The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens
Abstract
:1. Introduction
2. Results
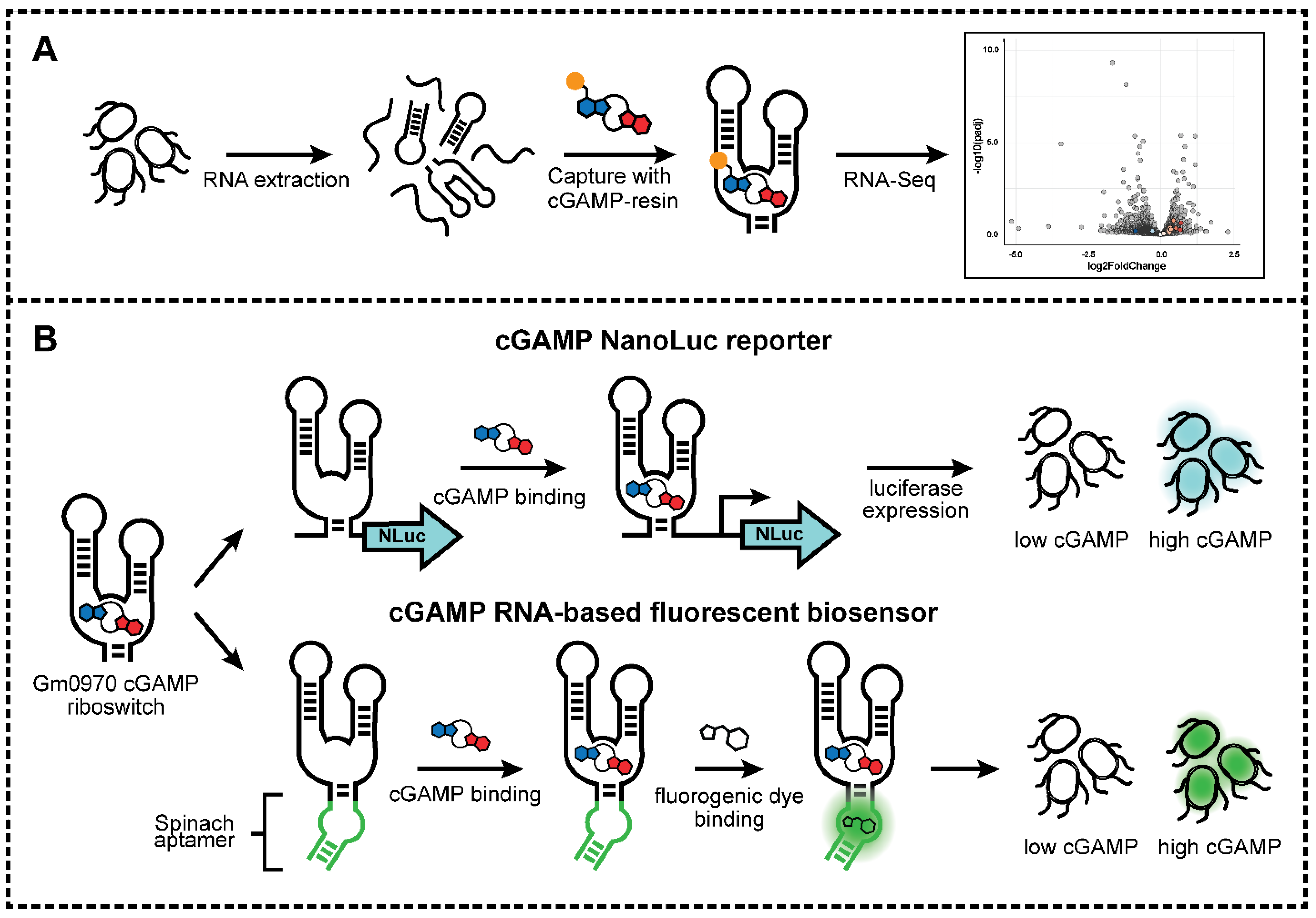
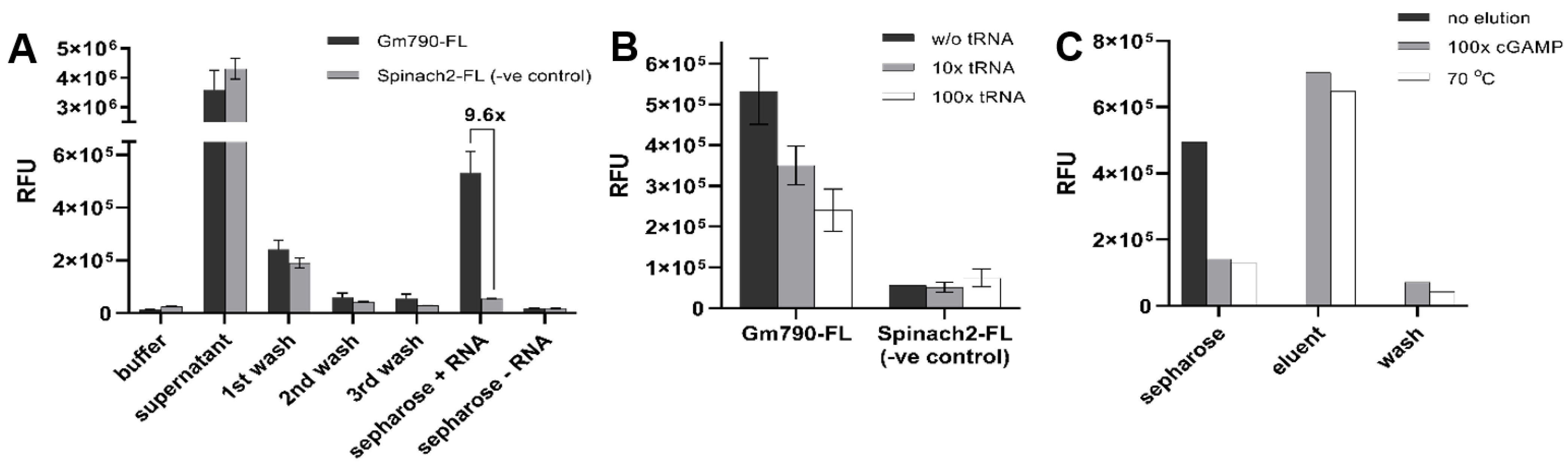
2.1. Riboswitch Analysis: Development of cGAMP Riboswitch Affinity Capture Method
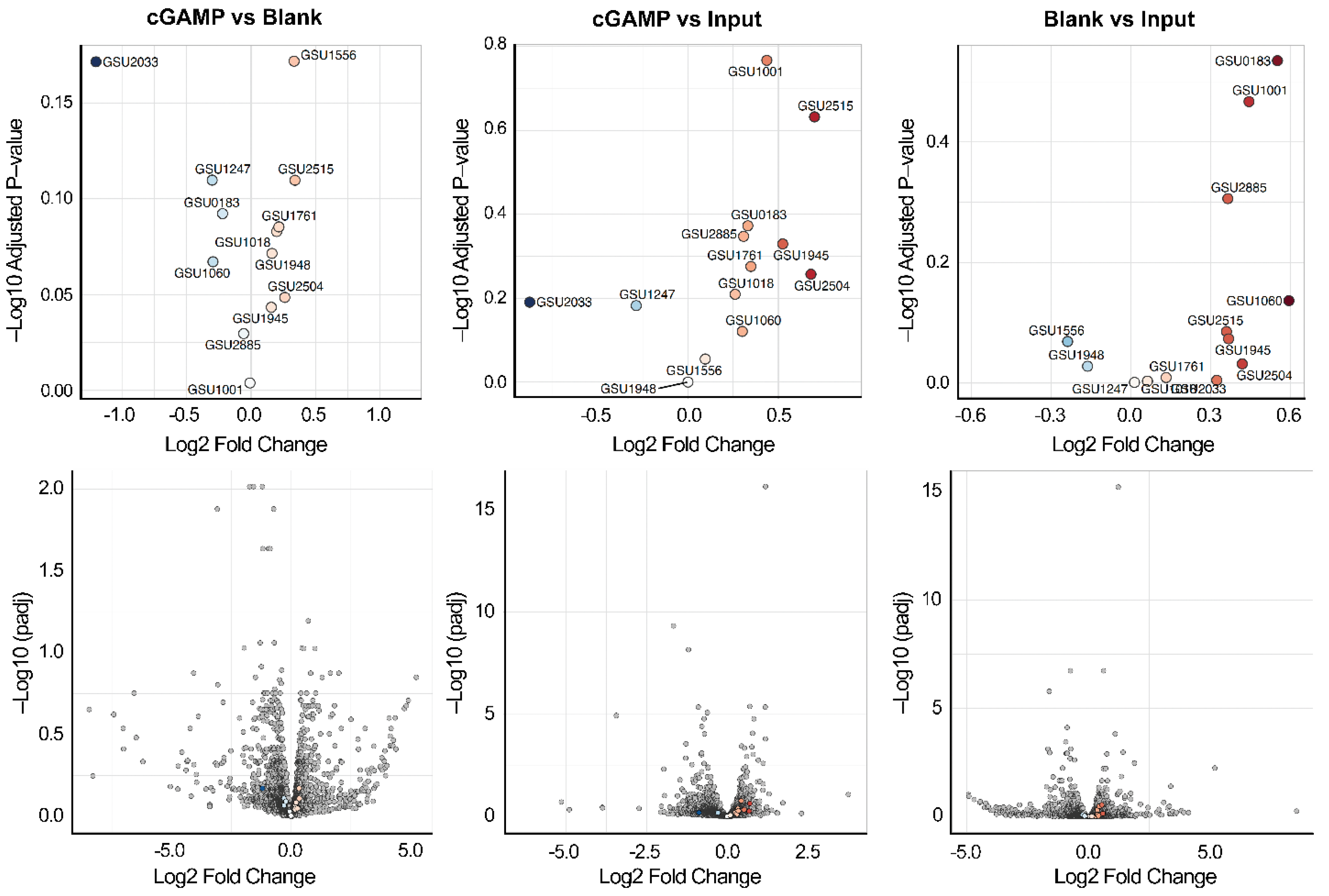
2.2. Riboswitch Analysis: Transcriptome Affinity Capture and RNA-Seq Analysis
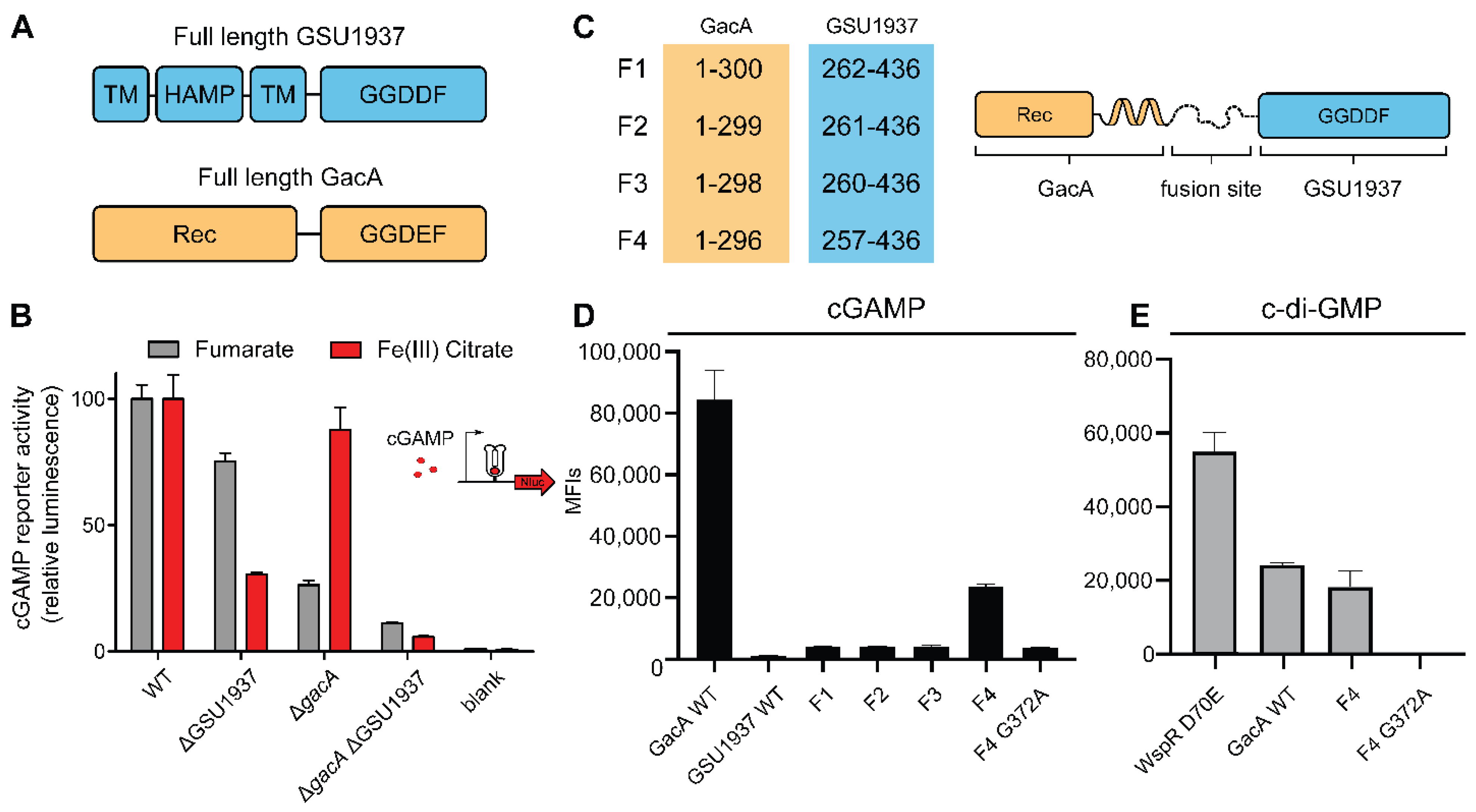
2.3. Riboswitch Application: Analysis of a Second Hypr GGDEF in Geobacter sulfurreducens
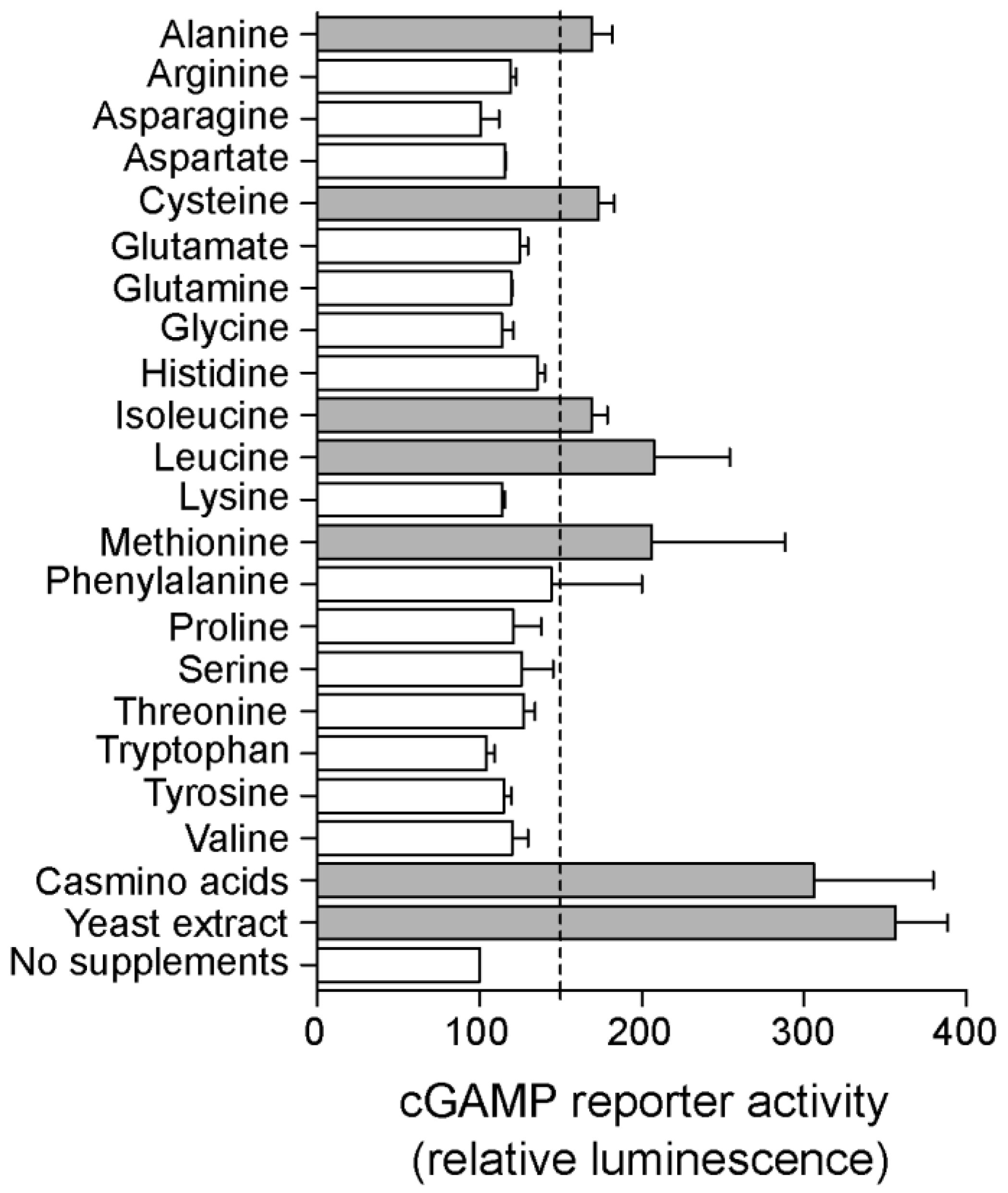
2.4. Riboswitch Application: Screening for New Conditions and Genes That Affect cGAMP Levels in Geobacter sulfurreducens
3. Materials and Methods
3.1. Preparation of cGAMP-Sepharose Capture Compound
3.2. RNA Labeling with Fluorescein
3.3. Binding of RNA Samples to cGAMP-Sepharose
3.4. Total RNA Isolation and Quality Check
3.5. rRNA Depletion
3.6. RNAseq
3.7. Growth and Transposon Library Generation
3.8. Deletion of GSU1937
3.9. Nanoluciferase (Nluc) Reporter Assay
3.10. Fluorescent Biosensor Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Römling, U.; Galperin, M.Y.; Gomelsky, M. Cyclic Di-GMP: The First 25 Years of a Universal Bacterial Second Messenger. Microbiol. Mol. Biol. Rev. 2013, 77, 1–52. [Google Scholar] [CrossRef] [Green Version]
- Witte, G.; Hartung, S.; Büttner, K.; Hopfner, K.P. Structural Biochemistry of a Bacterial Checkpoint Protein Reveals Diadenylate Cyclase Activity Regulated by DNA Recombination Intermediates. Mol. Cell 2008, 30, 167–178. [Google Scholar] [CrossRef]
