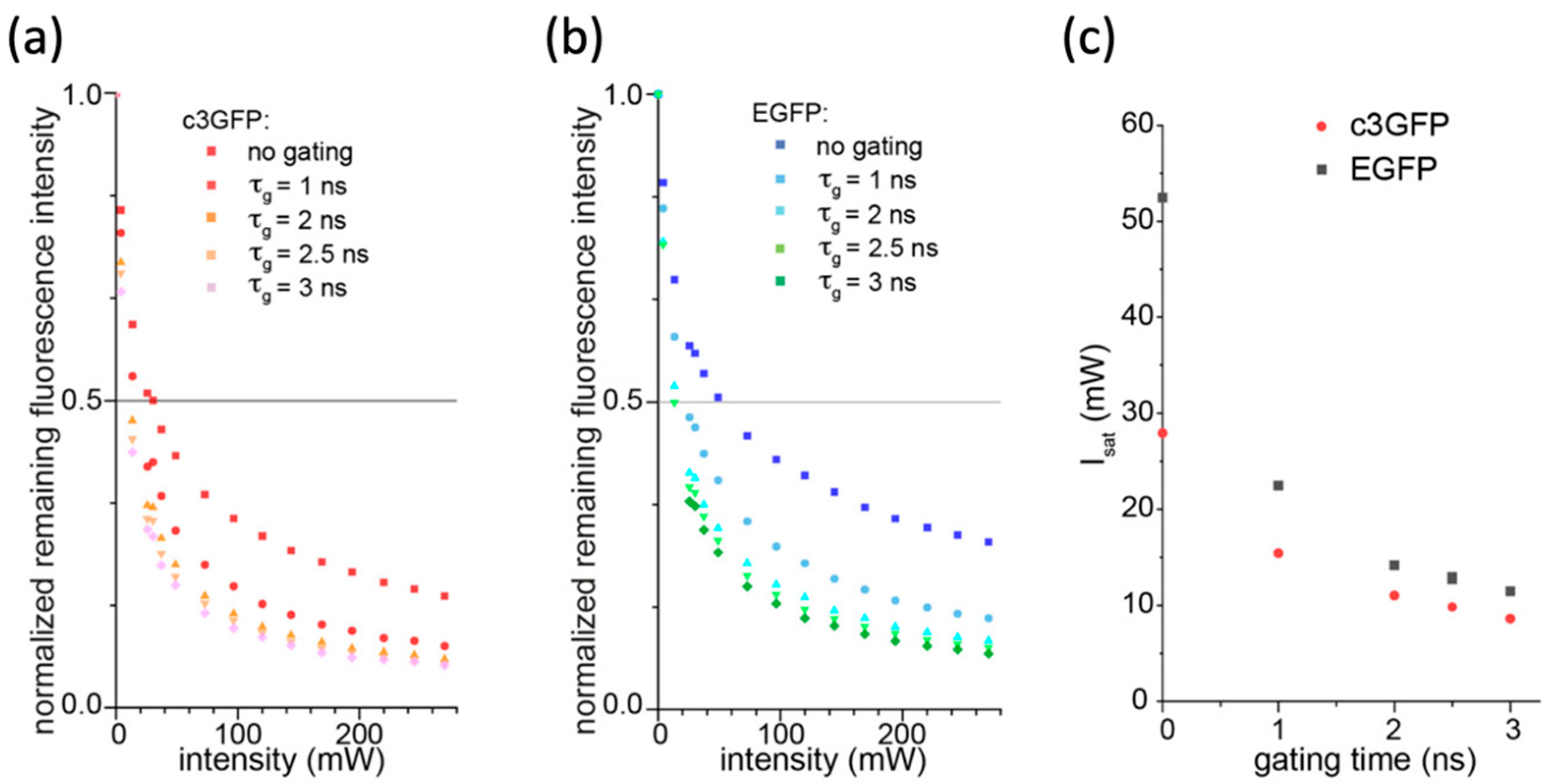
2.3. In Vitro Depletion Properties of c3GFP and Comparison with EGFP
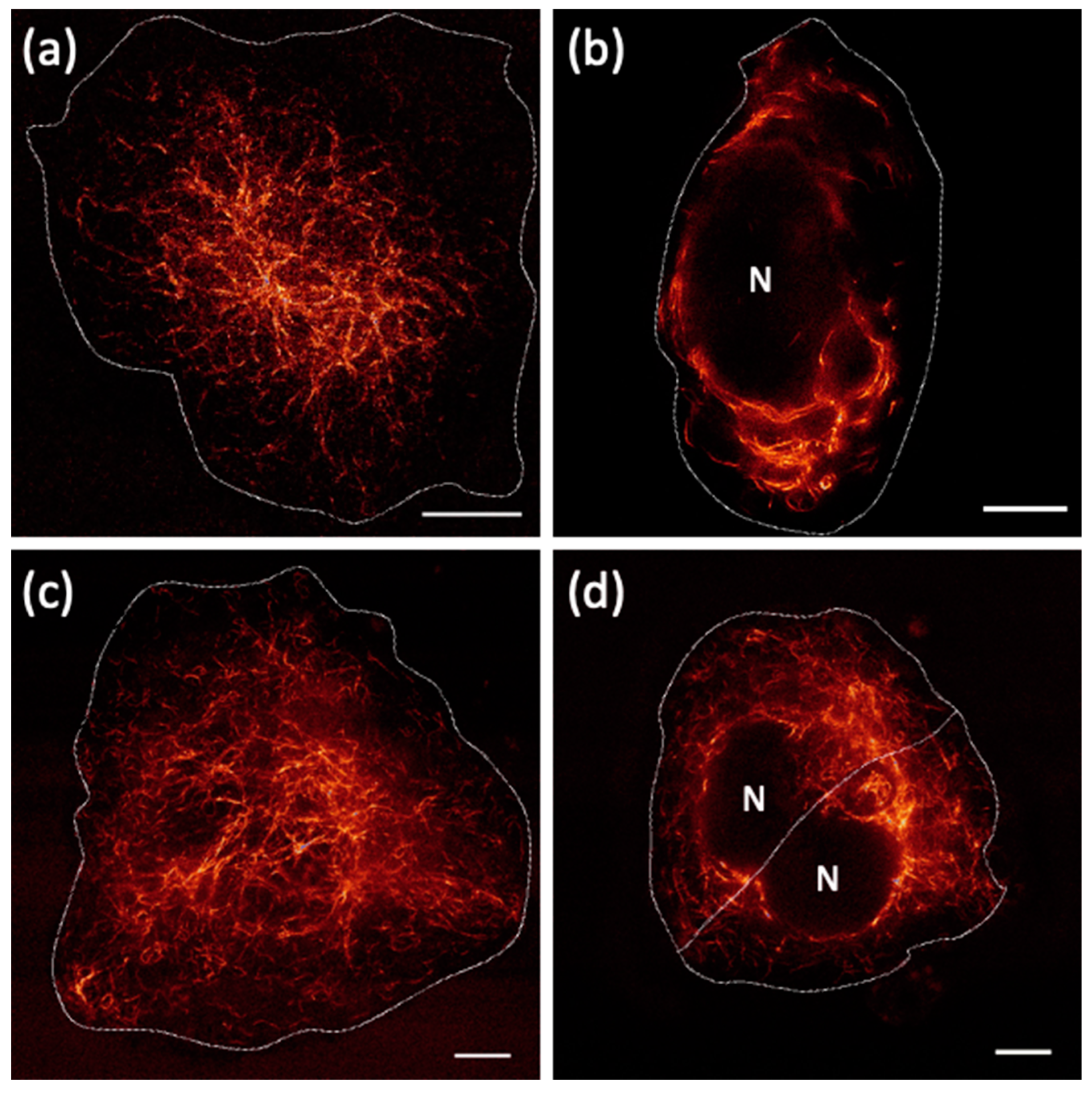
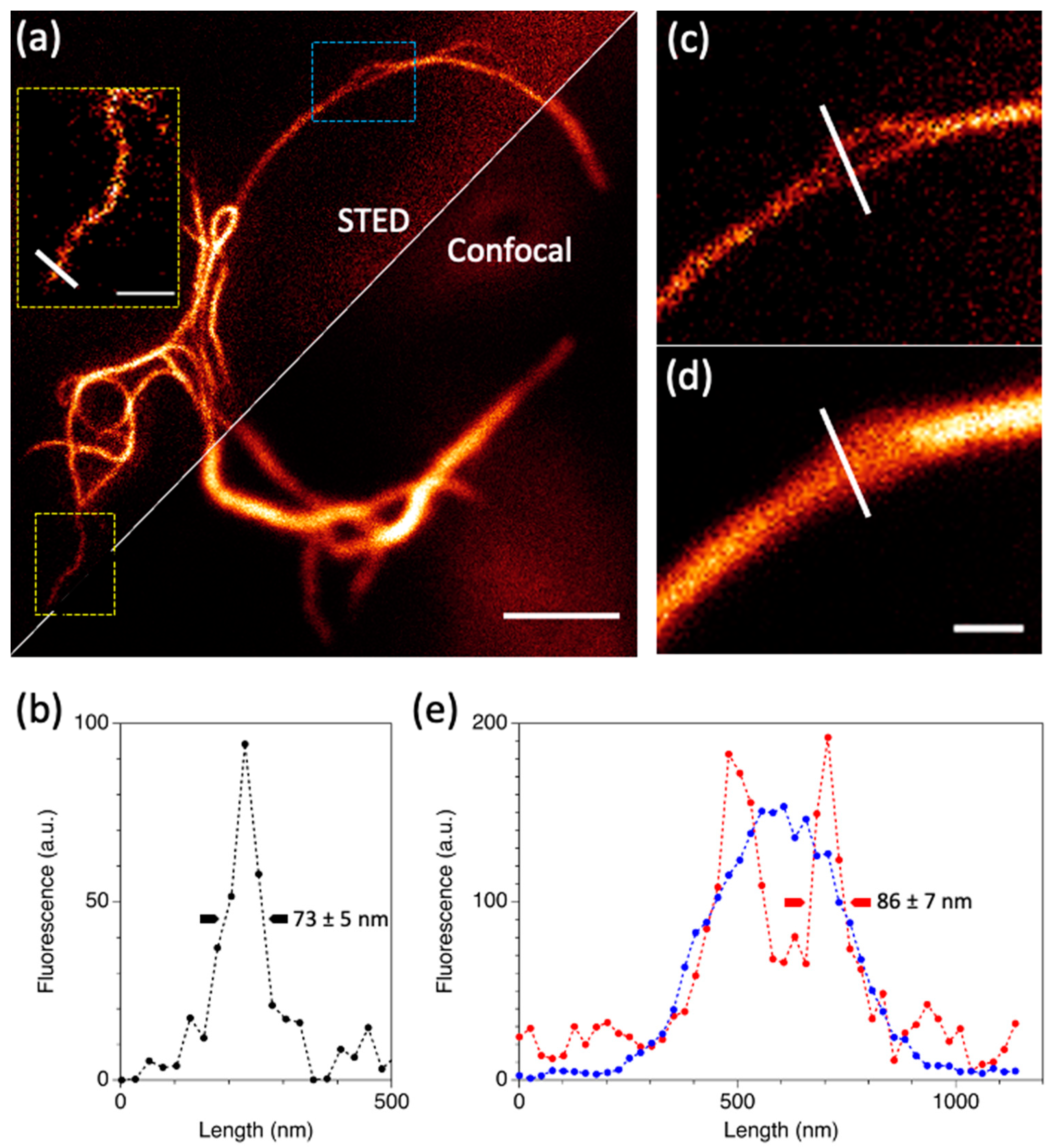
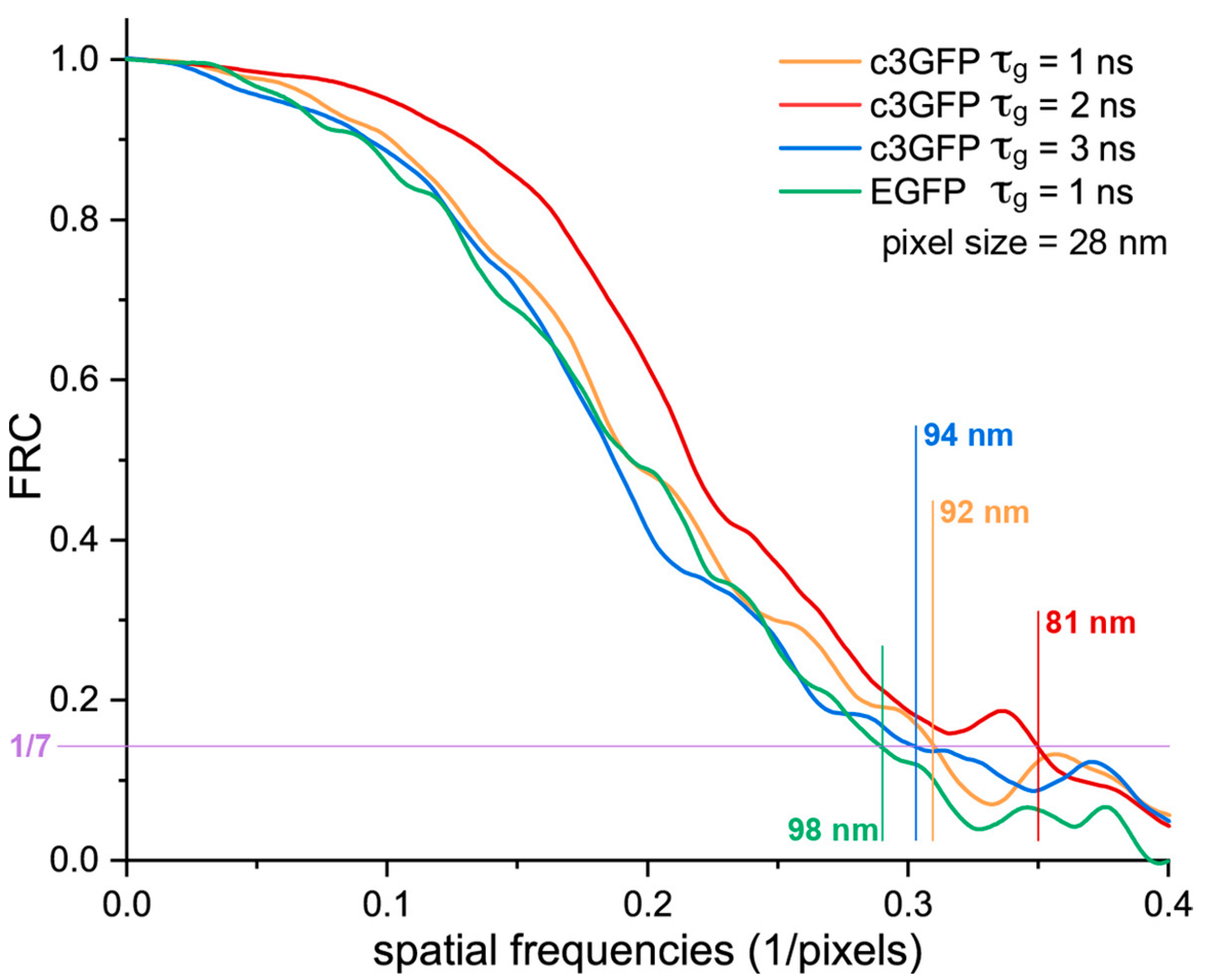
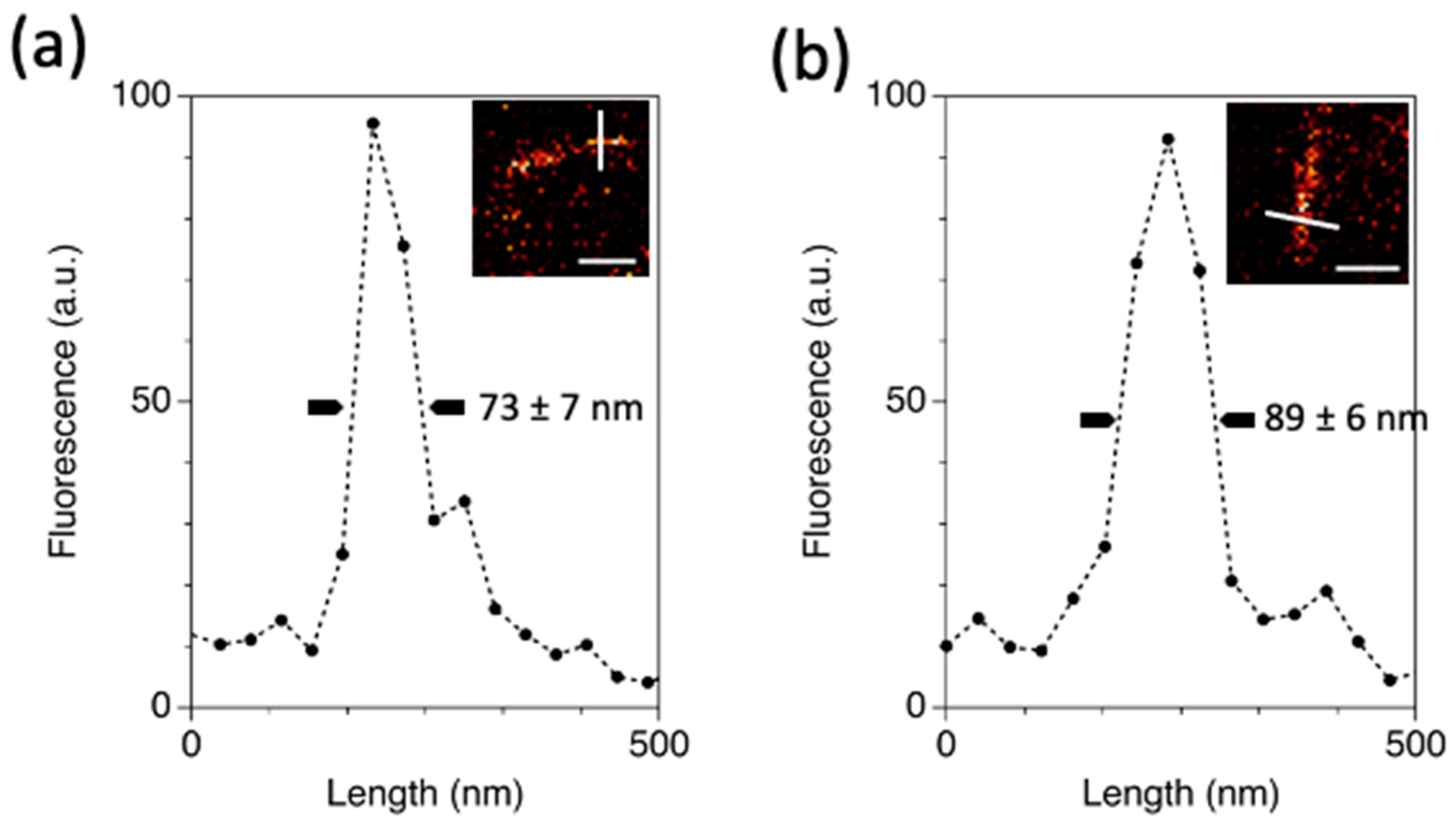
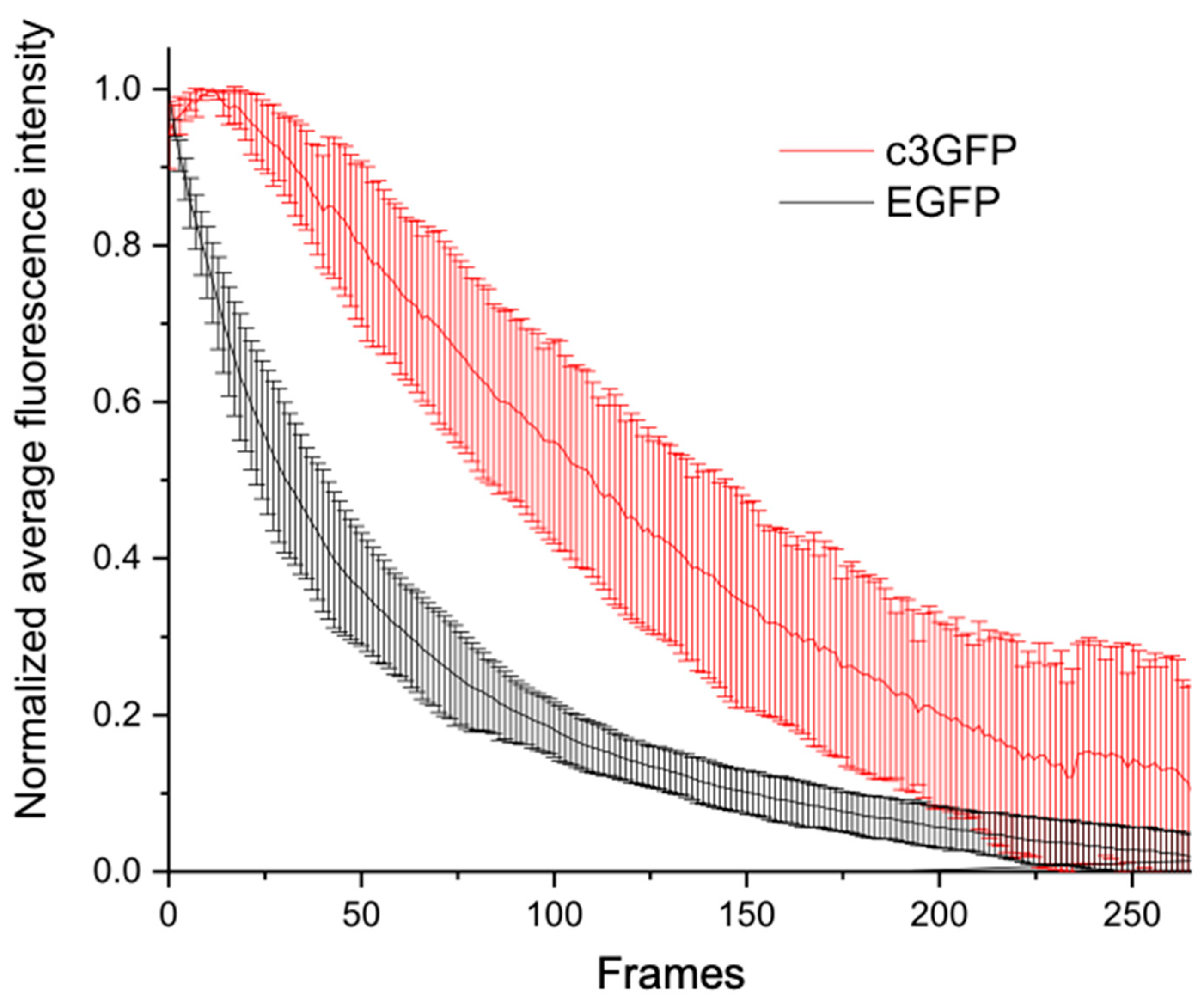
2.4. In Cellulo STED Imaging of c3GFP and Comparison with EGFP
3. Discussion
4. Materials and Methods
4.1. Solvents
4.2. Protein Construction and Expression
4.3. Protein for Eukaryotic Expression
4.4. Cell Cultures and Transfections
4.5. Cell Transfections
4.6. Steady-State Absorption and Fluorescence Spectra
4.7. Molar Absorption Coefficients and Quantum Yields
4.8. Fluorescence Lifetime Detection
4.9. Ultrafast Spectroscopy
4.10. g-STED Measurements
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Theoretical Description of In Vitro Depletion Curve
References
- Hell, S.W. Far-field optical nanoscopy. Science 2007, 316, 1153–1158. [Google Scholar] [CrossRef] [PubMed]
- Masters, B.R. Abbe’s Theory of Image Formation in the Microscope. In Superresolution Optical Microscopy; Rhodes, W.T., Ed.; Springer Series in Optical Sciences; Springer Nature Switzerland AG: Cham, Switzerland, 2020; pp. 65–108. [Google Scholar]
- Sahl, S.J.; Hell, S.W.; Jakobs, S. Fluorescence nanoscopy in cell biology. Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. [Google Scholar] [CrossRef] [PubMed]
- Stender, A.S.; Marchuk, K.; Liu, C.; Sander, S.; Meyer, M.W.; Smith, E.A.; Neupane, B.; Wang, G.F.; Li, J.J.; Cheng, J.X.; et al. Single Cell Optical Imaging and Spectroscopy. Chem. Rev. 2013, 113, 2469–2527. [Google Scholar] [CrossRef] [PubMed]
