Role of eIF5A in Mitochondrial Function
Abstract
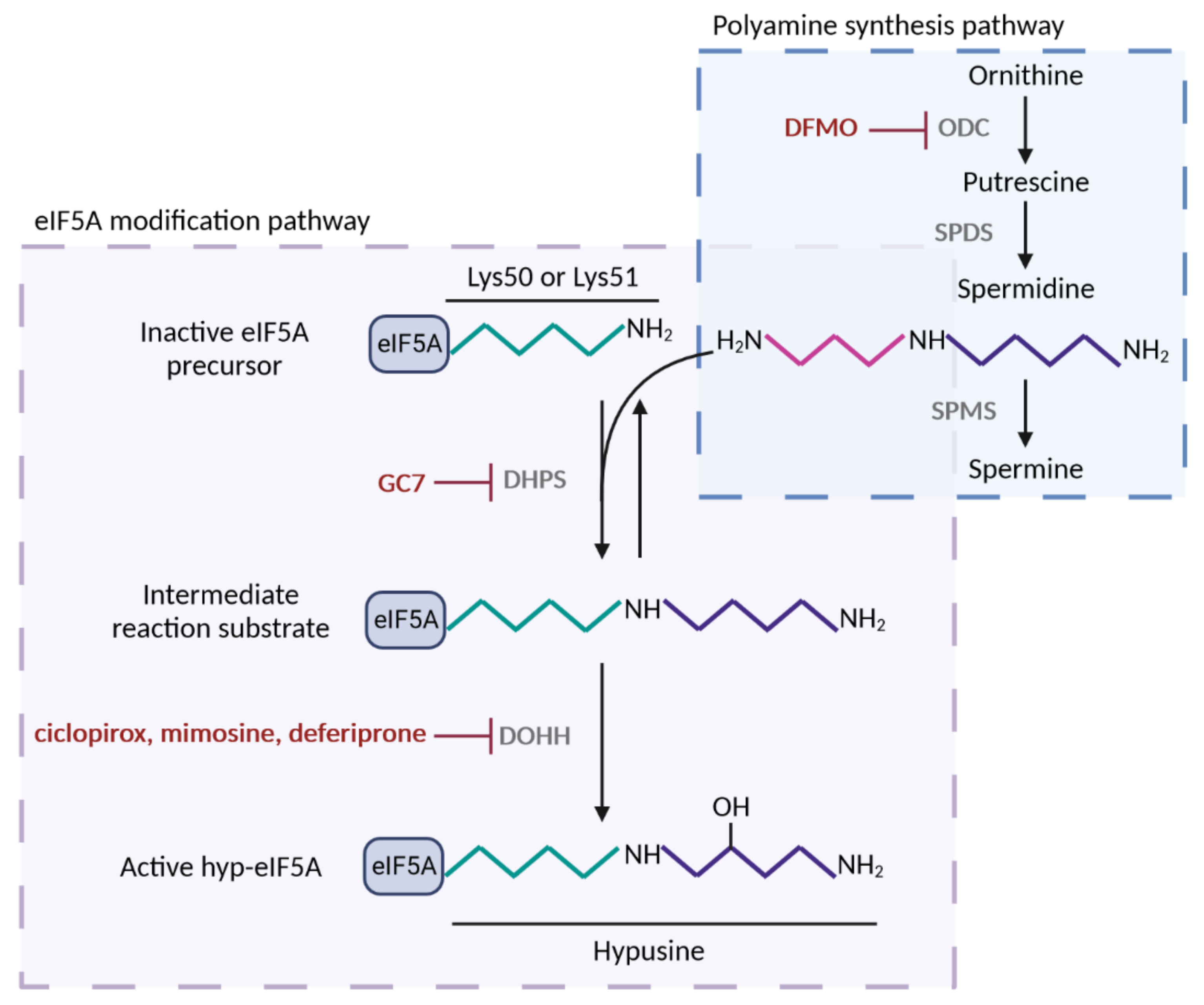
1. The Molecular Function of eIF5A
2. Mitochondrial Metabolism in Health and Disease
3. The Expression of eIF5A Isoforms Responds Differentially to the Cellular Metabolic State
4. Links between eIF5A and Mitochondrial Function
5. Polyamines Regulate Mitochondrial Function: eIF5A-Dependent and Independent Effects
6. Eukaryotic Translation Initiation Factor 5A’s Subcellular Localization and Its Association with Mitochondria
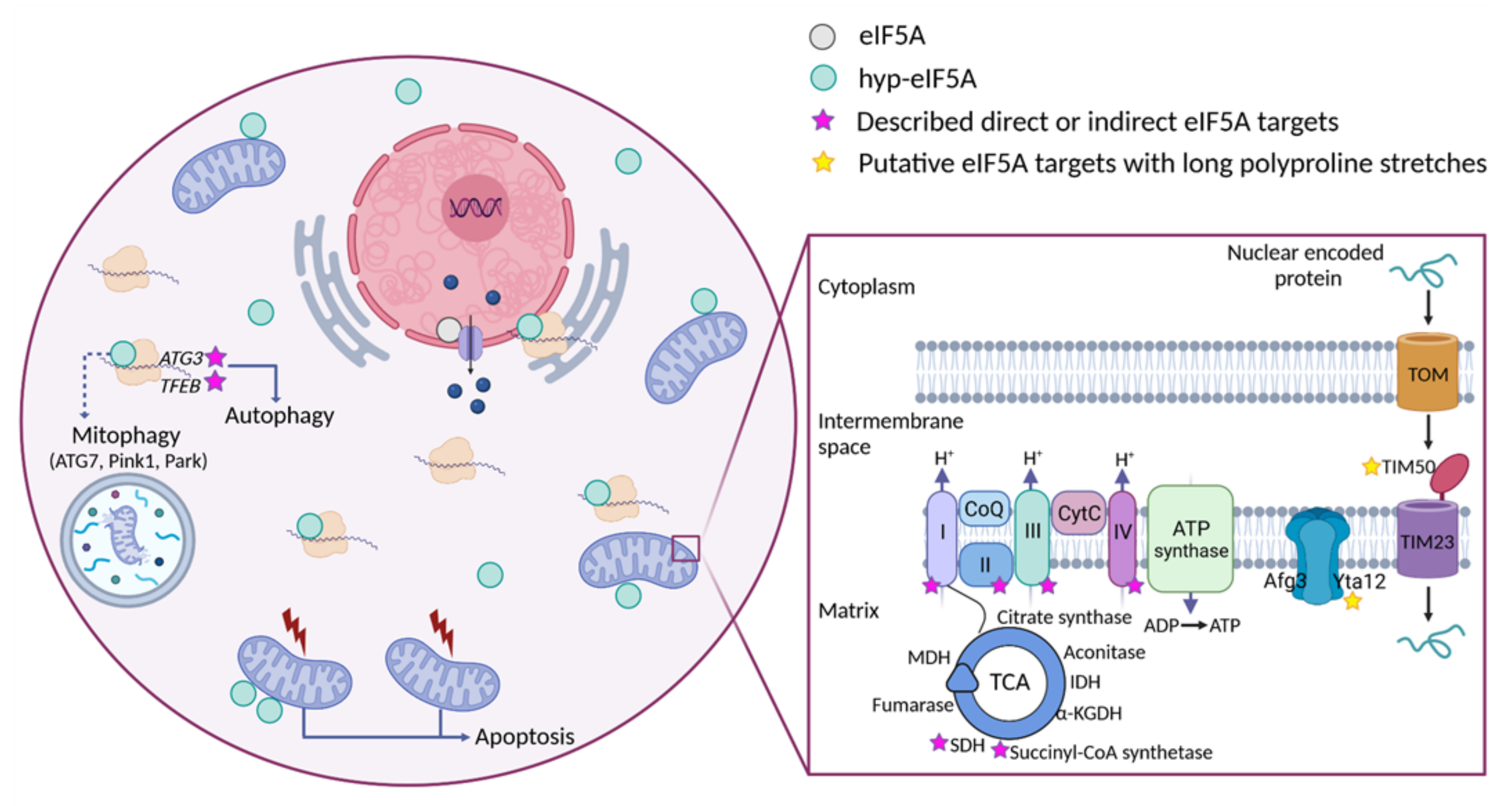
7. Eukaryotic Translation Initiation Factor 5A’s Molecular Roles in Mitochondrial Function
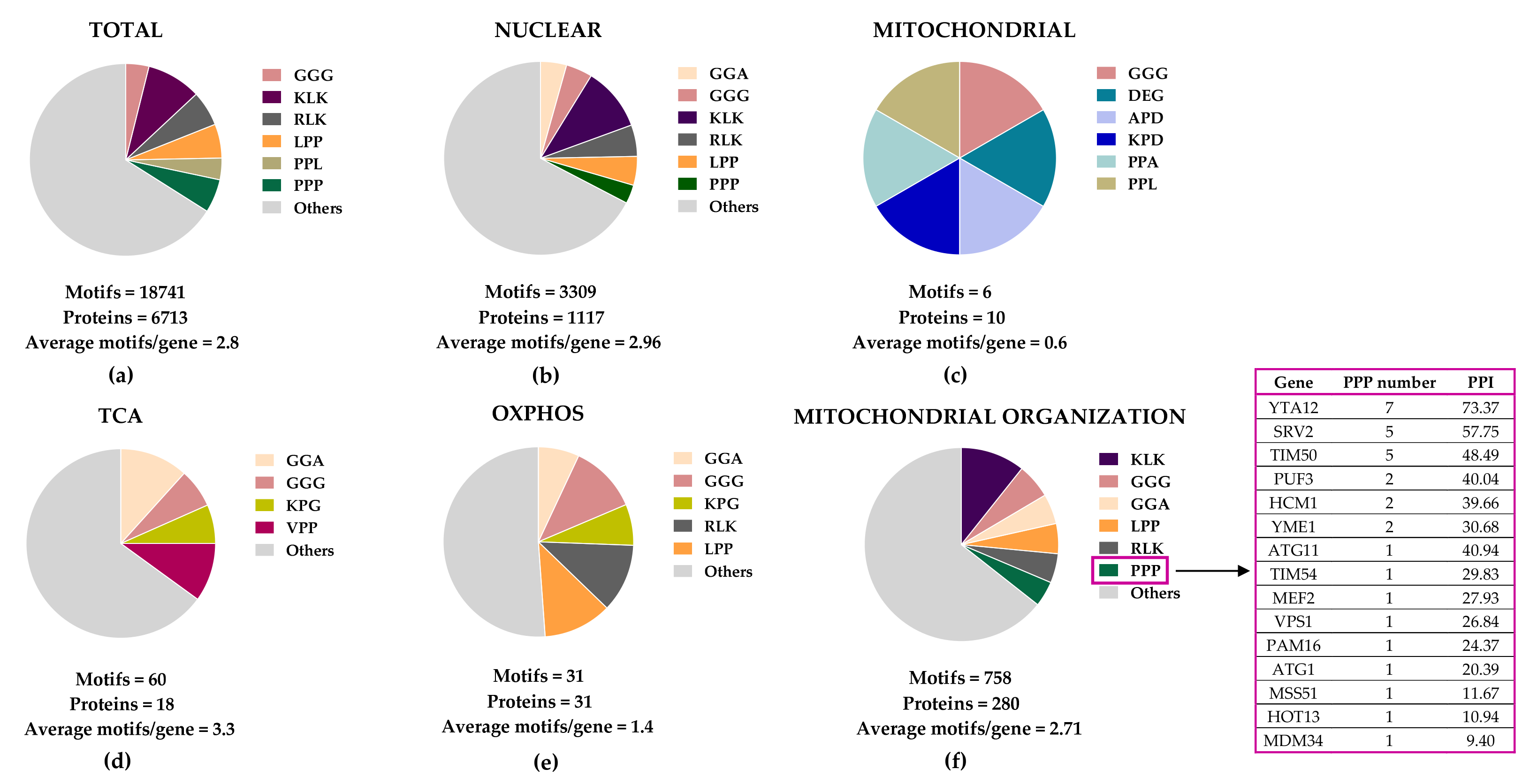
8. Perspectives on the Identification of Mitochondrial Processes and Targets under the Control of eIF5A
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Schnier, J.; Schwelberger, H.G.; Smit-McBride, Z.; Kang, H.A.; Hershey, J.W. Translation initiation factor 5A and its hypusine modification are essential for cell viability in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1991, 11, 3105–3114. [Google Scholar] [CrossRef]
- Kulak, N.A.; Pichler, G.; Paron, I.; Nagaraj, N.; Mann, M. Minimal, encapsulated proteomic-sample processing applied to copy-number estimation in eukaryotic cells. Nat. Methods 2014, 11, 319–324. [Google Scholar] [CrossRef] [PubMed]
- Kemper, W.M.; Berry, K.W.; Merrick, W.C. Purification and properties of rabbit reticulocyte protein synthesis initiation factors M2Bα and M2Bβ. J. Biol. Chem. 1976, 251, 5551–5557. [Google Scholar] [CrossRef]
- Benne, R.; Hershey, J.W.B. The mechanism of action of protein synthesis initiation factors from rabbit reticulocytes. J. Biol. Chem. 1978, 253, 3078–3087. [Google Scholar] [CrossRef]
- Jao, D.L.E.; Chen, K.Y. Tandem affinity purification revealed the hypusine-dependent binding of eukaryotic initiation factor 5A to the translating 80S ribosomal complex. J. Cell. Biochem. 2006, 97, 583–598. [Google Scholar] [CrossRef] [PubMed]
- Zanelli, C.F.; Maragno, A.L.C.; Gregio, A.P.B.; Komili, S.; Pandolfi, J.R.; Mestriner, C.A.; Lustri, W.R.; Valentini, S.R. eIF5A binds to translational machinery components and affects translation in yeast. Biochem. Biophys. Res. Commun. 2006, 348, 1358–1366. [Google Scholar] [CrossRef] [PubMed]
- Gregio, A.P.B.; Cano, V.P.S.; Avaca, J.S.; Valentini, S.R.; Zanelli, C.F. eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 2009, 308, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Saini, P.; Eyler, D.E.; Green, R.; Dever, T.E. Hypusine-containing protein eIF5A promotes translation elongation. Nature 2009, 459, 118–121. [Google Scholar] [CrossRef]
- Gutierrez, E.; Shin, B.S.; Woolstenhulme, C.J.; Kim, J.R.; Saini, P.; Buskirk, A.R.; Dever, T.E. eif5A promotes translation of polyproline motifs. Mol. Cell 2013, 51, 35–45. [Google Scholar] [CrossRef]
- Schuller, A.P.; Wu, C.C.C.; Dever, T.E.; Buskirk, A.R.; Green, R. eIF5A Functions Globally in Translation Elongation and Termination. Mol. Cell 2017, 66, 194–205. [Google Scholar] [CrossRef]
- Pelechano, V.; Alepuz, P. EIF5A facilitates translation termination globally and promotes the elongation of many non polyproline-specific tripeptide sequences. Nucleic Acids Res. 2017, 45, 7326–7338. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Wolff, E.C. Hypusine, a polyamine-derived amino acid critical for eukaryotic translation. J. Biol. Chem. 2018, 293, 18710–18718. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Lee, Y.B.; Joe, Y.A. Hypusine is essential for eukaryotic cell proliferation. NeuroSignals 1997, 6, 115–123. [Google Scholar] [CrossRef] [PubMed]
- Klier, H.; Csonga, R.; João, H.C.; Eckerskom, C.; Auer, M.; Lottspeich, F.; Eder, J. Isolation and Structural Characterization of Different Isoforms of the Hypusine-Containing Protein eIF-5A from HeLa Cells. Biochemistry 1995, 34, 14693–14702. [Google Scholar] [CrossRef]
- Schwelberger, H.G.; Kang, H.A.; Hershey, J.W.B. Translation initiation factor eIF-5A expressed from either of two yeast genes or from human cDNA. Functional identity under aerobic and anaerobic conditions. J. Biol. Chem. 1993, 268, 14018–14025. [Google Scholar] [CrossRef]
- Zarembinski, T.I.; Hung, L.I.W.; Mueller-Dieckmann, H.J.; Kim, K.K.; Yokota, M.; Kim, R.; Kim, S.H. Structure-based assignment of the biochemical function of a hypothetical protein: A test case of structural genomics. Proc. Natl. Acad. Sci. USA 1998, 95, 15189–15193. [Google Scholar] [CrossRef]
- Peat, T.S.; Newman, J.; Waldo, G.S.; Berendzen, J.; Terwilliger, T.C. Structure of translation initiation factor 5A from Pyrobaculum aerophilum at 1.75 Å resolution. Structure 1998, 6, 1207–1214. [Google Scholar] [CrossRef]
- Tong, Y.; Park, I.; Hong, B.S.; Nedyalkova, L.; Tempel, W.; Park, H.W. Crystal structure of human eIF5A1: Insight into functional similarity of human eIF5A1 and eIF5A2. Proteins Struct. Funct. Bioinforma. 2009, 75, 1040–1045. [Google Scholar] [CrossRef]
- Cooper, H.L.; Park, M.H.; Folk, J.E. Posttranslational formation of hypusine in a single major protein occurs generally in growing cells and is associated with activation of lymphocyte growth. Cell 1982, 29, 791–797. [Google Scholar] [CrossRef]
