Deposition of Lead Phosphate by Lead-Tolerant Bacteria Isolated from Fresh Water near an Abandoned Mine
Abstract
1. Introduction
2. Results
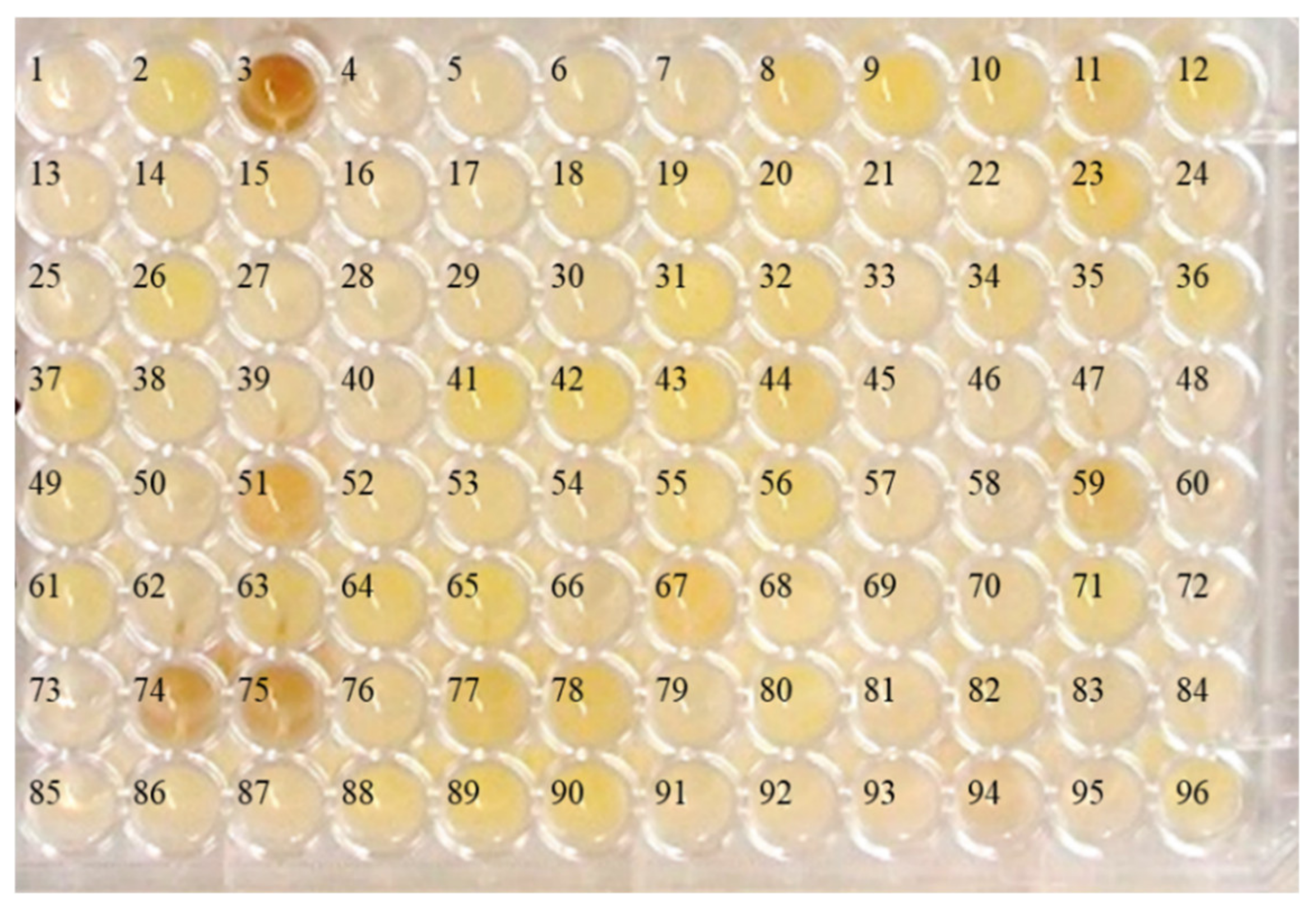
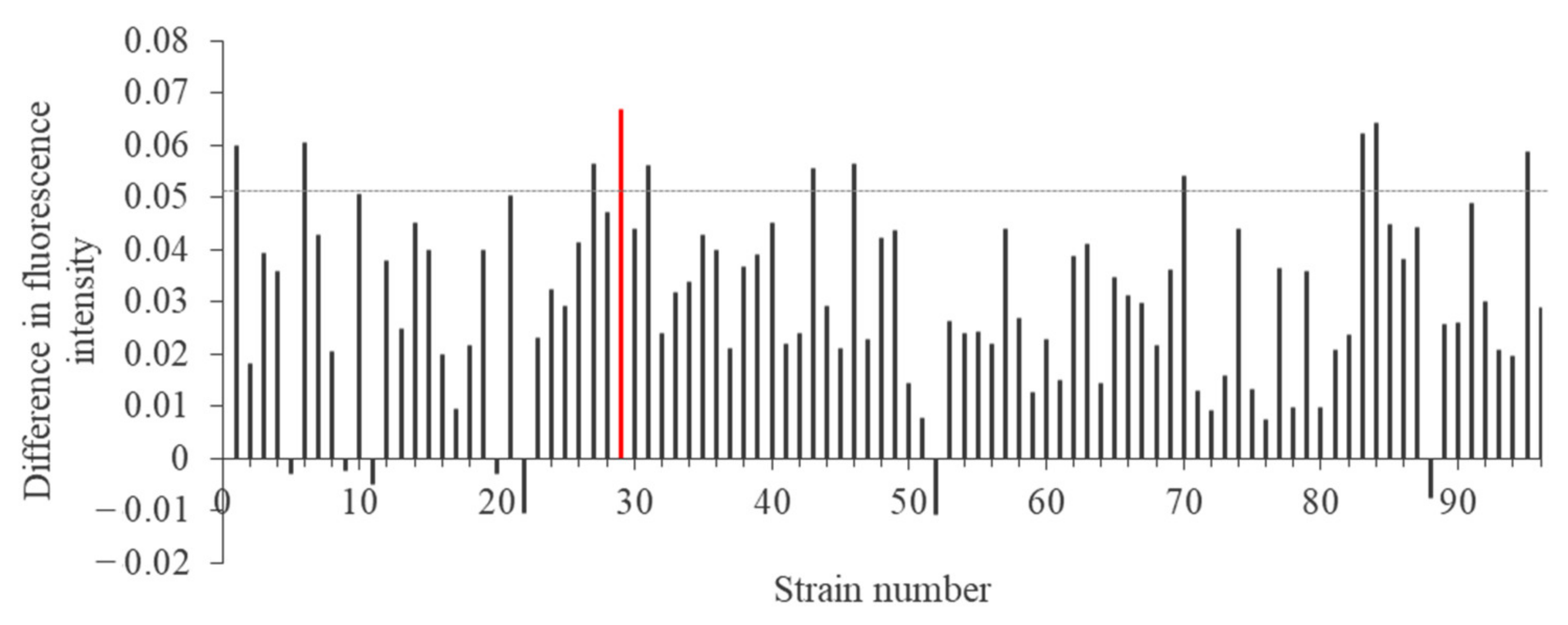
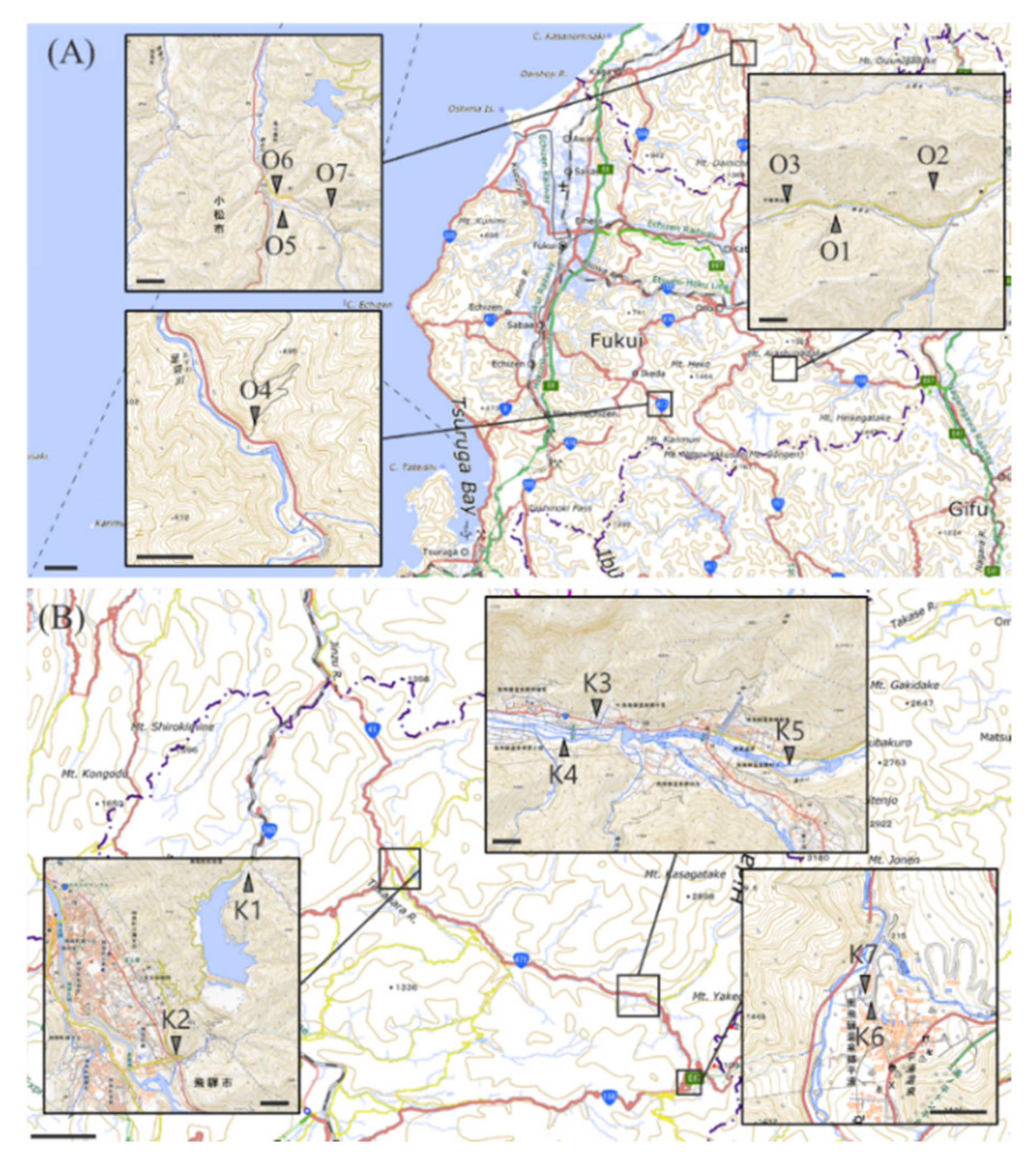
2.1. Isolation of Bacteria from an Abandoned Mine
2.2. Appropriate Conditions for Synthesis of Lead Nanoparticles
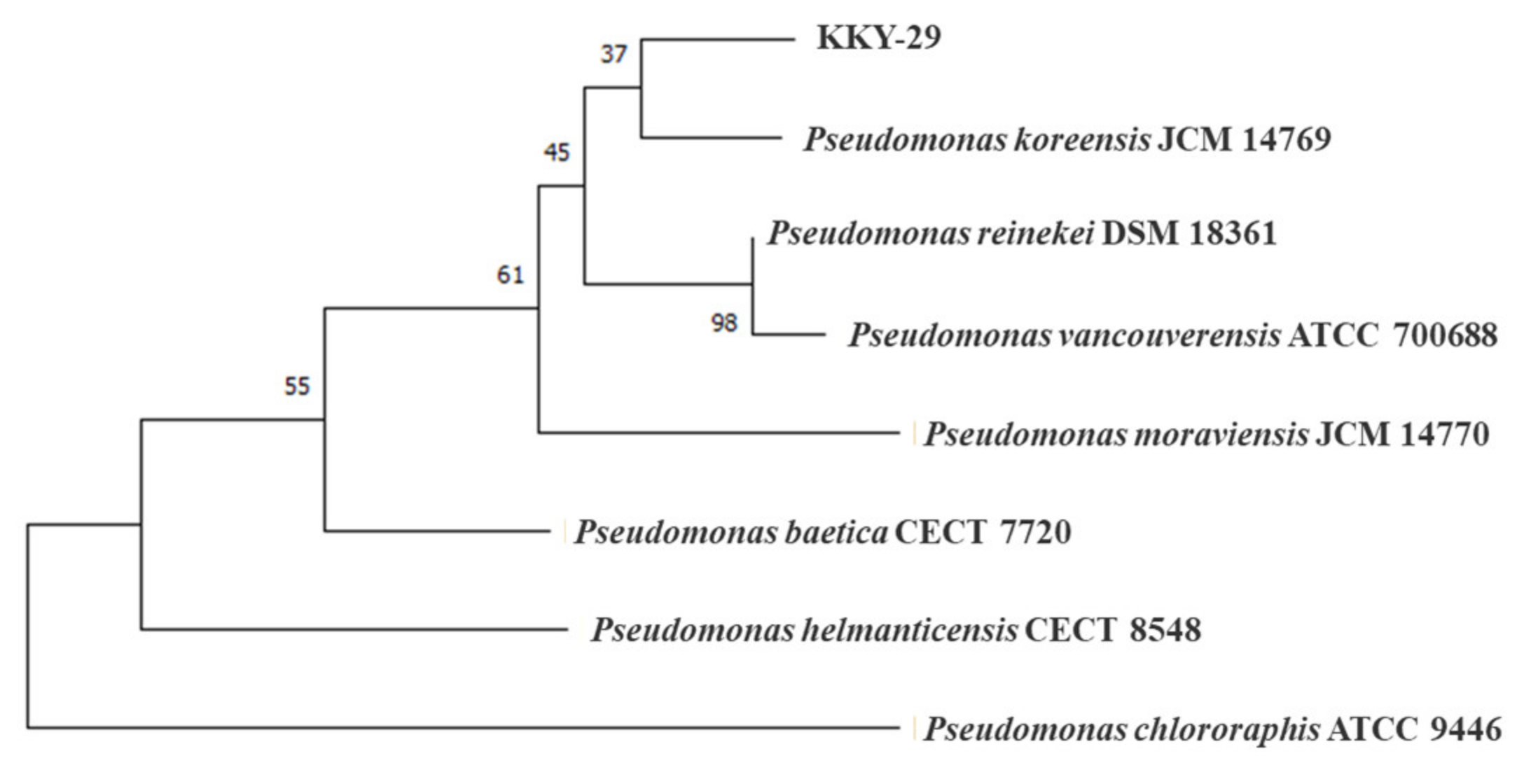
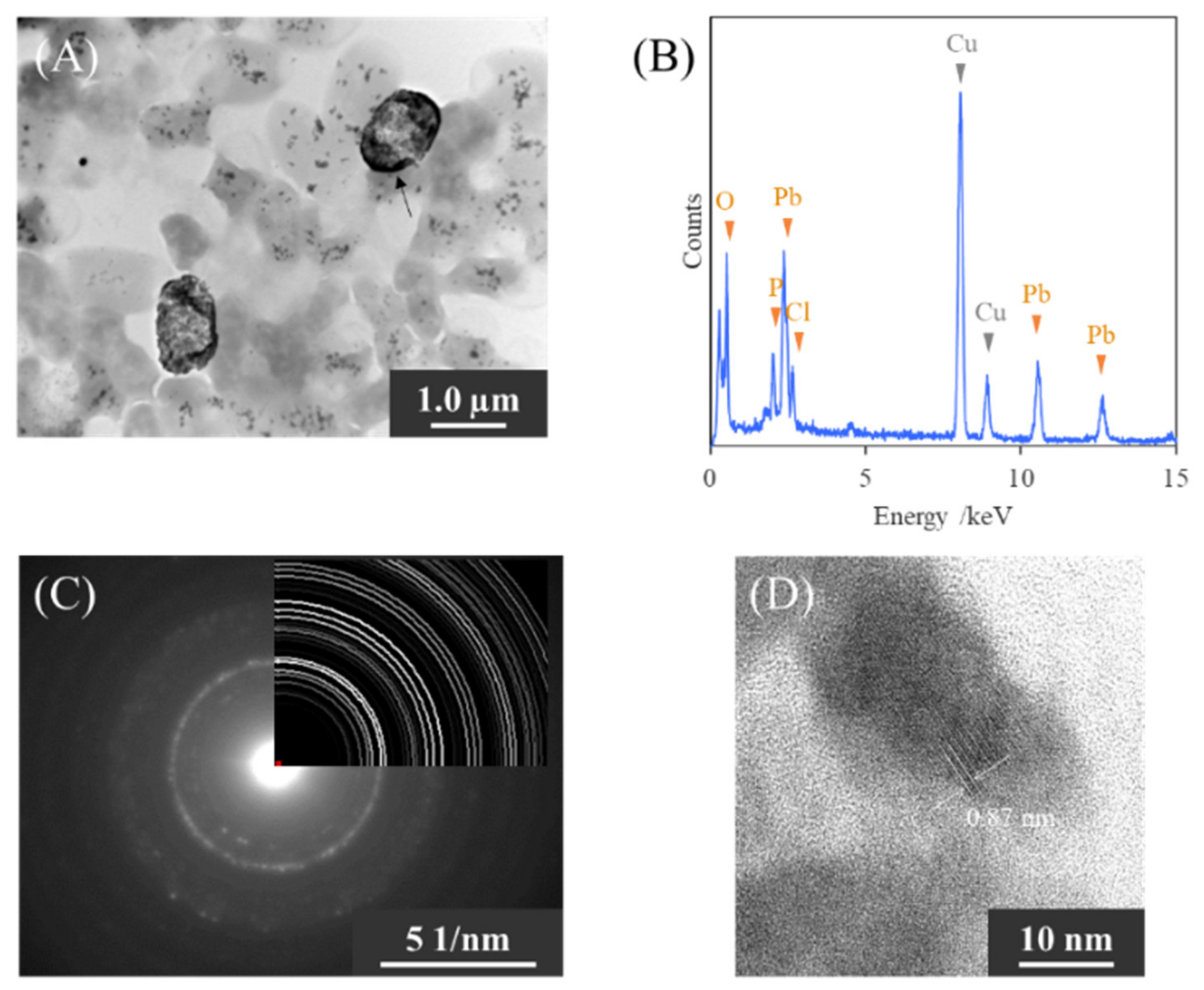
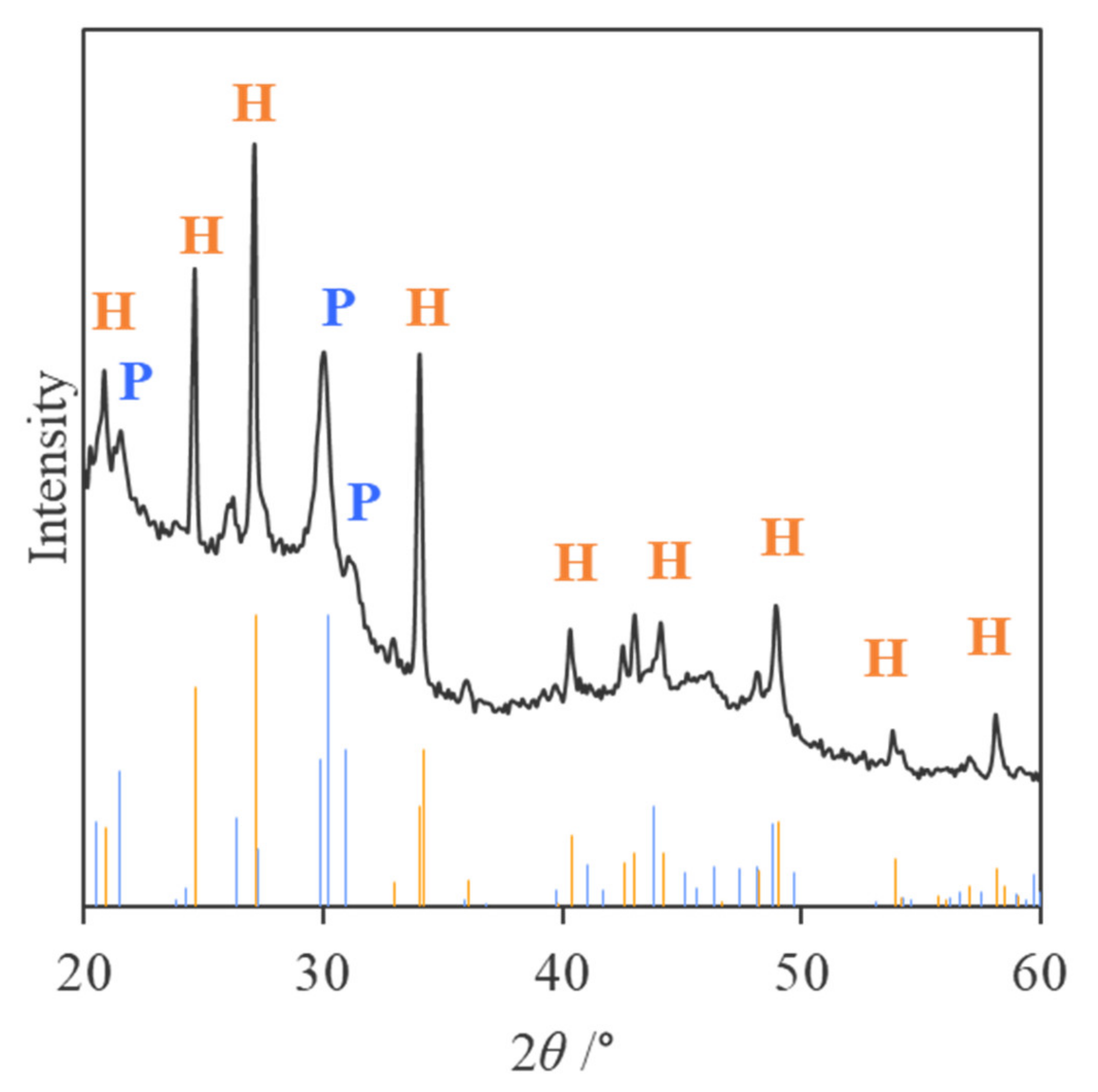
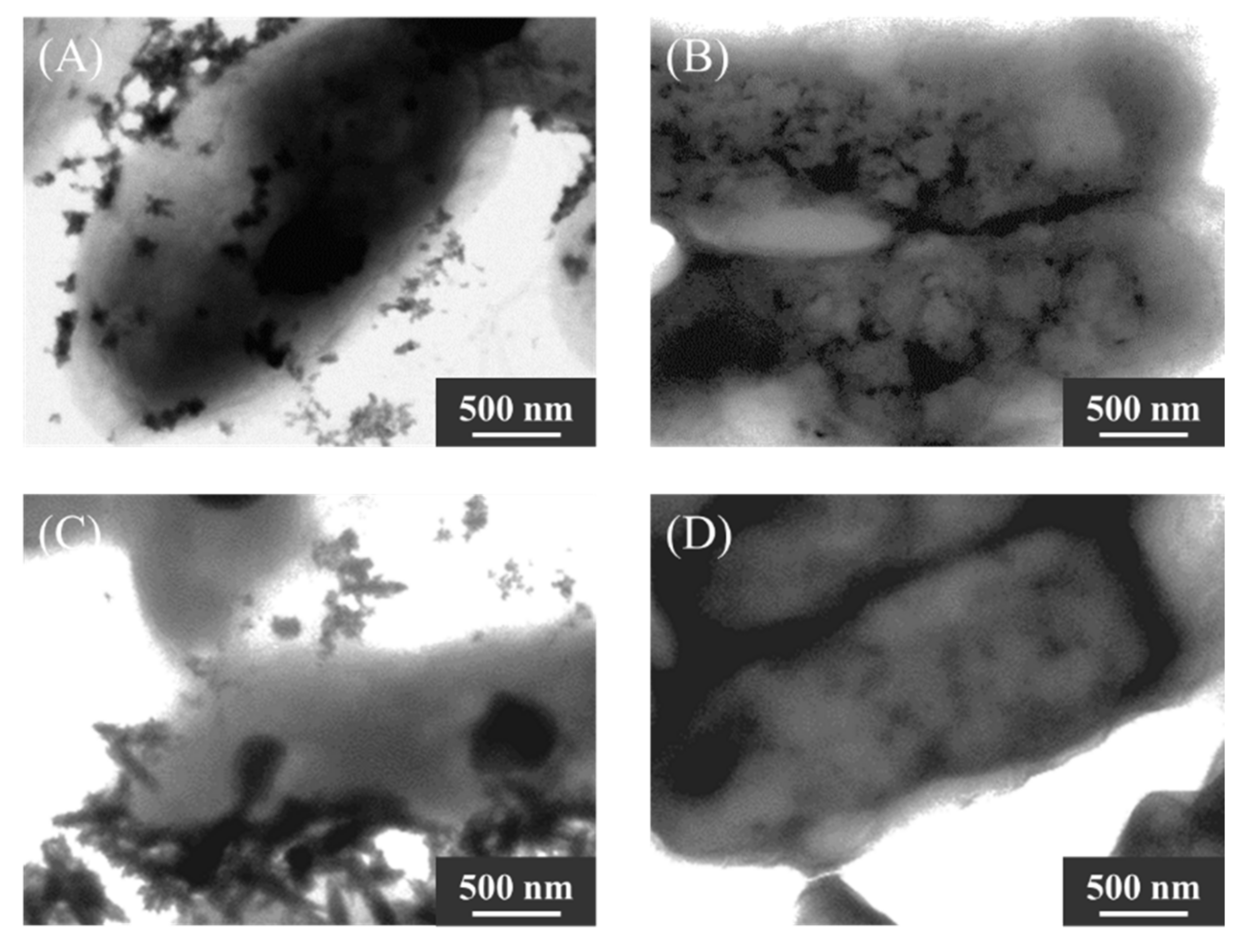
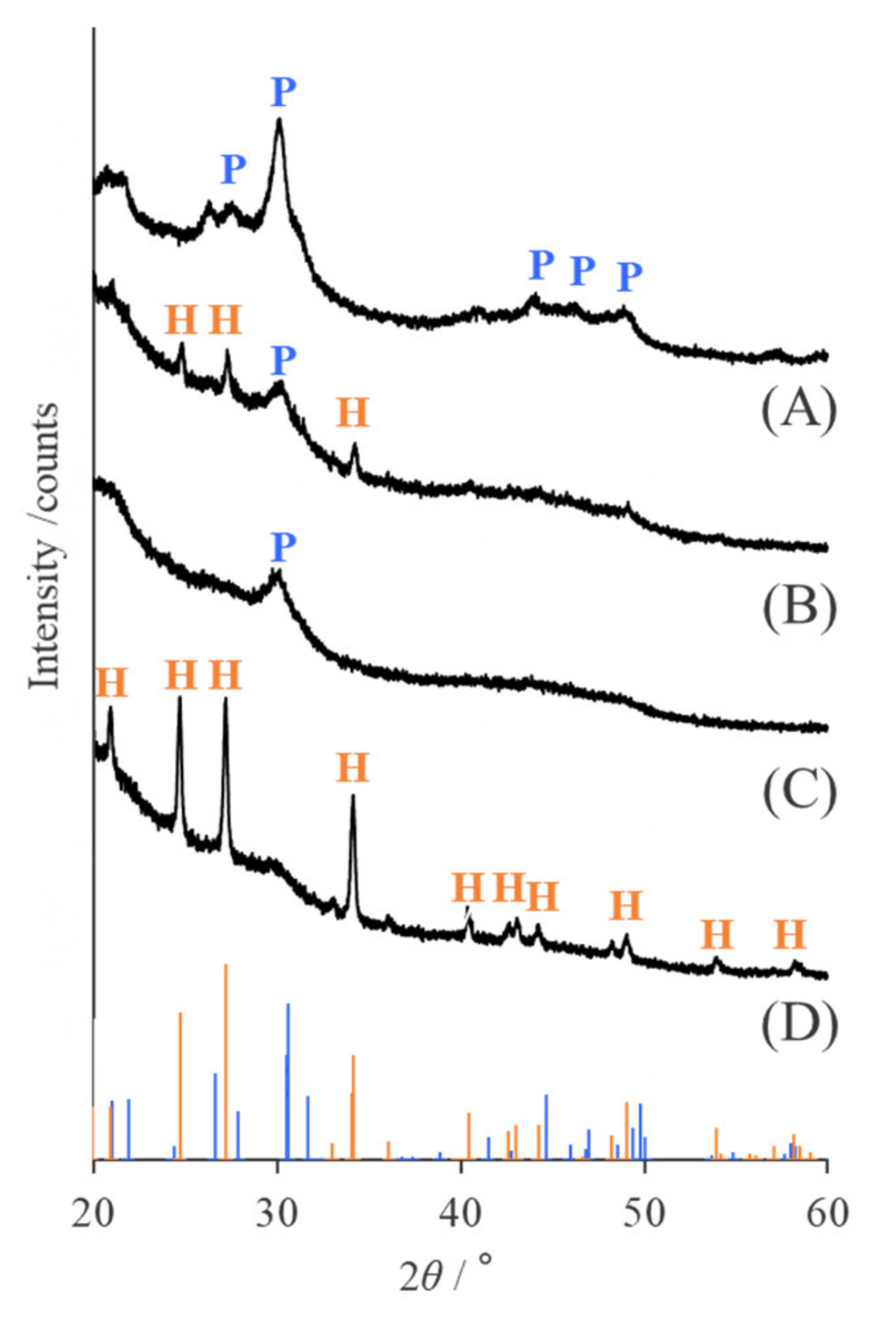
2.3. Identification of Mineral Species
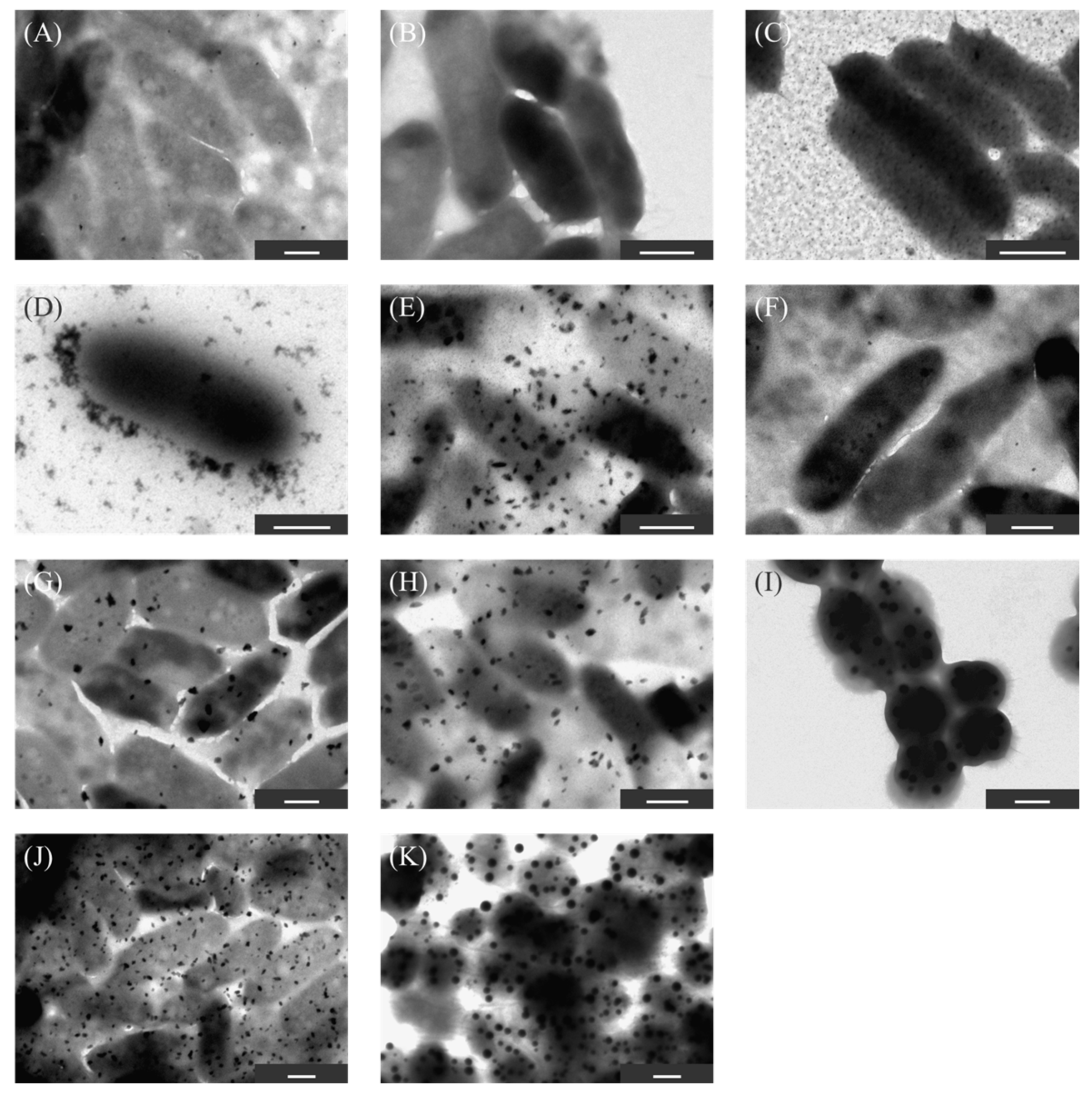
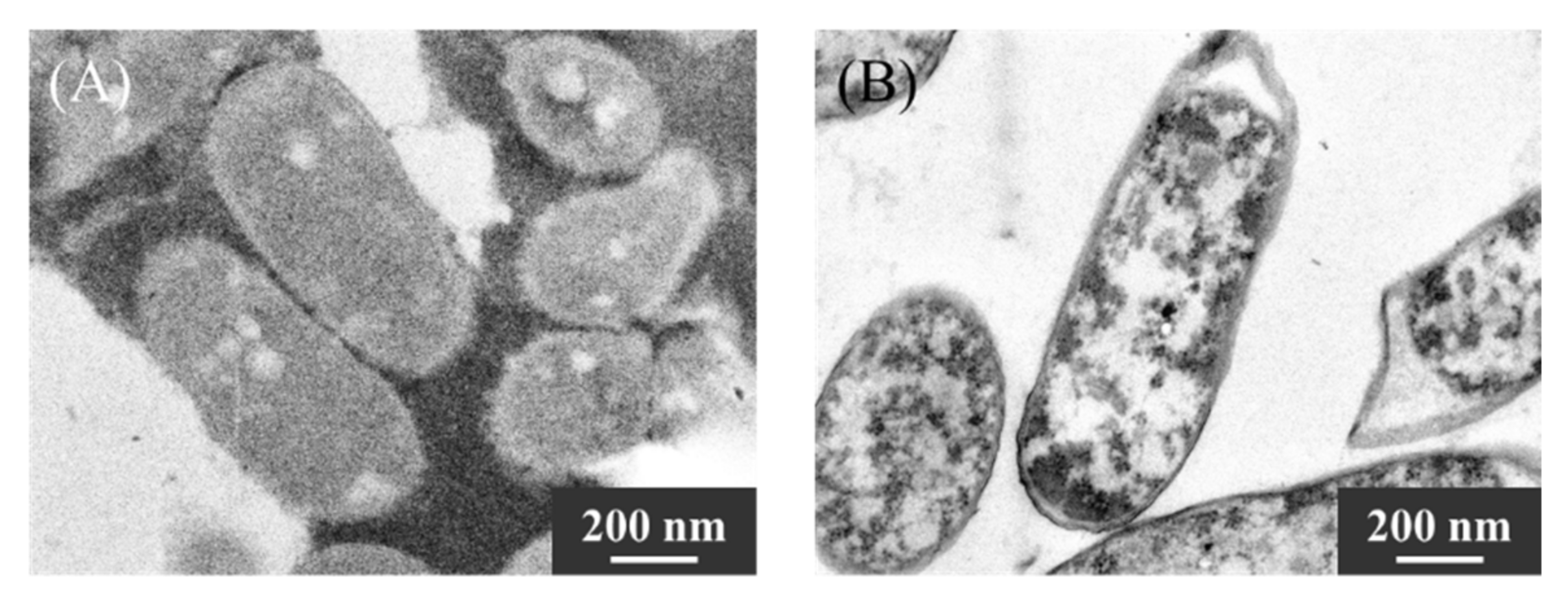
2.4. Evaluation of Ultra-Thin Cross-Sections of KKY-29
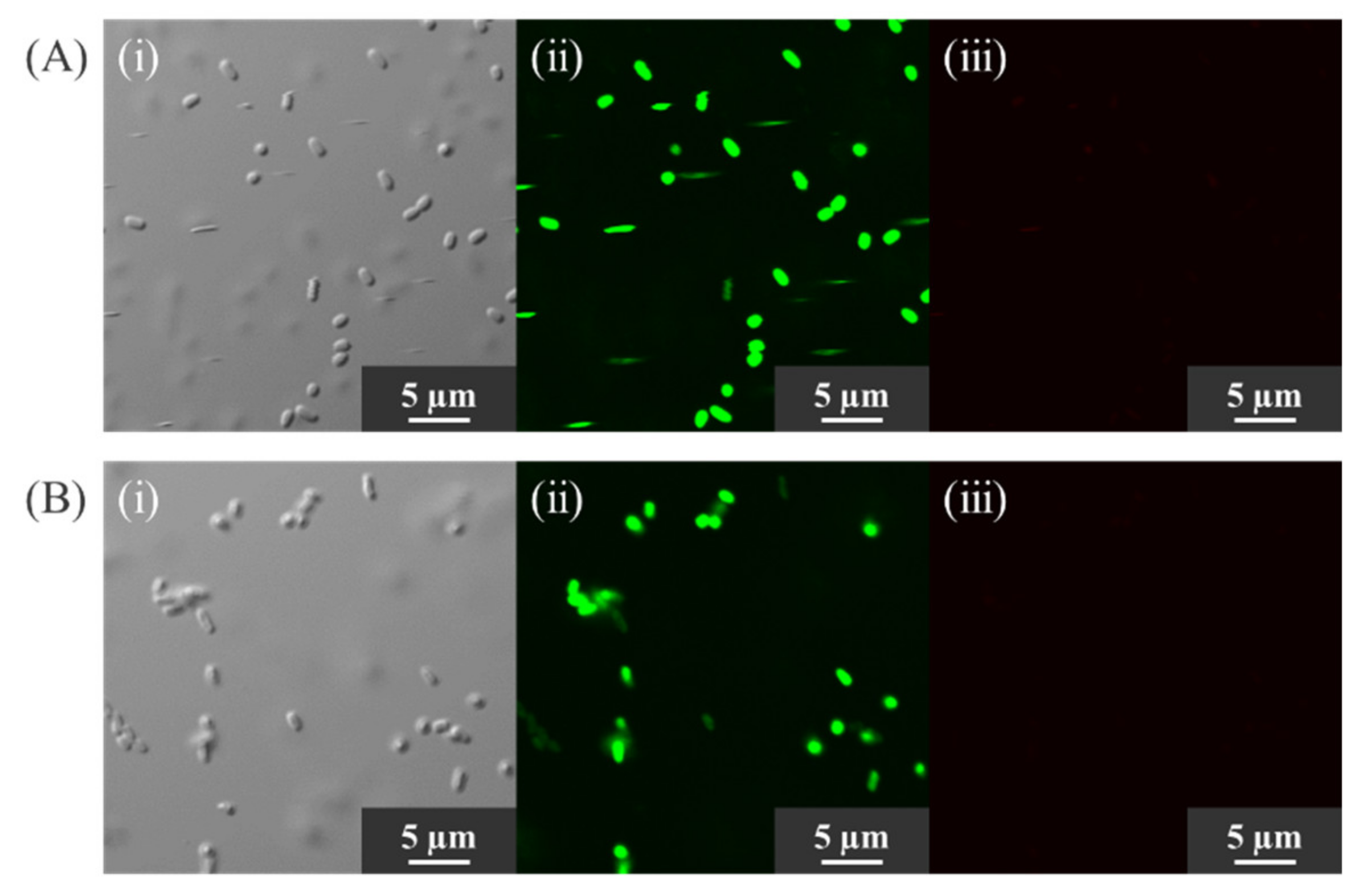
2.5. Confirmation of Cell Viability
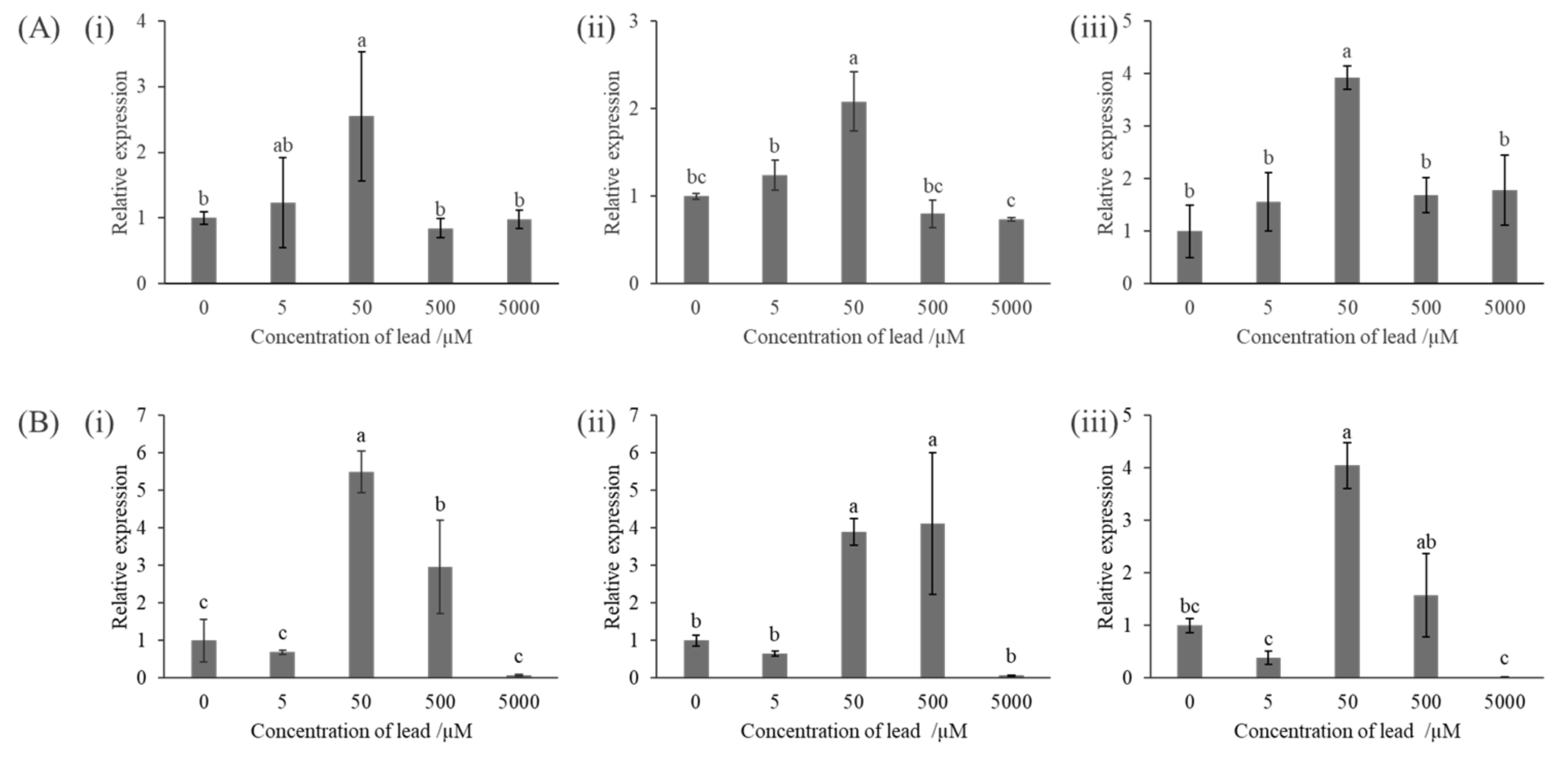
2.6. Evaluation of the Effect of Phosphate Concentration
2.7. Various Metal Nanoparticles Synthesized by KKY-29
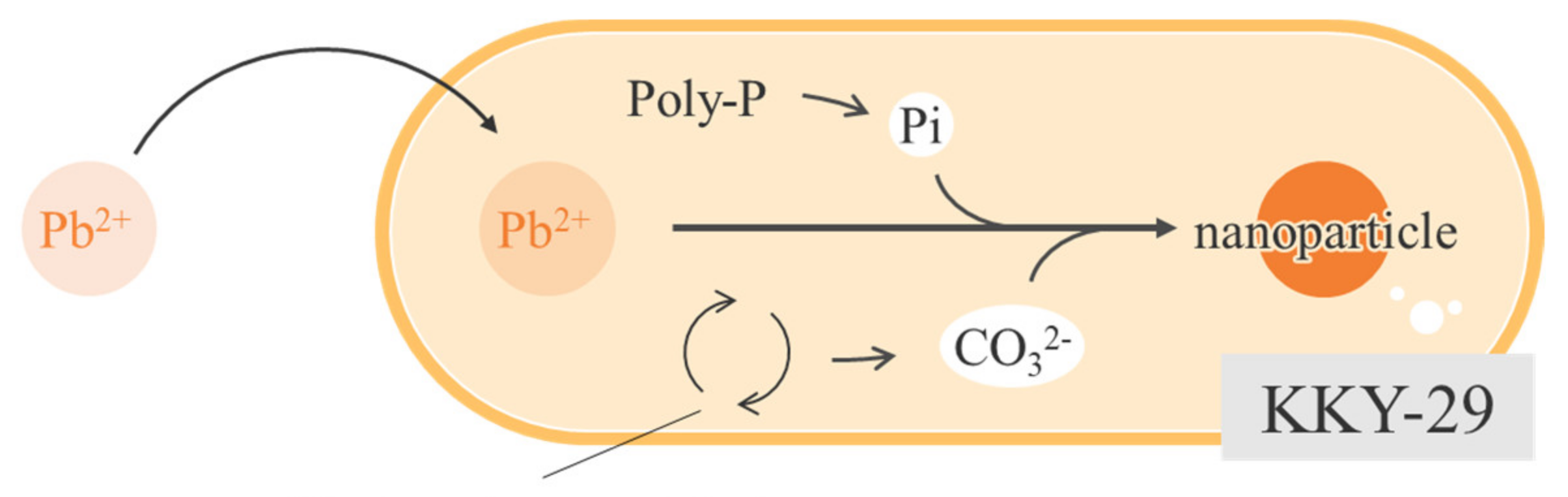
3. Discussion
4. Materials and Methods
4.1. Collection and Culture of Microorganisms
4.2. Screening
4.3. Identification of the Strain
4.4. Synthesis of Nanoparticles
4.5. Observation of Nanoparticles
4.6. Confirmation of Survival State of Bacterial Cells
4.7. Preparation of Ultra-Thin Cross-Sections of Bacterial Cells
4.8. Measurement of Lead Concentration by ICP-MS
4.9. Synthesis of Nanoparticles Other than Lead
4.10. Total DNA Isolation and Genome Sequencing
4.11. Quantitative PCR
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Blagodatskaya, E.V.; Pampura, T.V.; Dem’yanova, E.G.; Myakshina, T.N. Effect of Lead on Growth Characteristics of Microorganisms in Soil and Rhizosphere of Dactylis glomerata. Eurasian Soil Sci. 2006, 39, 653–660. [Google Scholar] [CrossRef]