- Chudakov, D.M.; Matz, M.V.; Lukyanov, S.; Lukyanov, K.A. Fluorescent proteins and their applications in imaging living cells and tissues. Physiol. Rev. 2010, 90, 1103–1163. [Google Scholar] [CrossRef] [PubMed]
- Diaspro, A.; Bianchini, P. Optical nanoscopy. Riv. Nuovo Cim. 2020, 43, 385–455. [Google Scholar] [CrossRef]
- Vicidomini, G.; Bianchini, P.; Diaspro, A. STED super-resolved microscopy. Nat. Methods 2018, 15, 173–182. [Google Scholar] [CrossRef]
- Hell, S.W. Microscopy and its focal switch. Nat. Methods 2009, 6, 24–32. [Google Scholar] [CrossRef]
- Leutenegger, M.; Eggeling, C.; Hell, S.W. Analytical description of STED microscopy performance. Opt. Express 2010, 18, 26417–26429. [Google Scholar] [CrossRef]
- Vicidomini, G.; Moneron, G.; Han, K.Y.; Westphal, V.; Ta, H.; Reuss, M.; Engelhardt, J.; Eggeling, C.; Hell, S.W. Sharper low-power STED nanoscopy by time gating. Nat. Methods 2011, 8, 571–573. [Google Scholar] [CrossRef] [PubMed]
- Jahr, W.; Velicky, P.; Danzl, J.G. Strategies to maximize performance in STimulated Emission Depletion (STED) nanoscopy of biological specimens. Methods 2020, 174, 27–41. [Google Scholar] [CrossRef]
- Hotta, J.; Fron, E.; Dedecker, P.; Janssen, K.P.; Li, C.; Mullen, K.; Harke, B.; Buckers, J.; Hell, S.W.; Hofkens, J. Spectroscopic rationale for efficient stimulated-emission depletion microscopy fluorophores. J. Am. Chem. Soc. 2010, 132, 5021–5023. [Google Scholar] [CrossRef] [PubMed]
- Sednev, M.V.; Belov, V.N.; Hell, S.W. Fluorescent dyes with large Stokes shifts for super-resolution optical microscopy of biological objects: A review. Methods Appl. Fluores. 2015, 3, 042004. [Google Scholar] [CrossRef] [PubMed]