- Davies, B.W.; Bogard, R.W.; Young, T.S.; Mekalanos, J.J. Coordinated Regulation of Accessory Genetic Elements Produces Cyclic Di-Nucleotides for V. Cholerae Virulence. Cell 2012, 149, 358–370. [Google Scholar] [CrossRef] [Green Version]
- Ye, Q.; Lau, R.K.; Mathews, I.T.; Birkholz, E.A.; Watrous, J.D.; Azimi, C.S.; Pogliano, J.; Jain, M.; Corbett, K.D. HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial CGAS-like Enzymes to Mediate Bacteriophage Immunity. Mol. Cell 2020, 77, 709. [Google Scholar] [CrossRef] [PubMed]
- Whiteley, A.T.; Eaglesham, J.B.; de Oliveira Mann, C.C.; Morehouse, B.R.; Lowey, B.; Nieminen, E.A.; Danilchanka, O.; King, D.S.; Lee, A.S.Y.; Mekalanos, J.J.; et al. Bacterial CGAS-like Enzymes Synthesize Diverse Nucleotide Signals. Nature 2019, 567, 194–199. [Google Scholar] [CrossRef] [PubMed]
- Kazlauskiene, M.; Kostiuk, G.; Venclovas, Č.; Tamulaitis, G.; Siksnys, V. A Cyclic Oligonucleotide Signaling Pathway in Type III CRISPR-Cas Systems. Science 2017, 357, 605–609. [Google Scholar] [CrossRef] [Green Version]
- Nelson, J.W.; Sudarsan, N.; Furukawa, K.; Weinberg, Z.; Wang, J.X.; Breaker, R.R. Riboswitches in Eubacteria Sense the Second Messenger C-Di-AMP. Nat. Chem. Biol. 2013, 9, 834–839. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, C.A.; Wilson, S.C.; Hickey, S.F.; Gonzalez, T.L.; Su, Y.; Hallberg, Z.F.; Brewer, T.F.; Iavarone, A.; Carlson, H.K.; Hsieh, Y.-F.; et al. GEMM-I Riboswitches from Geobacter Sense the Bacterial Second Messenger Cyclic AMP-GMP. Proc. Natl. Acad. Sci. USA 2015, 112, 5383–5388. [Google Scholar] [CrossRef] [Green Version]
- Sudarsan, N.; Lee, E.R.; Weinberg, Z.; Moy, R.H.; Kim, J.N.; Link, K.H.; Breaker, R.R. Riboswitches in Eubacteria Sense the Second Messenger Cyclic Di-GMP. Science 2008, 321, 411–413. [Google Scholar] [CrossRef] [Green Version]
- Kulshina, N.; Baird, N.J.; Ferré-D’Amaré, A.R. Recognition of the Bacterial Second Messenger Cyclic Diguanylate by Its Cognate Riboswitch. Nat. Struct. Mol. Biol. 2009, 16, 1212. [Google Scholar] [CrossRef]
- Gao, A.; Serganov, A. Structural Insights into Recognition of C-Di-AMP by the YdaO Riboswitch. Nat. Chem. Biol. 2014, 10, 787–792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, A.; Wang, X.C.; Kellenberger, C.A.; Rajashankar, K.R.; Jones, R.A.; Hammond, M.C.; Patel, D.J. Structural Basis for Molecular Discrimination by a 3′,3′-CGAMP Sensing Riboswitch. Cell Rep. 2015, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Hallberg, Z.F.; Wang, X.C.; Wright, T.A.; Nan, B.; Ad, O.; Yeo, J.; Hammond, M.C. Hybrid Promiscuous (Hypr) GGDEF Enzymes Produce Cyclic AMP-GMP (3′, 3′-CGAMP). Proc. Natl. Acad. Sci. USA 2016, 113, 1790–1795. [Google Scholar] [CrossRef] [Green Version]