- Flora, S.J.S.; Mittal, M.; Mehta, A. Heavy Metal Induced Oxidative Stress & Its Possible Reversal by Chelation Therapy. Indian J. Med Res. 2008, 128, 501–524. [Google Scholar] [PubMed]
- Stohs, S.J.; Bagchi, D. Oxidative Mechanisms in the Toxicity of Metal Ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef]
- Zheng, Z.; Kawakami, K.; Zhang, D.; Negishi, L.; Abomosallam, M.; Asakura, T.; Nagata, K.; Suzuki, M. Identification and Functional Analysis of Cadmium-Binding Protein in the Visceral Mass of Crassostrea gigas. Sci. Rep. 2021, 11, 11306. [Google Scholar] [CrossRef]
- May, G.J.; Davidson, A.; Monahov, B. Lead Batteries for Utility Energy Storage: A Review. J. Energy Storage 2018, 15, 145–157. [Google Scholar] [CrossRef]
- Batool, Z.; Yousafzai, N.A.; Murad, M.S.; Shahid, S.; Iqbal, A. Lead Toxicity and Evaluation of Oxidative Stress in Humans. PSM Biol. Res. 2017, 2, 79–82. [Google Scholar]
- De Schamphelaere, K.A.C.; Nys, C.; Janssen, C.R. Toxicity of Lead (Pb) to Freshwater Green Algae: Development and Validation of a Bioavailability Model and Inter-Species Sensitivity Comparison. Aquat. Toxicol. 2014, 155, 348–359. [Google Scholar] [CrossRef]
- Gillis, B.S.; Arbieva, Z.; Gavin, I.M. Analysis of Lead Toxicity in Human Cells. BMC Genom. 2012, 13, 344. [Google Scholar] [CrossRef]
- Needleman, H.L.; Tuncay, O.C.; Shapiro, I.M. Lead Levels in Deciduous Teeth of Urban and Suburban American Children. Nature 1972, 235, 111–112. [Google Scholar] [CrossRef]
- Papanikolaou, N.C.; Hatzidaki, E.G.; Belivanis, S.; Tzanakakis, G.N.; Tsatsakis, A.M. Lead Toxicity Update. A Brief Review. Med. Sci. Monit. 2005, 11, RA329–RA336. [Google Scholar]