- Muller, T.; Schumann, C.; Kraegeloh, A. STED Microscopy and its Applications: New Insights into Cellular Processes on the Nanoscale. Chemphyschem 2012, 13, 1986–2000. [Google Scholar] [CrossRef] [PubMed]
- Nienhaus, K.; Nienhaus, G.U. Fluorescent proteins for live-cell imaging with super-resolution. Chem. Soc. Rev. 2014, 43, 1088–1106. [Google Scholar] [CrossRef]
- Willig, K.I.; Kellner, R.R.; Medda, R.; Hein, B.; Jakobs, S.; Hell, S.W. Nanoscale resolution in GFP-based microscopy. Nat. Methods 2006, 3, 721–723. [Google Scholar] [CrossRef]
- Rankin, B.R.; Moneron, G.; Wurm, C.A.; Nelson, J.C.; Walter, A.; Schwarzer, D.; Schroeder, J.; Colon-Ramos, D.A.; Hell, S.W. Nanoscopy in a living multicellular organism expressing GFP. Biophys. J. 2011, 100, L63–L65. [Google Scholar] [CrossRef]
- Bianchini, P.; Cardarelli, F.; Di Luca, M.; Diaspro, A.; Bizzarri, R. Nanoscale Protein Diffusion by STED-Based Pair Correlation Analysis. PLoS ONE 2014, 9, e99619. [Google Scholar] [CrossRef]
- Nagerl, U.V.; Willig, K.I.; Hein, B.; Hell, S.W.; Bonhoeffer, T. Live-cell imaging of dendritic spines by STED microscopy. Proc. Natl. Acad. Sci. USA 2008, 105, 18982–18987. [Google Scholar] [CrossRef]
- Hein, B.; Willig, K.I.; Hell, S.W. Stimulated emission depletion (STED) nanoscopy of a fluorescent protein-labeled organelle inside a living cell. Proc. Natl. Acad. Sci. USA 2008, 105, 14271–14276. [Google Scholar] [CrossRef]
- Cardarelli, F.; Bizzarri, R.; Serresi, M.; Albertazzi, L.; Beltram, F. Probing nuclear localization signal-importin alpha binding equilibria in living cells. J. Biol. Chem. 2009, 284, 36638–36646. [Google Scholar] [CrossRef]