- Cohen, D.; Melamed, S.; Millman, A.; Shulman, G.; Oppenheimer-Shaanan, Y.; Kacen, A.; Doron, S.; Amitai, G.; Sorek, R. Cyclic GMP–AMP Signalling Protects Bacteria against Viral Infection. Nature 2019, 574, 691–695. [Google Scholar] [CrossRef] [PubMed]
- Hallberg, Z.F.; Chan, C.H.; Wright, T.A.; Kranzusch, P.J.; Doxzen, K.W.; Park, J.J.; Bond, D.R.; Hammond, M.C. Structure and Mechanism of a Hypr GGDEF Enzyme That Activates CGAMP Signaling to Control Extracellular Metal Respiration. Elife 2019, 8, e43959. [Google Scholar] [CrossRef] [PubMed]
- Nesper, J.; Reinders, A.; Glatter, T.; Schmidt, A.; Jenal, U. A Novel Capture Compound for the Identification and Analysis of Cyclic Di-GMP Binding Proteins. J. Proteom. 2012, 75, 4874–4878. [Google Scholar] [CrossRef] [Green Version]
- Sureka, K.; Choi, P.H.; Precit, M.; Delince, M.; Pensinger, D.A.; Huynh, T.A.N.; Jurado, A.R.; Goo, Y.A.; Sadilek, M.; Iavarone, A.T.; et al. The Cyclic Di-Nucleotide c-Di-AMP Is an Allosteric Regulator of Metabolic Enzyme Function. Cell 2014, 158, 1389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, C.; Paul, R.; Samoray, D.; Amiot, N.C.; Giese, B.; Jenal, U.; Schirmer, T. Structural Basis of Activity and Allosteric Control of Diguanylate Cyclase. Proc. Natl. Acad. Sci. USA 2004, 101, 17084–17089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christen, B.; Christen, M.; Paul, R.; Schmid, F.; Folcher, M.; Jenoe, P.; Meuwly, M.; Jenal, U. Allosteric Control of Cyclic Di-GMP Signaling. J. Biol. Chem. 2006, 281, 32015–32024. [Google Scholar] [CrossRef]