- Rossi, D.; Barbosa, N.M.; Galvão, F.C.; Boldrin, P.E.G.; Hershey, J.W.B.; Zanelli, C.F.; Fraser, C.S.; Valentini, S.R. Evidence for a negative cooperativity between eIF5A and eEF2 on binding to the ribosome. PLoS ONE 2016, 11, e0154205. [Google Scholar] [CrossRef]
- Dias, C.A.O.; Gregio, A.P.B.; Rossi, D.; Galvão, F.C.; Watanabe, T.F.; Park, M.H.; Valentini, S.R.; Zanelli, C.F. EIF5A interacts functionally with eEF2. Amino Acids 2012, 42, 697–702. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, C.; Becker, T.; Heuer, A.; Braunger, K.; Shanmuganathan, V.; Pech, M.; Berninghausen, O.; Wilson, D.N.; Beckmann, R. Structure of the hypusinylated eukaryotic translation factor eIF-5A bound to the ribosome. Nucleic Acids Res. 2015, 44, 1944–1951. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.P.; Yin, K.C.; Zimolo, Z.A.; Stern, A.M.; Waxman, L. The subcellular distribution of eukaryotic translation initiation factor, eIF-5A, in cultured cells. Exp. Cell Res. 1996, 225, 348–356. [Google Scholar] [CrossRef] [PubMed]
- Valentini, S.R.; Casolari, J.M.; Oliveira, C.C.; Silver, P.A.; McBride, A.E. Genetic interactions of yeast eukaryotic translation initiation factor 5a (eIF5A) reveal connections to poly(A)-binding protein and protein kinase C signaling. Genetics 2002, 160, 393–405. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Galvão, F.C.; Bellato, H.M.; Boldrin, P.E.G.; Andrews, B.J.; Valentini, S.R.; Zanelli, C.F. eIF5A has a function in the cotranslational translocation of proteins into the ER. Amino Acids 2014, 46, 645–653. [Google Scholar] [CrossRef]
- Barba-Aliaga, M.; Mena, A.; Espinoza, V.; Apostolova, N.; Costell, M.; Alepuz, P. Hypusinated eIF5A is required for the translation of collagen. J. Cell Sci. 2021, 134, jcs258643. [Google Scholar] [CrossRef]
- Mandal, A.; Mandal, S.; Park, M.H. Global quantitative proteomics reveal up-regulation of endoplasmic reticulum stress response proteins upon depletion of eIF5A in HeLa cells. Sci. Rep. 2016, 6, 25795. [Google Scholar] [CrossRef]
- Xu, A.; Chen, K.Y. Hypusine Is Required for a Sequence-specific Interaction of Eukaryotic Initiation Factor 5A with Postsystematic Evolution of Ligands by Exponential Enrichment RNA. J. Biol. Chem. 2001, 276, 2555–2561. [Google Scholar] [CrossRef]
- Xu, A.; Jao, D.L.E.; Chen, K.Y. Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem. J. 2004, 384, 585–590. [Google Scholar] [CrossRef]
- Aksu, M.; Trakhanov, S.; Görlich, D. Structure of the exportin Xpo4 in complex with RanGTP and the hypusine-containing translation factor eIF5A. Nat. Commun. 2016, 7, 11952. [Google Scholar] [CrossRef]
- Zuk, D.; Jacobson, A. A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 1998, 17, 2914–2925. [Google Scholar] [CrossRef] [PubMed]
- Schrader, R.; Young, C.; Kozian, D.; Hoffmann, R.; Lottspeich, F. Temperature-sensitive eIF5A mutant accumulates transcripts targeted to the nonsense-mediated decay pathway. J. Biol. Chem. 2006, 281, 35336–35346. [Google Scholar] [CrossRef] [PubMed]
- Bassani, F.; Zink, I.A.; Pribasnig, T.; Wolfinger, M.T.; Romagnoli, A.; Resch, A.; Schleper, C.; Bläsi, U.; La Teana, A. Indications for a moonlighting function of translation factor aIF5A in the crenarchaeum Sulfolobus solfataricus. RNA Biol. 2019, 16, 675–685. [Google Scholar] [CrossRef]
- Park, M.H.; Joe, Y.A.; Kang, K.R.; Lee, Y.B.; Wolff, E.C. The polyamine-derived amino acid hypusine: Its post-translational formation in eIF-5A and its role in cell proliferation. Amino Acids 1996, 10, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Hanauske-Abel, H.M.; Slowinska, B.; Zagulska, S.; Wilson, R.C.; Staiano-Coico, L.; Hanauske, A.R.; McCaffrey, T.; Szabo, P. Detection of a sub-set of polysomal mRNAs associated with modulation of hypusine formation at the G1-S boundary Proposal of a role for eIF-5A in onset of DNA replication. FEBS Lett. 1995, 366, 92–98. [Google Scholar] [CrossRef]
- Chen, Z.P.; Yan, Y.P.; Ding, Q.J.; Knapp, S.; Potenza, J.A.; Schugar, H.J.; Chen, K.Y. Effects of inhibitors of deoxyhypusine synthase on the differentiation of mouse neuroblastoma and erythroleukemia cells. Cancer Lett. 1996, 105, 233–239. [Google Scholar] [CrossRef]
- Clement, P.M.; Hanauske-Abel, H.M.; Wolff, E.C.; Kleinman, H.K.; Park, M.H. The antifungal drug ciclopirox inhibits deoxyhypusine and proline hydroxylation, endothelial cell growth and angiogenesis in vitro. Int. J. Cancer 2002, 100, 491–498. [Google Scholar] [CrossRef]
- Nishimura, K.; Lee, S.B.; Park, J.H.; Park, M.H. Essential role of eIF5A-1 and deoxyhypusine synthase in mouse embryonic development. Amino Acids 2012, 42, 703–710. [Google Scholar] [CrossRef]
- Sievert, H.; Pällmann, N.; Miller, K.K.; Hermans-Borgmeyer, I.; Venz, S.; Sendoel, A.; Preukschas, M.; Schweizer, M.; Boettcher, S.; Janiesch, P.C.; et al. A novel mouse model for inhibition of DOHH-mediated hypusine modification reveals a crucial function in embryonic development, proliferation and oncogenic transformation. DMM Dis. Model. Mech. 2014, 7, 963–976. [Google Scholar] [CrossRef]
- Padgett, L.R.; Robertson, M.A.; Anderson-Baucum, E.K.; Connors, C.T.; Wu, W.; Mirmira, R.G.; Mastracci, T.L. Deoxyhypusine synthase, an essential enzyme for hypusine biosynthesis, is required for proper exocrine pancreas development. FASEB J. 2021, 35, e21473. [Google Scholar] [CrossRef]