- Heikal, A.A.; Hess, S.T.; Baird, G.S.; Tsien, R.Y.; Webb, W.W. Molecular spectroscopy and dynamics of intrinsically fluorescent proteins: Coral red (dsRed) and yellow (Citrine). Proc. Natl. Acad. Sci. USA 2000, 97, 11996–12001. [Google Scholar] [CrossRef] [PubMed]
- Schwille, P.; Kummer, S.; Heikal, A.A.; Moerner, W.E.; Webb, W.W. Fluorescence correlation spectroscopy reveals fast optical excitation-driven intramolecular dynamics of yellow fluorescent proteins. Proc. Natl. Acad. Sci. USA 2000, 97, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Jacchetti, E.; Gabellieri, E.; Cioni, P.; Bizzarri, R.; Nifosi, R. Temperature and pressure effects on GFP mutants: Explaining spectral changes by molecular dynamics simulations and TD-DFT calculations. Phys. Chem. Chem. Phys. 2016, 18, 12828–12838. [Google Scholar] [CrossRef] [PubMed]
- Bizzarri, R.; Nifosi, R.; Abbruzzetti, S.; Rocchia, W.; Guidi, S.; Arosio, D.; Garau, G.; Campanini, B.; Grandi, E.; Ricci, F.; et al. Green Fluorescent Protein Ground States: The Influence of a Second Protonation Site near the Chromophore. Biochemistry 2007, 46, 5494–5504. [Google Scholar] [CrossRef]
- Wiehler, J.; Jung, G.; Seebacher, C.; Zumbusch, A.; Steipe, B. Mutagenic stabilization of the photocycle intermediate of green fluorescent protein (GFP). Chembiochem 2003, 4, 1164–1171. [Google Scholar] [CrossRef]
- Chattoraj, M.; King, B.A.; Bublitz, G.U.; Boxer, S.G. Ultra-fast excited state dynamics in green fluorescent protein: Multiple states and proton transfer. Proc. Natl. Acad. Sci. USA 1996, 93, 8362–8367. [Google Scholar] [CrossRef]
- Voityuk, A.A.; Michel-Beyerle, M.E.; Rosch, N. Quantum chemical modeling of structure and absorption spectra of the chromophore in green fluorescent proteins. Chem. Phys. 1998, 231, 13–25. [Google Scholar] [CrossRef]