- Galperin, M.Y. A Census of Membrane-Bound and Intracellular Signal Transduction Proteins in Bacteria: Bacterial IQ, Extroverts and Introverts. BMC Microbiol. 2005, 5, 35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, Y.; Srikanth, V.; Salazar-Morales, A.I.; Jain, R.; O’Brien, J.P.; Yi, S.M.; Soni, R.K.; Samatey, F.A.; Yalcin, S.E.; Malvankar, N.S. Structure of Geobacter Pili Reveals Secretory Rather than Nanowire Behaviour. Nature 2021, 597, 430–434. [Google Scholar] [CrossRef] [PubMed]
- Kilmury, S.L.N.; Burrows, L.L. Type IV Pilins Regulate Their Own Expression via Direct Intramembrane Interactions with the Sensor Kinase PilS. Proc. Natl. Acad. Sci. USA 2016, 113, 6017–6022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, W.; Yang, Z.; Lux, R.; Zhao, M.; Wang, J.; He, X.; Shi, W. Direct Visualization of the Interaction between Pilin and Exopolysaccharides of Myxococcus Xanthus with EGFP-Fused PilA Protein. FEMS Microbiol. Lett. 2012, 326, 23–30. [Google Scholar] [CrossRef]
- Wright, T.A.; Jiang, L.; Park, J.J.; Anderson, W.A.; Chen, G.; Hallberg, Z.F.; Nan, B.; Hammond, M.C. Second Messengers and Divergent HD-GYP Phosphodiesterases Regulate 3′,3′-CGAMP Signaling. Mol. Microbiol. 2020, 113, 222–236. [Google Scholar] [CrossRef]