- Chen, X.G.; Poretz, R.D. Lead Causes Human Fibroblasts to Mis-Sort Arylsulfatase A. Toxicology 2001, 163, 107–114. [Google Scholar] [CrossRef]
- Bahadir, T.; Bakan, G.; Altas, L.; Buyukgungor, H. The Investigation of Lead Removal by Biosorption: An Application at Storage Battery Industry Wastewaters. Enzym. Microb. Technol. 2007, 41, 98–102. [Google Scholar] [CrossRef]
- Esther, J.; Sukla, L.B.; Pradhan, N.; Panda, S. Fe (III) Reduction Strategies of Dissimilatory Iron Reducing Bacteria. Korean J. Chem. Eng. 2015, 32, 1–14. [Google Scholar] [CrossRef]
- Arakaki, A.; Yamagishi, A.; Matsunaga, T. Protein-Mediated Morphological Regulation of Magnetite Crystal in Magnetotactic Bacteria. J. Jpn. Soc. Powder Powder Metall. 2014, 61, S99–S103. [Google Scholar] [CrossRef]
- Blakemore, R. Magnetotactic Bacteria. Science 1975, 190, 377–379. [Google Scholar] [CrossRef]
- Amini, S.M. Preparation of Antimicrobial Metallic Nanoparticles with Bioactive Compounds. Mater. Sci. Eng. C 2019, 103, 109809. [Google Scholar] [CrossRef]
- García-Domínguez, P.; Nevado, C. Au–Pd Bimetallic Catalysis: The Importance of Anionic Ligands in Catalyst Speciation. J. Am. Chem. Soc. 2016, 138, 3266–3269. [Google Scholar] [CrossRef]
- Li, X.; Xu, H.; Chen, Z.-S.; Chen, G. Biosynthesis of Nanoparticles by Microorganisms and Their Applications. J. Nanomater. 2011, 2011, 1–16. [Google Scholar] [CrossRef]
- Lohse, S.E.; Murphy, C.J. Applications of Colloidal Inorganic Nanoparticles: From Medicine to Energy. J. Am. Chem. Soc. 2012, 134, 15607–15620. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, W.; Cai, Z.; Han, B.; Qian, T.; Zhao, D. An Overview of Preparation and Applications of Stabilized Zero-Valent Iron Nanoparticles for Soil and Groundwater Remediation. Water Res. 2016, 100, 245–266. [Google Scholar] [CrossRef]