- Creemers, T.M.; Lock, A.J.; Subramaniam, V.; Jovin, T.M.; Volker, S. Three photoconvertible forms of green fluorescent protein identified by spectral hole-burning. Nat. Struct. Biol. 1999, 6, 557–560. [Google Scholar] [CrossRef][Green Version]
- Abbruzzetti, S.; Grandi, E.; Viappiani, C.; Bologna, S.; Campanini, B.; Raboni, S.; Bettati, S.; Mozzarelli, A. Kinetics of acid-induced spectral changes in the GFPmut2 chromophore. J. Am. Chem. Soc. 2005, 127, 626–635. [Google Scholar] [CrossRef]
- Morise, H.; Shimomura, O.; Johnson, F.H.; Winant, J. Intermolecular energy transfer in the bioluminescent system of Aequorea. Biochemistry 1974, 13, 2656–2662. [Google Scholar] [CrossRef]
- Patterson, G.H.; Knobel, S.M.; Sharif, W.D.; Kain, S.R.; Piston, D.W. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys. J. 1997, 73, 2782–2790. [Google Scholar] [CrossRef]
- Stefl, M.; James, N.G.; Ross, J.A.; Jameson, D.M. Applications of phasors to in vitro time-resolved fluorescence measurements. Anal. Biochem. 2011, 410, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Kennis, J.T.; Larsen, D.S.; van Stokkum, I.H.; Vengris, M.; van Thor, J.J.; van Grondelle, R. Uncovering the hidden ground state of green fluorescent protein. Proc. Natl. Acad. Sci. USA 2004, 101, 17988–17993. [Google Scholar] [CrossRef] [PubMed]
- Gather, M.C.; Yun, S.H. Bio-optimized energy transfer in densely packed fluorescent protein enables near-maximal luminescence and solid-state lasers. Nat. Commun. 2014, 5, 5722. [Google Scholar] [CrossRef]