- Gavira, J.A.; Gumerov, V.M.; Rico-Jiménez, M.; Petukh, M.; Upadhyay, A.A.; Ortega, A.; Matilla, M.A.; Zhulin, I.B.; Krell, T. How Bacterial Chemoreceptors Evolve Novel Ligand Specificities. MBio 2020, 11, e03066-19. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, S.I.; Takahashi, Y.; Yamamoto, K.; Suzuki, D.; Itoh, Y.; Sumita, K.; Uchida, Y.; Homma, M.; Imada, K.; Kawagishi, I. Identification of a Vibrio Cholerae Chemoreceptor That Senses Taurine and Amino Acids as Attractants. Sci. Rep. 2016, 6, 20866. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.F.; Machuca, M.A.; Rahman, M.M.; Koç, C.; Norton, R.S.; Smith, B.J.; Roujeinikova, A. Structure-Activity Relationship Study Reveals the Molecular Basis for Specific Sensing of Hydrophobic Amino Acids by the Campylobacter Jejuni Chemoreceptor Tlp3. Biomolecules 2020, 10, 744. [Google Scholar] [CrossRef]
- Minkyung, B.; Frank, D.; Ivan, A.; Justas, D.; Sergey, O.; Rie, L.G.; Jue, W.; Qian, C.; Kinch, L.N.; Schaeffer, R.D.; et al. Accurate Prediction of Protein Structures and Interactions Using a Three-Track Neural Network. Science 2021, 373, 871–876. [Google Scholar] [CrossRef]
- Le Guilloux, V.; Schmidtke, P.; Tuffery, P. Fpocket: An Open Source Platform for Ligand Pocket Detection. BMC Bioinform. 2009, 10, 168. [Google Scholar] [CrossRef] [Green Version]