- Zhang, D.; Yamamoto, T.; Tang, D.; Kato, Y.; Horiuchi, S.; Ogawa, S.; Yoshimura, E.; Suzuki, M. Enhanced Biosynthesis of CdS Nanoparticles through Arabidopsis thaliana Phytochelatin Synthase-Modified Escherichia coli with Fluorescence Effect in Detection of Pyrogallol and Gallic Acid. Talanta 2019, 195, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhou, L.; Xu, X.; Ai, C.; Zhao, P.; Yan, L.; Jiang, C.; Shi, J. Recovery of Ag+ by Cyclic Lipopeptide Iturin A and Corresponding Chain Peptide: Reaction Mechanisms, Kinetics, Toxicity Reduction, and Applications. Sci. Total Environ. 2021, 763, 142988. [Google Scholar] [CrossRef] [PubMed]
- Diba, H.; Cohan, R.A.; Salimian, M.; Mirjani, R.; Soleimani, M.; Khodabakhsh, F. Isolation and Characterization of Halophilic Bacteria with the Ability of Heavy Metal Bioremediation and Nanoparticle Synthesis from Khara Salt Lake in Iran. Arch. Microbiol. 2021, 203, 3893–3903. [Google Scholar] [CrossRef]
- Pinel-Cabello, M.; Chapon, V.; Ruiz-Fresneda, M.A.; Alpha-Bazin, B.; Berthomieu, C.; Armengaud, J.; Merroun, M.L. Delineation of Cellular Stages and Identification of Key Proteins for Reduction and Biotransformation of Se(IV) by Stenotrophomonas bentonitica BII-R7. J. Hazard. Mater. 2021, 418, 126150. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Suzuki, M. Metal Accumulation Using a Bacterium (K-142) Identified from Environmental Microorganisms by the Screening of Au Nanoparticles Synthesis. Materials 2020, 13, 4922. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Qi, X.; Zeng, X.; Lu, Y.; Zhou, J.; Cui, K.; Zhang, L. A Newly Isolated Bacterium Comamonas sp. XL8 Alleviates the Toxicity of Cadmium Exposure in Rice Seedlings by Accumulating Cadmium. J. Hazard. Mater. 2021, 403, 123824. [Google Scholar] [CrossRef]
- Kato, Y.; Suzuki, M. Synthesis of Metal Nanoparticles by Microorganisms. Crystals 2020, 10, 589. [Google Scholar] [CrossRef]
- Iwao, S.; Sugita, M.; Tsuchiya, K. Some Metabolic Interrelationships Among Cadmium, Lead, Copper and Zinc: Results from a Field Survey in Cd-Polluted Areas in Japan Part One: Dietary Intake of the Heavy Metals. Keio J. Med. 1981, 30, 17–36. [Google Scholar] [CrossRef][Green Version]
- Yoshida, F.; Hata, A.; Tonegawa, H. Itai-Itai Disease and the Countermeasures against Cadmium Pollution by the Kamioka Mine. Environ. Econ. Policy Stud. 1999, 2, 215–229. [Google Scholar] [CrossRef]
- Mills, S.J.; Ferraris, G.; Kampf, A.R.; Favreau, G. Twinning in Pyromorphite: The First Documented Occurrence of Twinning by Merohedry in the Apatite Supergroup. Am. Mineral. 2012, 97, 415–418. [Google Scholar] [CrossRef]
- Martinetto, P.; Anne, M.; Dooryhée, E.; Walter, P.; Tsoucaris, G. Synthetic Hydro cerussite, 2PbCO3·Pb(OH)2, by X-ray Powder Diffraction. Acta Crystallogr. C 2002, 58, i82–i84. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, P.; Kowshik, M. Fluorescent Lead(IV) Sulfide Nanoparticles Synthesized by Idiomarina sp. Strain PR58-8 for Bioimaging Applications. Appl. Environ. Microbiol. 2017, 83, e03091-16. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Zhang, C.; Zheng, W.; Zhang, Q.; Wan, Q.; Kong, L.; Li, L. Synthesis of Lead Halide Perovskite Nanocrystals by Melt Crystallization in Halide Salts. Chem. Commun. 2020, 56, 11291–11294. [Google Scholar] [CrossRef] [PubMed]