- Oh, H.J.; Gather, M.C.; Song, J.J.; Yun, S.H. Lasing from fluorescent protein crystals. Opt. Express 2014, 22, 31411–31416. [Google Scholar] [CrossRef]
- Bianchini, P.; Harke, B.; Galiani, S.; Vicidomini, G.; Diaspro, A. Single-wavelength two-photon excitation-stimulated emission depletion (SW2PE-STED) superresolution imaging. Proc. Natl. Acad. Sci. USA 2012, 109, 6390–6393. [Google Scholar] [CrossRef]
- Galiani, S.; Harke, B.; Vicidomini, G.; Lignani, G.; Benfenati, F.; Diaspro, A.; Bianchini, P. Strategies to maximize the performance of a STED microscope. Opt. Express 2012, 20, 7362–7374. [Google Scholar] [CrossRef]
- Tortarolo, G.; Castello, M.; Diaspro, A.; Koho, S.; Vicidomini, G. Evaluating image resolution in stimulated emission depletion microscopy. Optica 2018, 5, 32–35. [Google Scholar] [CrossRef]
- Kreplak, L.; Bar, H.; Leterrier, J.F.; Herrmann, H.; Aebi, U. Exploring the mechanical behavior of single intermediate filaments. J. Mol. Biol. 2005, 354, 569–577. [Google Scholar] [CrossRef]
- Lowery, J.; Kuczmarski, E.R.; Herrmann, H.; Goldman, R.D. Intermediate Filaments Play a Pivotal Role in Regulating Cell Architecture and Function. J. Biol. Chem. 2015, 290, 17145–17153. [Google Scholar] [CrossRef] [PubMed]
- Chung, B.M.; Rotty, J.D.; Coulombe, P.A. Networking galore: Intermediate filaments and cell migration. Curr. Opin. Cell Biol. 2013, 25, 600–612. [Google Scholar] [CrossRef]
- Danielsson, F.; Peterson, M.K.; Araujo, H.C.; Lautenschlager, F.; Gad, A.K.B. Vimentin Diversity in Health and Disease. Cells 2018, 7, 147. [Google Scholar] [CrossRef] [PubMed]
- Grotjohann, T.; Testa, I.; Reuss, M.; Brakemann, T.; Eggeling, C.; Hell, S.W.; Jakobs, S. rsEGFP2 enables fast RESOLFT nanoscopy of living cells. eLife 2012, 1, e00248. [Google Scholar] [CrossRef] [PubMed]