- Simonovsky, M.; Meyers, J. DeeplyTough: Learning Structural Comparison of Protein Binding Sites. J. Chem. Inf. Model. 2020, 60, 2356–2366. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.H.; Levar, C.E.; Zacharoff, L.; Badalamenti, J.P.; Bond, D.R. Scarless Genome Editing and Stable Inducible Expression Vectors for Geobacter Sulfurreducens. Appl. Environ. Microbiol. 2015, 81, 7178–7186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rollefson, J.B.; Levar, C.E.; Bond, D.R. Identification of Genes Involved in Biofilm Formation and Respiration via Mini-Himar Transposon Mutagenesis of Geobacter Sulfurreducens. J. Bacteriol. 2009, 191, 4207–4217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouhenni, R.; Gehrke, A.; Saffarini, D. Identification of Genes Involved in Cytochrome c Biogenesis in Shewanella Oneidensis, Using a Modified Mariner Transposon. Appl. Environ. Microbiol. 2005, 71, 4935–4937. [Google Scholar] [CrossRef] [Green Version]
- Ellison, C.K.; Kan, J.; Dillard, R.S.; Kysela, D.T.; Ducret, A.; Berne, C.; Hampton, C.M.; Ke, Z.; Wright, E.R.; Biais, N.; et al. Obstruction of Pilus Retraction Stimulates Bacterial Surface Sensing. Science 2017, 358, 535–538. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.K.; de Anda, J.; Baker, A.E.; Bennett, R.R.; Luo, Y.; Lee, E.Y.; Keefe, J.A.; Helali, J.S.; Ma, J.; Zhao, K.; et al. Multigenerational Memory and Adaptive Adhesion in Early Bacterial Biofilm Communities. Proc. Natl. Acad. Sci. USA 2018, 115, 4471–4476. [Google Scholar] [CrossRef] [Green Version]