- Levasseur, E.M.; Yamada, K.; Piñeros, A.R.; Wu, W.; Syed, F.; Orr, K.S.; Anderson-Baucum, E.; Mastracci, T.L.; Maier, B.; Mosley, A.L.; et al. Hypusine biosynthesis in β cells links polyamine metabolism to facultative cellular proliferation to maintain glucose homeostasis. Sci. Signal. 2019, 12. [Google Scholar] [CrossRef] [PubMed]
- Lubas, M.; Harder, L.M.; Kumsta, C.; Tiessen, I.; Hansen, M.; Andersen, J.S.; Lund, A.H.; Frankel, L.B. eIF 5A is required for autophagy by mediating ATG 3 translation. EMBO Rep. 2018, 19, e46072. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Alsaleh, G.; Feltham, J.; Sun, Y.; Napolitano, G.; Riffelmacher, T.; Charles, P.; Frau, L.; Hublitz, P.; Yu, Z.; et al. Polyamines Control eIF5A Hypusination, TFEB Translation, and Autophagy to Reverse B Cell Senescence. Mol. Cell 2019, 76, 110–125. [Google Scholar] [CrossRef] [PubMed]
- Zanelli, C.F.; Valentini, S.R. Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics 2005, 171, 1571–1581. [Google Scholar] [CrossRef]
- Chatterjee, I.; Gross, S.R.; Kinzy, T.G.; Chen, K.Y. Rapid depletion of mutant eukaryotic initiation factor 5A at restrictive temperature reveals connections to actin cytoskeleton and cell cycle progression. Mol. Genet. Genomics 2006, 275, 264–276. [Google Scholar] [CrossRef]
- Nguyen, S.; Leija, C.; Kinch, L.; Regmi, S.; Li, Q.; Grishin, N.V.; Phillips, M.A. Deoxyhypusine modification of eukaryotic translation initiation factor 5A (eIF5A) is essential for Trypanosoma brucei growth and for expression of polyprolyl-containing proteins. J. Biol. Chem. 2015, 290, 19987–19988. [Google Scholar] [CrossRef]
- Li, T.; Belda-Palazón, B.; Ferrando, A.; Alepuz, P. Fertility and polarized cell growth depends on eIF5A for translation of polyproline-rich formins in Saccharomyces cerevisiae. Genetics 2014, 197, 1191–1200. [Google Scholar] [CrossRef]
- Muñoz-Soriano, V.; Domingo-Muelas, A.; Li, T.; Gamero, E.; Bizy, A.; Fariñas, I.; Alepuz, P.; Paricio, N. Evolutionary conserved role of eukaryotic translation factor eIF5A in the regulation of actin-nucleating formins. Sci. Rep. 2017, 7, 9580. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, K.; Choi, S.; Wyse, M.; Strnadel, J.; Wright, T.; Klemke, R. Eukaryotic translation initiation factor 5A (EIF5A) regulates pancreatic cancer metastasis by modulating RhoA and rho-associated kinase (ROCK) protein expression levels. J. Biol. Chem. 2015, 290, 29907–29919. [Google Scholar] [CrossRef]
- Coni, S.; Serrao, S.M.; Yurtsever, Z.N.; Di Magno, L.; Bordone, R.; Bertani, C.; Licursi, V.; Ianniello, Z.; Infante, P.; Moretti, M.; et al. Blockade of EIF5A hypusination limits colorectal cancer growth by inhibiting MYC elongation. Cell Death Dis. 2020, 11, 1045. [Google Scholar] [CrossRef]
- Ning, L.; Wang, L.; Zhang, H.; Jiao, X.; Chen, D. Eukaryotic translation initiation factor 5A in the pathogenesis of cancers (Review). Oncol. Lett. 2020, 20, 81. [Google Scholar] [CrossRef] [PubMed]
- Li, A.L.; Li, H.Y.; Jin, B.F.; Ye, Q.N.; Zhou, T.; Yu, X.D.; Pan, X.; Man, J.H.; He, K.; Yu, M.; et al. A novel eIF5A complex functions as a regulator of p53 and p53-dependent apoptosis. J. Biol. Chem. 2004, 279, 49251–49258. [Google Scholar] [CrossRef] [PubMed]
- Caraglia, M.; Park, M.H.; Wolff, E.C.; Marra, M.; Abbruzzese, A. EIF5A isoforms and cancer: Two brothers for two functions? Amino Acids 2013, 44, 103–109. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Wang, D.B.; Lu, X.; Wei, H.; Zhu, R.; Zhu, S.S.; Jiang, H.; Yang, Z.J. Doxorubicin induces apoptosis in H9c2 cardiomyocytes: Role of overexpressed Eukaryotic translation initiation factor 5A. Biol. Pharm. Bull. 2010, 33, 1666–1672. [Google Scholar] [CrossRef]
- Martella, M.; Catalanotto, C.; Talora, C.; La Teana, A.; Londei, P.; Benelli, D. Inhibition of eukaryotic translation initiation factor 5a (Eif5a) hypusination suppress p53 translation and alters the association of eif5a to the ribosomes. Int. J. Mol. Sci. 2020, 21, 4583. [Google Scholar] [CrossRef] [PubMed]
- Ruhl, M.; Himmelspach, M.; Bahr, G.M.; Hammerschmid, F.; Jaksche, H.; Wolff, B.; Aschauer, H.; Farrington, G.K.; Probst, H.; Bevec, D.; et al. Eukaryotic initiation factor 5A is a cellular target of the human immunodeficiency virus type 1 Rev activation domain mediating trans- activation. J. Cell Biol. 1993, 123, 1309–1320. [Google Scholar] [CrossRef]
- Rosorius, O.; Reichart, B.; Krätzer, F.; Heger, P.; Dabauvalle, M.C.; Hauber, J. Nuclear pore localization and nucleocytoplasmic transport of eIF-5A: Evidence for direct interaction with the export receptor CRM1. J. Cell Sci. 1999, 112, 2369–2380. [Google Scholar] [CrossRef]
- Olsen, M.E.; Connor, J.H. Hypusination of eIF5A as a Target for Antiviral Therapy. DNA Cell Biol. 2017, 36, 198–201. [Google Scholar] [CrossRef]
- Tersey, S.A.; Colvin, S.C.; Maier, B.; Mirmira, R.G. Protective effects of polyamine depletion in mouse models of type 1 diabetes: Implications for therapy. Amino Acids 2014, 46, 633–642. [Google Scholar] [CrossRef]
- Maier, B.; Ogihara, T.; Trace, A.P.; Tersey, S.A.; Robbins, R.D.; Chakrabarti, S.K.; Nunemaker, C.S.; Stull, N.D.; Taylor, C.A.; Thompson, J.E.; et al. The unique hypusine modification of eIF5A promotes islet β cell inflammation and dysfunction in mice. J. Clin. Investig. 2010, 120, 2156–2170. [Google Scholar] [CrossRef]