- Crameri, A.; Whitehorn, E.A.; Tate, E.; Stemmer, W.P.C. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat. Biotechnol. 1996, 14, 315–319. [Google Scholar] [CrossRef] [PubMed]
- Battistutta, R.; Negro, A.; Zanotti, G. Crystal structure and refolding properties of the mutant F99S/M153T/V163A of the Green Fluorescent Protein. Proteins-Struct. Funct. Bioinform. 2000, 41, 429–437. [Google Scholar] [CrossRef]
- Borst, J.W.; Hink, M.A.; van Hoek, A.; Visser, A.J.W.G. Effects of refractive index and viscosity on fluorescence and anisotropy decays of enhanced cyan and yellow fluorescent proteins. J. Fluoresc. 2005, 15, 153–160. [Google Scholar] [CrossRef]
- Battisti, A.; Digman, M.A.; Gratton, E.; Storti, B.; Beltram, F.; Bizzarri, R. Intracellular pH measurements made simple by fluorescent protein probes and the phasor approach to fluorescence lifetime imaging. Chem. Commun. 2012, 48, 5127–5129. [Google Scholar] [CrossRef]
- Storti, B.; Margheritis, E.; Abbandonato, G.; Domenichini, G.; Dreier, J.; Testa, I.; Garau, G.; Nifosi, R.; Bizzarri, R. Role of Gln222 in Photoswitching of Aequorea Fluorescent Proteins: A Twisting and H-Bonding Affair? ACS Chem. Biol. 2018, 13, 2082–2093. [Google Scholar] [CrossRef]
- Rout, Y.; Montanari, C.; Pasciucco, E.; Misra, R.; Carlotti, B. Tuning the Fluorescence and the Intramolecular Charge Transfer of Phenothiazine Dipolar and Quadrupolar Derivatives by Oxygen Functionalization. J. Am. Chem. Soc. 2021, 143, 9933–9943. [Google Scholar] [CrossRef]