- Wolfbeis, O.S. An Overview of Nanoparticles Commonly Used in Fluorescent Bioimaging. Chem. Soc. Rev. 2015, 44, 4743–4768. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Tsuruda, Y.; Furukawa, T.; Negishi, L.; Imura, Y.; Sakuda, S.; Yoshimura, E.; Suzuki, M. Synthesis of CdSe Quantum Dots Using Fusarium oxysporum. Materials 2016, 9, 855. [Google Scholar] [CrossRef] [PubMed]
- Kaur, A.; Gupta, U. A Review on Applications of Nanoparticles for the Preconcentration of Environmental Pollutants. J. Mater. Chem. 2009, 19, 8279. [Google Scholar] [CrossRef]
- Babu, A.G.; Shea, P.J.; Sudhakar, D.; Jung, I.-B.; Oh, B.-T. Potential Use of Pseudomonas koreensis AGB-1 in Association with Miscanthus Sinensis to Remediate Heavy Metal(Loid)-Contaminated Mining Site Soil. J. Environ. Manag. 2015, 151, 160–166. [Google Scholar] [CrossRef]
- Fulladosa, E.; Murat, J.C.; Martínez, M.; Villaescusa, I. Patterns of Metals and Arsenic Poisoning in Vibrio fischeri Bacteria. Chemosphere 2005, 60, 43–48. [Google Scholar] [CrossRef]
- Sobolev, D.; Begonia, M.F.T. Effects of Heavy Metal Contamination upon Soil Microbes: Lead-Induced Changes in General and Denitrifying Microbial Communities as Evidenced by Molecular Markers. Int. J. Environ. Res. Public Health 2008, 5, 450–456. [Google Scholar] [CrossRef]
- Gu, T.; Qin, S.; Wu, X. Thermal Behavior of Pyromorphite (Pb10(PO4)6Cl2): In Situ High Temperature Powder X-Ray Diffraction Study. Crystals 2020, 10, 1070. [Google Scholar] [CrossRef]
- Chen, X.; Wright, J.V.; Conca, J.L.; Peurrung, L.M. Evaluation of Heavy Metal Remediation Using Mineral Apatite. Water Air Soil Pollut. 1997, 98, 57–78. [Google Scholar] [CrossRef]