- Jenkins, Z.A.; Hååg, P.G.; Johansson, H.E. Human EIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 2001, 71, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Clement, P.M.J.; Henderson, C.A.; Jenkins, Z.A.; Smit-McBride, Z.; Wolff, E.C.; Hershey, J.W.B.; Park, M.H.; Johansson, H.E. Identification and characterization of eukaryotic initiation factor 5A-2. Eur. J. Biochem. 2003, 270, 4254–4263. [Google Scholar] [CrossRef] [PubMed]
- Park, M.H.; Kar, R.K.; Banka, S.; Ziegler, A.; Chung, W.K. Post-translational formation of hypusine in eIF5A: Implications in human neurodevelopment. Amino Acids 2021, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Hyvönen, M.T.; Keinänen, T.A.; Khomutov, M.; Simonian, A.; Vepsäläinen, J.; Park, J.H.; Khomutov, A.R.; Alhonen, L.; Park, M.H. Effects of novel C-methylated spermidine analogs on cell growth via hypusination of eukaryotic translation initiation factor 5A. Amino Acids 2012, 42, 685–695. [Google Scholar] [CrossRef] [PubMed]
- Mounce, B.C.; Cesaro, T.; Moratorio, G.; Hooikaas, P.J.; Yakovleva, A.; Werneke, S.W.; Smith, E.C.; Poirier, E.Z.; Simon-Loriere, E.; Prot, M.; et al. Inhibition of Polyamine Biosynthesis Is a Broad-Spectrum Strategy against RNA Viruses. J. Virol. 2016, 90, 9683–9692. [Google Scholar] [CrossRef]
- Li, J.; Meng, Y.; Wu, X.; Sun, Y. Polyamines and related signaling pathways in cancer. Cancer Cell Int. 2020, 20, 1–16. [Google Scholar] [CrossRef]
- Wolff, E.C.; Kang, K.R.; Kim, Y.S.; Park, M.H. Posttranslational synthesis of hypusine: Evolutionary progression and specificity of the hypusine modification. Amino Acids 2007, 33, 341–350. [Google Scholar] [CrossRef]
- Lou, B.; Fan, J.; Wang, K.; Chen, W.; Zhou, X.; Zhang, J.; Lin, S.; Lv, F.; Chen, Y. N1-guanyl-1,7-diaminoheptane (GC7) enhances the therapeutic efficacy of doxorubicin by inhibiting activation of eukaryotic translation initiation factor 5A2 (eIF5A2) and preventing the epithelial-mesenchymal transition in hepatocellular carcinoma cells. Exp. Cell Res. 2013, 319, 2708–2717. [Google Scholar] [CrossRef]
- Bandino, A.; Geerts, D.; Koster, J.; Bachmann, A.S. Deoxyhypusine synthase (DHPS) inhibitor GC7 induces p21/Rb-mediated inhibition of tumor cell growth and DHPS expression correlates with poor prognosis in neuroblastoma patients. Cell. Oncol. 2014, 37, 387–398. [Google Scholar] [CrossRef]
- Guerville, F.; De Souto Barreto, P.; Ader, I.; Andrieu, S.; Casteilla, L.; Dray, C.; Fazilleau, N.; Guyonnet, S.; Langin, D.; Liblau, R.; et al. Revisiting the Hallmarks of Aging to Identify Markers of Biological Age. J. Prev. Alzheimer’s Dis. 2020, 7, 56–64. [Google Scholar] [CrossRef]
- Nazaret, C.; Heiske, M.; Thurley, K.; Mazat, J.P. Mitochondrial energetic metabolism: A simplified model of TCA cycle with ATP production. J. Theor. Biol. 2009, 258, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Kocher, T.D.; Thomas, W.K.; Meyer, A.; Edwards, S.V.; Paabo, S.; Villablanca, F.X.; Wilson, A.C. Dynamics of mitochondrial DNA evolution in animals: Amplification and sequencing with conserved primers. Proc. Natl. Acad. Sci. USA 1989, 86, 6196–6200. [Google Scholar] [CrossRef] [PubMed]
- Wiedemann, N.; Pfanner, N. Mitochondrial machineries for protein import and assembly. Annu. Rev. Biochem. 2017, 86, 685–714. [Google Scholar] [CrossRef]
- Kembro, J.M.; Cortassa, S.; Aon, M.A. Complex oscillatory redox dynamics with signaling potential at the edge between normal and pathological mitochondrial function. Front. Physiol. 2014, 5, 257. [Google Scholar] [CrossRef] [PubMed]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef] [PubMed]
- Wallace, D.C.; Fan, W. Energetics, epigenetics, mitochondrial genetics. Mitochondrion 2010, 10, 12–31. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.P. How mitochondria produce reactive oxygen species. Biochem. J. 2009, 417, 1–13. [Google Scholar] [CrossRef]
- Stowe, D.F.; Camara, A.K.S. Mitochondrial reactive oxygen species production in excitable cells: Modulators of mitochondrial and cell function. Antioxid. Redox Signal. 2009, 11, 1373–1414. [Google Scholar] [CrossRef]
- Anderson, E.J.; Lustig, M.E.; Boyle, K.E.; Woodlief, T.L.; Kane, D.A.; Lin, C.T.; Price, J.W.; Kang, L.; Rabinovitch, P.S.; Szeto, H.H.; et al. Mitochondrial H2O2 emission and cellular redox state link excess fat intake to insulin resistance in both rodents and humans. J. Clin. Investig. 2009, 119, 573–581. [Google Scholar] [CrossRef]
- Liu, T.; Rourke, B.O. Enhancing mitochondrial Ca2+ uptake in myocytes from failing hearts restores energy supply and demand matching. Circ. Res. 2008, 103, 279–288. [Google Scholar] [CrossRef]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Biol. 2011, 12, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Archer, S.L. Mitochondrial Dynamics—Mitochondrial Fission and Fusion in Human Diseases. N. Engl. J. Med. 2013, 369, 2236–2251. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, S. The mitochondrial basis of aging and age-related disorders. Genes 2017, 8, 398. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chen, X.J. A cytosolic network suppressing mitochondria-mediated proteostatic stress and cell death. Nature 2015, 524, 481–484. [Google Scholar] [CrossRef]