- Dai, W.; Bianconi, T.; Ferraguzzi, E.; Wu, X.; Lei, Y.; Shi, J.; Tong, B.; Carlotti, B.; Cai, Z.; Dong, Y. Excited-State Modulation of Aggregation-Induced Emission Molecules for High-Efficiency Triplet Exciton Generation. ACS Mater. Lett. 2021, 3, 1767–1777. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Storti, B.; Carlotti, B.; Chiellini, G.; Ruglioni, M.; Salvadori, T.; Scotto, M.; Elisei, F.; Diaspro, A.; Bianchini, P.; Bizzarri, R. An Efficient Aequorea victoria Green Fluorescent Protein for Stimulated Emission Depletion Super-Resolution Microscopy. Int. J. Mol. Sci. 2022, 23, 2482. https://doi.org/10.3390/ijms23052482
Storti B, Carlotti B, Chiellini G, Ruglioni M, Salvadori T, Scotto M, Elisei F, Diaspro A, Bianchini P, Bizzarri R. An Efficient Aequorea victoria Green Fluorescent Protein for Stimulated Emission Depletion Super-Resolution Microscopy. International Journal of Molecular Sciences. 2022; 23(5):2482. https://doi.org/10.3390/ijms23052482
Chicago/Turabian StyleStorti, Barbara, Benedetta Carlotti, Grazia Chiellini, Martina Ruglioni, Tiziano Salvadori, Marco Scotto, Fausto Elisei, Alberto Diaspro, Paolo Bianchini, and Ranieri Bizzarri. 2022. "An Efficient Aequorea victoria Green Fluorescent Protein for Stimulated Emission Depletion Super-Resolution Microscopy" International Journal of Molecular Sciences 23, no. 5: 2482. https://doi.org/10.3390/ijms23052482
APA StyleStorti, B., Carlotti, B., Chiellini, G., Ruglioni, M., Salvadori, T., Scotto, M., Elisei, F., Diaspro, A., Bianchini, P., & Bizzarri, R. (2022). An Efficient Aequorea victoria Green Fluorescent Protein for Stimulated Emission Depletion Super-Resolution Microscopy. International Journal of Molecular Sciences, 23(5), 2482. https://doi.org/10.3390/ijms23052482