- Rhee, Y.J.; Hillier, S.; Gadd, G.M. Lead Transformation to Pyromorphite by Fungi. Curr. Biol. 2012, 22, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Ma, L.Q.; Singh, S.P.; Cao, R.X.; Melamed, R. Field Demonstration of in Situ Immobilization of Soil Pb Using P Amendments. Adv. Environ. Res. 2003, 8, 93–102. [Google Scholar] [CrossRef]
- Ma, Q.Y.; Traina, S.J.; Logan, T.J.; Ryan, J.A. In Situ Lead Immobilization by Apatite. Environ. Sci. Technol. 1993, 27, 1803–1810. [Google Scholar] [CrossRef]
- Miretzky, P.; Fernandez-Cirelli, A. Phosphates for Pb Immobilization in Soils: A Review. Environ. Chem. Lett. 2008, 6, 121–133. [Google Scholar] [CrossRef]
- Hashimoto, Y.; Takaoka, M.; Oshita, K.; Tanida, H. Incomplete Transformations of Pb to Pyromorphite by Phosphate-Induced Immobilization Investigated by X-ray Absorption Fine Structure (XAFS) Spectroscopy. Chemosphere 2009, 76, 616–622. [Google Scholar] [CrossRef]
- Wang, X.; Phillips, B.L.; Boily, J.-F.; Hu, Y.; Hu, Z.; Yang, P.; Feng, X.; Xu, W.; Zhu, M. Phosphate Sorption Speciation and Precipitation Mechanisms on Amorphous Aluminum Hydroxide. Soil Syst. 2019, 3, 20. [Google Scholar] [CrossRef]
- Zhang, L.; Song, X.; Shao, X.; Wu, Y.; Zhang, X.; Wang, S.; Pan, J.; Hu, S.; Li, Z. Lead Immobilization Assisted by Fungal Decomposition of Organophosphate under Various PH Values. Sci. Rep. 2019, 9, 13353. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
- Kogure, T. A Program to Assist Kikuchi Pattern Analyses. J. Crystallogr. Soc. Jpn. 2003, 45, 391–395. [Google Scholar] [CrossRef]

| The Concentration of Metal Salt/mM | Ca | Fe | Ni | Cu | Zn | Pd | Ag | Cd | Pt | Au |