- Jovaisaite, V.; Auwerx, J. The mitochondrial unfolded protein response-synchronizing genomes. Curr. Opin. Cell Biol. 2015, 33, 74–81. [Google Scholar] [CrossRef]
- Nunnari, J.; Suomalainen, A. Mitochondria: In sickness and in health. Cell 2012, 148, 1145–1159. [Google Scholar] [CrossRef]
- Pandya, J.D.; Royland, J.E.; MacPhail, R.C.; Sullivan, P.G.; Kodavanti, P.R.S. Age- and brain region-specific differences in mitochondrial bioenergetics in Brown Norway rats. Neurobiol. Aging 2016, 42, 25–34. [Google Scholar] [CrossRef]
- Pollard, A.K.; Craig, E.L.; Chakrabarti, L. Mitochondrial complex 1 activity measured by spectrophotometry is reduced across all brain regions in ageing and more specifically in neurodegeneration. PLoS ONE 2016, 11, e0157405. [Google Scholar] [CrossRef]
- Yao, J.; Hamilton, R.T.; Cadenas, E.; Brinton, R.D. Decline in mitochondrial bioenergetics and shift to ketogenic profile in brain during reproductive senescence. Biochim. Biophys. Acta Gen. Subj. 2010, 1800, 1121–1126. [Google Scholar] [CrossRef] [PubMed]
- Boengler, K.; Kosiol, M.; Mayr, M.; Schulz, R.; Rohrbach, S. Mitochondria and ageing: Role in heart, skeletal muscle and adipose tissue. J. Cachexia. Sarcopenia Muscle 2017, 8, 349–369. [Google Scholar] [CrossRef]
- Moreno-Ulloa, A.; Nogueira, L.; Rodriguez, A.; Barboza, J.; Hogan, M.C.; Ceballos, G.; Villarreal, F.; Ramirez-Sanchez, I. Recovery of Indicators of Mitochondrial Biogenesis, Oxidative Stress, and Aging With (-)-Epicatechin in Senile Mice. J. Gerontol. Ser. A Biol. Sci. Med. Sci. 2015, 70, 1370–1378. [Google Scholar] [CrossRef]
- Preston, C.C.; Oberlin, A.S.; Holmuhamedov, E.L.; Gupta, A.; Sagar, S.; Syed, R.H.K.; Siddiqui, S.A.; Raghavakaimal, S.; Terzic, A.; Jahangir, A. Aging-induced alterations in gene transcripts and functional activity of mitochondrial oxidative phosphorylation complexes in the heart. Mech. Ageing Dev. 2008, 129, 304–312. [Google Scholar] [CrossRef] [PubMed]
- Short, K.R.; Bigelow, M.L.; Kahl, J.; Singh, R.; Coenen-Schimke, J.; Raghavakaimal, S.; Nair, K.S. Decline in skeletal muscle mitochondrial function with aging in humans. Proc. Natl. Acad. Sci. USA 2005, 102, 5618–5623. [Google Scholar] [CrossRef] [PubMed]
- García-Prat, L.; Martínez-Vicente, M.; Perdiguero, E.; Ortet, L.; Rodríguez-Ubreva, J.; Rebollo, E.; Ruiz-Bonilla, V.; Gutarra, S.; Ballestar, E.; Serrano, A.L.; et al. Autophagy maintains stemness by preventing senescence. Nature 2016, 529, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Chistiakov, D.A.; Shkurat, T.P.; Melnichenko, A.A.; Grechko, A.V.; Orekhov, A.N. The role of mitochondrial dysfunction in cardiovascular disease: A brief review. Ann. Med. 2018, 50, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Srinivas, U.S.; Tan, B.W.Q.; Vellayappan, B.A.; Jeyasekharan, A.D. ROS and the DNA damage response in cancer. Redox Biol. 2019, 25, 101084. [Google Scholar] [CrossRef] [PubMed]
- Masschelin, P.M.; Cox, A.R.; Chernis, N.; Hartig, S.M. The Impact of Oxidative Stress on Adipose Tissue Energy Balance. Front. Physiol. 2020, 10, 1638. [Google Scholar] [CrossRef] [PubMed]
- Vafai, S.B.; Mootha, V.K. Mitochondrial disorders as windows into an ancient organelle. Nature 2012, 491, 374–383. [Google Scholar] [CrossRef]
- Stanga, S.; Caretto, A.; Boido, M.; Vercelli, A. Mitochondrial dysfunctions: A red thread across neurodegenerative diseases. Int. J. Mol. Sci. 2020, 21, 3719. [Google Scholar] [CrossRef]
- Cassim, S.; Vučetić, M.; Ždralević, M.; Pouyssegur, J. Warburg and beyond: The power of mitochondrial metabolism to collaborate or replace fermentative glycolysis in cancer. Cancers 2020, 12, 1119. [Google Scholar] [CrossRef]
- Schüller, H.J. Transcriptional control of nonfermentative metabolism in the yeast Saccharomyces cerevisiae. Curr. Genet. 2003, 43, 139–160. [Google Scholar] [CrossRef] [PubMed]
- Rolland, F.; Winderickx, J.; Thevelein, J.M. Glucose-sensing and -signalling mechanisms in yeast. FEMS Yeast Res. 2002, 2, 183–201. [Google Scholar] [CrossRef]
- Kayikci, Ö.; Nielsen, J. Glucose repression in Saccharomyces cerevisiae. FEMS Yeast Res. 2015, 15. [Google Scholar] [CrossRef] [PubMed]
- Cardenas, M.E.; Cutler, N.S.; Lorenz, M.C.; Di Como, C.J.; Heitman, J. The TOR signaling cascade regulates gene expression in response to nutrients. Genes Dev. 1999, 13, 3271–3327. [Google Scholar] [CrossRef]
- Hardwick, J.S.; Kuruvilla, F.G.; Tong, J.K.; Shamji, A.F.; Schreiber, S.L. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 1999, 96, 14866–14870. [Google Scholar] [CrossRef]
- Proud, C.G. Regulation of mammalian translation factors by nutrients. Eur. J. Biochem. 2002, 269, 5338–5349. [Google Scholar] [CrossRef]
- Barba-Aliaga, M.; Villarroel-Vicente, C.; Stanciu, A.; Corman, A.; Martínez-Pastor, M.T.; Alepuz, P. Yeast translation elongation factor eif5a expression is regulated by nutrient availability through different signalling pathways. Int. J. Mol. Sci. 2021, 22, 219. [Google Scholar] [CrossRef]
- Fendt, S.M.; Sauer, U. Transcriptional regulation of respiration in yeast metabolizing differently repressive carbon substrates. BMC Syst. Biol. 2010, 4, 1–11. [Google Scholar] [CrossRef]
- Galdieri, L.; Mehrotra, S.; Yu, S.; Vancura, A. Transcriptional regulation in yeast during diauxic shift and stationary phase. Omi. A J. Integr. Biol. 2010, 14, 629–638. [Google Scholar] [CrossRef]
- Zhang, L.; Hach, A. Molecular mechanism of heme signaling in yeast: The transcriptional activator Hap1 serves as the key mediator. Cell. Mol. Life Sci. 1999, 56, 415–426. [Google Scholar] [CrossRef]
- Forsburg, S.L.; Guarente, L. Identification and characterization of HAP4: A third component of the CCAAT-bound HAP2/HAP3 heteromer. Genes Dev. 1989, 3, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Zaman, S.; Lippman, S.I.; Schneper, L.; Slonim, N.; Broach, J.R. Glucose regulates transcription in yeast through a network of signaling pathways. Mol. Syst. Biol. 2009, 5, 245. [Google Scholar] [CrossRef]
- Schneider, J.C.; Guarente, L. Regulation of the yeast CYT1 gene encoding cytochrome c1 by HAP1 and HAP2/3/4. Mol. Cell. Biol. 1991, 11, 4934–4942. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Zhang, T.; Bu, P.; Zeng, J.; Vancura, A. Increased heme synthesis in yeast induces a metabolic switch from fermentation to respiration even under conditions of glucose repression. J. Biol. Chem. 2017, 292, 16942–16954. [Google Scholar] [CrossRef] [PubMed]
- Zitomer, R.S.; Lowry, C.V. Regulation of gene expression by oxygen in Saccharomyces cerevisiae. Microbiol. Rev. 1992, 56, 1–11. [Google Scholar] [CrossRef]
- Kastaniotis, A.J.; Mennella, T.A.; Konrad, C.; Torres, A.M.R.; Zitomer, R.S. Roles of Transcription Factor Mot3 and Chromatin in Repression of the Hypoxic Gene ANB1 in Yeast. Mol. Cell. Biol. 2000, 20, 7088–7098. [Google Scholar] [CrossRef]
- Klinkenberg, L.G.; Mennella, T.A.; Luetkenhaus, K.; Zitomer, R.S. Combinatorial repression of the hypoxic genes of Saccharomyces cerevisiae by DNA binding proteins Rox1 and Mot3. Eukaryot. Cell 2005, 4, 649–660. [Google Scholar] [CrossRef]
- Sertil, O.; Kapoor, R.; Cohen, B.D.; Abramova, N.; Lowry, C.V. Synergistic repression of anaerobic genes by Mot3 and Rox1 in Saccharomyces cerevisiae. Nucleic Acids Res. 2003, 31, 5831–5837. [Google Scholar] [CrossRef][Green Version]
- Tariq, M.; Ito, A.; Ishfaq, M.; Bradshaw, E.; Yoshida, M. Eukaryotic translation initiation factor 5A (eIF5A) is essential for HIF-1α activation in hypoxia. Biochem. Biophys. Res. Commun. 2016, 470, 417–424. [Google Scholar] [CrossRef]
- Hickman, M.J.; Winston, F. Heme Levels Switch the Function of Hap1 of Saccharomyces cerevisiae between Transcriptional Activator and Transcriptional Repressor. Mol. Cell. Biol. 2007, 27, 7414–7424. [Google Scholar] [CrossRef]
- Cao, T.T.; Lin, S.H.; Fu, L.; Tang, Z.; Che, C.M.; Zhang, L.Y.; Ming, X.Y.; Liu, T.F.; Tang, X.M.; Tan, B.B.; et al. Eukaryotic translation initiation factor 5A2 promotes metabolic reprogramming in hepatocellular carcinoma cells. Carcinogenesis 2017, 38, 94–104. [Google Scholar] [CrossRef] [PubMed]
- San-Millán, I.; Brooks, G.A. Reexamining cancer metabolism: Lactate production for carcinogenesis could be the purpose and explanation of the Warburg Effect. Carcinogenesis 2017, 38, 119–133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.Y.; Tu, C.Y.; Shao, C.X.; Wang, W.K.; Zhu, J.D.; Cai, Y.; Mao, J.Y.; Chen, W. GC7 blocks epithelial-mesenchymal transition and reverses hypoxia-induced chemotherapy resistance in hepatocellular carcinoma cells. Am. J. Transl. Res. 2017, 9, 2608. [Google Scholar] [PubMed]
- Sun, Z.; Cheng, Z.; Taylor, C.A.; Mcconkey, B.J.; Thompson, J.E. Apoptosis induction by eIF5A1 involves activation of the intrinsic mitochondrial pathway. J. Cell. Physiol. 2010, 223, 798–809. [Google Scholar] [CrossRef]
- Weir, B.A.; Yaffe, M.P. Mmd1p, a Novel, Conserved Protein Essential for Normal Mitochondrial Morphology and Distribution in the Fission Yeast Schizosaccharomyces pombe. Mol. Biol. Cell 2004, 15, 1656–1665. [Google Scholar] [CrossRef]
- Melis, N.; Rubera, I.; Cougnon, M.; Giraud, S.; Mograbi, B.; Belaid, A.; Pisani, D.F.; Huber, S.M.; Lacas-Gervais, S.; Fragaki, K.; et al. Targeting eIF5A hypusination prevents anoxic cell death through mitochondrial silencing and improves kidney transplant outcome. J. Am. Soc. Nephrol. 2017, 28, 811–822. [Google Scholar] [CrossRef]
- Giraud, S.; Kerforne, T.; Zely, J.; Ameteau, V.; Couturier, P.; Tauc, M.; Hauet, T. The inhibition of eIF5A hypusination by GC7, a preconditioning protocol to prevent brain death-induced renal injuries in a preclinical porcine kidney transplantation model. Am. J. Transplant. 2020, 20, 3326–3340. [Google Scholar] [CrossRef]
- Bourourou, M.; Gouix, E.; Melis, N.; Friard, J.; Heurteaux, C.; Tauc, M.; Blondeau, N. Inhibition of eIF5A hypusination pathway as a new pharmacological target for stroke therapy. J. Cereb. Blood Flow Metab. 2021, 41, 1080–1090. [Google Scholar] [CrossRef]
- Kaiser, A.; Heiss, K.; Mueller, A.K.; Fimmers, R.; Matthes, J.; Njuguna, J.T. Inhibition of EIF-5A prevents apoptosis in human cardiomyocytes after malaria infection. Amino Acids 2020, 52, 693–710. [Google Scholar] [CrossRef]
- Igarashi, K.; Kashiwagi, K. Functional roles of polyamines and their metabolite acrolein in eukaryotic cells. Amino Acids 2021, 53, 1473–1492. [Google Scholar] [CrossRef]
- Pegg, A.E. Mammalian polyamine metabolism and function. IUBMB Life 2009, 61, 880–894. [Google Scholar] [CrossRef] [PubMed]