|---|---|---|---|---|---|---|---|---|---|---|
| 5.0 | 65.5 ± 10.9 | - | - | - | - | - | 24.8 ± 5.66 | - | - | - |
| 0.5 | 81.2 ± 15.5 | - | 41.9 ± 8.88 | - | - | - | 7.93 ± 6.74 | 40.1 ± 5.81 | - | 27.8 ± 6.45 |
| 0.05 | 74.8 ± 14.8 | 65.0 ± 14.0 | - | - | - | >100 | - | - | 56.5 ± 14.3 | - |
| Primer Name | Sequences |
|---|---|
| ppX_F | AGTGAATGCCGAAGGTCTGG |
| ppX_R | CAGTGATCCATGCGTTGCAG |
| ppk_F | TGGTGCACGGTCATGTAGAC |
| ppk_R | CGATCTCGTCGCGGAAGTAA |
| phoD_F | GTGCGCAATTTCGTGTTCCT |
| phoD_R | AGGTCTTGTCCAGCGGATTG |
| gyrB_F | TCCGAACTGTACCTCGTGGA |
| gyrB_R | GCGCAACTTGTCGATGTTGT |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, Y.; Kimura, S.; Kogure, T.; Suzuki, M. Deposition of Lead Phosphate by Lead-Tolerant Bacteria Isolated from Fresh Water near an Abandoned Mine. Int. J. Mol. Sci. 2022, 23, 2483. https://doi.org/10.3390/ijms23052483
Kato Y, Kimura S, Kogure T, Suzuki M. Deposition of Lead Phosphate by Lead-Tolerant Bacteria Isolated from Fresh Water near an Abandoned Mine. International Journal of Molecular Sciences. 2022; 23(5):2483. https://doi.org/10.3390/ijms23052483
Chicago/Turabian StyleKato, Yugo, Satoshi Kimura, Toshihiro Kogure, and Michio Suzuki. 2022. "Deposition of Lead Phosphate by Lead-Tolerant Bacteria Isolated from Fresh Water near an Abandoned Mine" International Journal of Molecular Sciences 23, no. 5: 2483. https://doi.org/10.3390/ijms23052483
APA StyleKato, Y., Kimura, S., Kogure, T., & Suzuki, M. (2022). Deposition of Lead Phosphate by Lead-Tolerant Bacteria Isolated from Fresh Water near an Abandoned Mine. International Journal of Molecular Sciences, 23(5), 2483. https://doi.org/10.3390/ijms23052483