- Agostinelli, E.; Marques, M.P.M.; Calheiros, R.; Gil, F.P.S.C.; Tempera, G.; Viceconte, N.; Battaglia, V.; Grancara, S.; Toninello, A. Polyamines: Fundamental characters in chemistry and biology. Amino Acids 2010, 38, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Grancara, S.; Dalla Via, L.; García-Argáez, A.N.; Ohkubo, S.; Pacella, E.; Manente, S.; Bragadin, M.; Toninello, A.; Agostinelli, E. Spermine cycling in mitochondria is mediated by adenine nucleotide translocase activity: Mechanism and pathophysiological implications. Amino Acids 2016, 48, 2327–2337. [Google Scholar] [CrossRef] [PubMed]
- Tassani, V.; Campagnolo, M.; Toninello, A.; Siliprandi, D. The contribution of endogenous polyamines to the permeability transition of rat liver mitochondria. Biochem. Biophys. Res. Commun. 1996, 226, 850–854. [Google Scholar] [CrossRef]
- Uemura, T.; Higashi, K.; Takigawa, M.; Toida, T.; Kashiwagi, K.; Igarashi, K. Polyamine modulon in yeast-Stimulation of COX4 synthesis by spermidine at the level of translation. Int. J. Biochem. Cell Biol. 2009, 41, 2538–2545. [Google Scholar] [CrossRef]
- Sriram, A.; Bohlen, J.; Teleman, A.A. Translation acrobatics: How cancer cells exploit alternate modes of translational initiation. EMBO Rep. 2018, 19, e45947. [Google Scholar] [CrossRef]
- Joaquín Cáceres, C.; Angulo, J.; Lowy, F.; Contreras, N.; Walters, B.; Olivares, E.; Allouche, D.; Merviel, A.; Pino, K.; Sargueil, B.; et al. Non-canonical translation initiation of the spliced mRNA encoding the human T-cell leukemia virus type 1 basic leucine zipper protein. Nucleic Acids Res. 2018, 46, 11030–11047. [Google Scholar] [CrossRef]
- Manjunath, H.; Zhang, H.; Rehfeld, F.; Han, J.; Chang, T.C.; Mendell, J.T. Suppression of Ribosomal Pausing by eIF5A Is Necessary to Maintain the Fidelity of Start Codon Selection. Cell Rep. 2019, 29, 3134–3146. [Google Scholar] [CrossRef]
- Ivanov, I.P.; Shin, B.S.; Loughran, G.; Tzani, I.; Young-Baird, S.K.; Cao, C.; Atkins, J.F.; Dever, T.E. Polyamine Control of Translation Elongation Regulates Start Site Selection on Antizyme Inhibitor mRNA via Ribosome Queuing. Mol. Cell 2018, 70, 254–264. [Google Scholar] [CrossRef]
- Lipowsky, G.; Bischoff, F.R.; Schwarzmaier, P.; Kraft, R.; Kostka, S.; Hartmann, E.; Kutay, U.; Görlich, D. Exportin 4: A mediator of a novel nuclear export pathway in higher eukaryotes. EMBO J. 2000, 19, 4362–4371. [Google Scholar] [CrossRef]
- Turpaev, K.T. Translation Factor eIF5A, Modification with Hypusine and Role in Regulation of Gene Expression. eIF5A as a Target for Pharmacological Interventions. Biochemistry 2018, 83, 863–873. [Google Scholar] [CrossRef] [PubMed]
- Jao, D.L.E.; Chen, K.Y. Subcellular localization of the hypusine-containing eukaryotic initiation factor 5A by immunofluorescent staining and green fluorescent protein tagging. J. Cell. Biochem. 2002, 86, 590–600. [Google Scholar] [CrossRef] [PubMed]
- Miyake, T.; Pradeep, S.; Wu, S.Y.; Rupaimoole, R.; Zand, B.; Wen, Y.; Gharpure, K.M.; Nagaraja, A.S.; Hu, W.; Cho, M.S.; et al. XPO1/CRM1 inhibition causes antitumor effects by mitochondrial accumulation of eIF5A. Clin. Cancer Res. 2015, 21, 3286–3297. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhan, X.; Li, M.; Li, G.; Zhang, P.; Xiao, Z.; Shao, M.; Peng, F.; Hu, R.; Chen, Z. Mitochondrial proteomics of nasopharyngeal carcinoma metastasis. BMC Med. Genomics 2012, 5, 1–17. [Google Scholar] [CrossRef]
- Pereira, K.D.; Tamborlin, L.; Meneguello, L.; de Proença, A.R.G.; Almeida, I.C.D.P.A.; Lourenço, R.F.; Luchessi, A.D. Alternative Start Codon Connects eIF5A to Mitochondria. J. Cell. Physiol. 2016, 231, 2682–2689. [Google Scholar] [CrossRef]
- Pereira, K.D.; Tamborlin, L.; de Lima, T.I.; Consonni, S.R.; Silveira, L.R.; Luchessi, A.D. Alternative human eIF5A protein isoform plays a critical role in mitochondria. J. Cell. Biochem. 2021, 122, 549–561. [Google Scholar] [CrossRef]
- Puleston, D.J.; Buck, M.D.; Klein Geltink, R.I.; Kyle, R.L.; Caputa, G.; O’Sullivan, D.; Cameron, A.M.; Castoldi, A.; Musa, Y.; Kabat, A.M.; et al. Polyamines and eIF5A Hypusination Modulate Mitochondrial Respiration and Macrophage Activation. Cell Metab. 2019, 30, 1–12. [Google Scholar] [CrossRef]
- Cougnon, M.; Carcy, R.; Melis, N.; Rubera, I.; Duranton, C.; Dumas, K.; Tanti, J.F.; Pons, C.; Soubeiran, N.; Shkreli, M.; et al. Inhibition of eIF5A hypusination reprogrammes metabolism and glucose handling in mouse kidney. Cell Death Dis. 2021, 12, 283. [Google Scholar] [CrossRef]
- Ma, D.; Zheng, B.; Liu, H.L.; Zhao, Y.B.; Liu, X.; Zhang, X.H.; Li, Q.; Shi, W.B.; Suzuki, T.; Wen, J.K. Klf5 down-regulation induces vascular senescence through eIF5a depletion and mitochondrial fission. PLoS Biol. 2020, 18, e3000808. [Google Scholar] [CrossRef]
- Adebayo, M.; Singh, S.; Singh, A.P.; Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 2021, 35, e21620. [Google Scholar] [CrossRef]
- Scalabrino, G.; Ferioli, M.E. Polyamines in mammalian ageing: An oncological problem, too? A review. Mech. Ageing Dev. 1984, 26, 149–164. [Google Scholar] [CrossRef]
- Minois, N.; Carmona-Gutierrez, D.; Madeo, F. Polyamines in aging and disease. Aging 2011, 3, 716–732. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Knauer, H.; Schauer, A.; Büttner, S.; Ruckenstuhl, C.; Carmona-Gutierrez, D.; Ring, J.; Schroeder, S.; Magnes, C.; Antonacci, L.; et al. Induction of autophagy by spermidine promotes longevity. Nat. Cell Biol. 2009, 11, 1305–1314. [Google Scholar] [CrossRef] [PubMed]
- Gupta, V.K.; Scheunemann, L.; Eisenberg, T.; Mertel, S.; Bhukel, A.; Koemans, T.S.; Kramer, J.M.; Liu, K.S.Y.; Schroeder, S.; Stunnenberg, H.G.; et al. Restoring polyamines protects from age-induced memory impairment in an autophagy-dependent manner. Nat. Neurosci. 2013, 16, 1453–1460. [Google Scholar] [CrossRef] [PubMed]
- Eisenberg, T.; Abdellatif, M.; Schroeder, S.; Primessnig, U.; Stekovic, S.; Pendl, T.; Harger, A.; Schipke, J.; Zimmermann, A.; Schmidt, A.; et al. Cardioprotection and lifespan extension by the natural polyamine spermidine. Nat. Med. 2016, 22, 1428–1438. [Google Scholar] [CrossRef] [PubMed]
- Minois, N.; Carmona-Gutierrez, D.; Bauer, M.A.; Rockenfeller, P.; Eisenberg, T.; Brandhorst, S.; Sigrist, S.J.; Kroemer, G.; Madeo, F. Spermidine promotes stress resistance in Drosophila melanogaster through autophagy-dependent and -independent pathways. Cell Death Dis. 2012, 11, e401. [Google Scholar] [CrossRef]
- Madeo, F.; Eisenberg, T.; Pietrocola, F.; Kroemer, G. Spermidine in health and disease. Science 2018, 359. [Google Scholar] [CrossRef]
- Hersch, S.J.; Elgamal, S.; Katz, A.; Ibba, M.; Navarre, W.W. Translation initiation rate determines the impact of ribosome stalling on bacterial protein synthesis. J. Biol. Chem. 2014, 289, 28160–28171. [Google Scholar] [CrossRef]
- Schroeder, S.; Hofer, S.J.; Zimmermann, A.; Pechlaner, R.; Dammbrueck, C.; Pendl, T.; Marcello, G.M.; Pogatschnigg, V.; Bergmann, M.; Müller, M.; et al. Dietary spermidine improves cognitive function. Cell Rep. 2021, 35, 108985. [Google Scholar] [CrossRef]
- Zhang, J.; Culp, M.L.; Craver, J.G.; Darley-Usmar, V. Mitochondrial function and autophagy: Integrating proteotoxic, redox, and metabolic stress in Parkinson’s disease. J. Neurochem. 2018, 144, 691–709. [Google Scholar] [CrossRef]
- Yang, X.; Zhang, M.; Dai, Y.; Sun, Y.; Aman, Y.; Xu, Y.; Yu, P.; Zheng, Y.; Yang, J.; Zhu, X. Spermidine inhibits neurodegeneration and delays aging via the PINK1-PDR1-dependent mitophagy pathway in C. elegans. Aging 2020, 12, 16852–16866. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.T.; Piao, C.; Beuschel, C.B.; Toppe, D.; Kollipara, L.; Bogdanow, B.; Maglione, M.; Lützkendorf, J.; See, J.C.K.; Huang, S.; et al. eIF5A hypusination, boosted by dietary spermidine, protects from premature brain aging and mitochondrial dysfunction. Cell Rep. 2021, 35, 108941. [Google Scholar] [CrossRef] [PubMed]
- Partridge, L.; Fuentealba, M.; Kennedy, B.K. The quest to slow ageing through drug discovery. Nat. Rev. Drug Discov. 2020, 19, 513–532. [Google Scholar] [CrossRef] [PubMed]
- Tain, L.S.; Jain, C.; Nespital, T.; Froehlich, J.; Hinze, Y.; Grönke, S.; Partridge, L. Longevity in response to lowered insulin signaling requires glycine N-methyltransferase-dependent spermidine production. Aging Cell 2020, 19, e13043. [Google Scholar] [CrossRef] [PubMed]
- Ganapathi, M.; Padgett, L.R.; Yamada, K.; Devinsky, O.; Willaert, R.; Person, R.; Au, P.Y.B.; Tagoe, J.; McDonald, M.; Karlowicz, D.; et al. Recessive Rare Variants in Deoxyhypusine Synthase, an Enzyme Involved in the Synthesis of Hypusine, Are Associated with a Neurodevelopmental Disorder. Am. J. Hum. Genet. 2019, 104, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Smith, S.B.; Yoon, Y. The short variant of the mitochondrial dynamin OPA1 maintains mitochondrial energetics and cristae structure. J. Biol. Chem. 2017, 292, 7115–7130. [Google Scholar] [CrossRef]
- Arlt, H.; Steglich, G.; Perryman, R.; Guiard, B.; Neupert, W.; Langer, T. The formation of respiratory chain complexes in mitochondria is under the proteolytic control of the m-AAA protease. EMBO J. 1998, 17, 4837–4847. [Google Scholar] [CrossRef]
- Chen, Y.C.; Cheng, T.H.; Lin, W.L.; Chen, C.L.; Yang, W.Y.; Blackstone, C.; Chang, C.R. Srv2 Is a Pro-fission Factor that Modulates Yeast Mitochondrial Morphology and Respiration by Regulating Actin Assembly. iScience 2019, 11, 305–317. [Google Scholar] [CrossRef]
- Hansen, K.G.; Herrmann, J.M. Transport of Proteins into Mitochondria. Protein J. 2019, 38, 330–342. [Google Scholar] [CrossRef]
- Hofer, S.J.; Liang, Y.T.; Zimmermann, A.; Schroeder, S.; Dengjel, J.; Kroemer, G.; Eisenberg, T.; Sigrist, S.J.; Madeo, F. Spermidine-induced hypusination preserves mitochondrial and cognitive function during aging. Autophagy 2021, 17, 2037–2039. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barba-Aliaga, M.; Alepuz, P. Role of eIF5A in Mitochondrial Function. Int. J. Mol. Sci. 2022, 23, 1284. https://doi.org/10.3390/ijms23031284
Barba-Aliaga M, Alepuz P. Role of eIF5A in Mitochondrial Function. International Journal of Molecular Sciences. 2022; 23(3):1284. https://doi.org/10.3390/ijms23031284
Chicago/Turabian StyleBarba-Aliaga, Marina, and Paula Alepuz. 2022. "Role of eIF5A in Mitochondrial Function" International Journal of Molecular Sciences 23, no. 3: 1284. https://doi.org/10.3390/ijms23031284
APA StyleBarba-Aliaga, M., & Alepuz, P. (2022). Role of eIF5A in Mitochondrial Function. International Journal of Molecular Sciences, 23(3), 1284. https://doi.org/10.3390/ijms23031284