| Strain or Transposon ID | Relative Luminescence 1 | Disrupted Gene, Transposon Location and Description |
|---|---|---|
| WT | 100 ± 27.9 | |
| ΔgacA | 26.6 ± 6.5 | GSU1658; Hypr GGDEF |
| ΔesnD | 154.1 ± 32.8 | GSU3376; Diguanylate cyclase |
| empty | 1.3 ± 1.3 | |
| 6G9 | 27.3 ± 11.6 | GSU1508; xapK; 463/1119 nt |
| 3F4 | 35.7 ± 4.1 | GSU1508; xapK; 730/1119 nt |
| 2G8 | 41.7 ± 15 | GSU1512; Hypothetical |
| 2F9 | 46.2 ± 44.4 | GSU1497; pilA-C |
| 1C4 | 56.4 ± 11.3 | 11 bp deletion in GSU0946 (Sensor Diguanylate cyclase/Phosphodiesterase EAL, HAMP-; PAS-containing also Intergenic insertion, negative strand; GSU0538 (hspA-1 ATP dependent chaperone)—GSU0539 (hypothetical). |
| 4F11 | 189.5 ± 107.6 | GSU1491: pilB; 424/1707 nt |
| 9C1 | 204.1 ± 52.5 | GSU2220; esnB; CheW-like protein |
| 4E6 | 266.5 ± 41.4 | GSU1491; pilB; 1522/1707 nt |
| Gene Name | Catalytic Domain | Known Cache-Ligand Structures | |||||
|---|---|---|---|---|---|---|---|
| - | - | PctA: AA | Mlp24: AA | Tlp3: AA | CitA: | DcuS: | TlpB: |
| Citrate | Malate | Pyruvate | |||||
| GSU0400 | MCP | 0.17 | 0.18 | 0.22 | 0.26 | 0.29 | 0.28 |
| GUS0962 | HisK | 0.25 | 0.28 | 0.24 | 0.23 | 0.22 | 0.21 |
| GSU1030 | MCP | 0.6 | 0.63 | 0.79 | 0.82 | 0.92 | 0.91 |
| GSU1033 | MCP | 0.61 | 0.64 | 0.82 | 0.84 | 0.85 | 0.84 |
| GSU1303 | MCP | 0.62 | 0.66 | 0.80 | 0.81 | 0.90 | 0.89 |
| GSU2297 | HisK | 0.5 | 0.52 | 0.54 | 0.56 | 0.57 | 0.57 |
| GSU2388 | HisK | 0.7 | 0.79 | 0.99 | 0.99 | 0.89 | 0.89 |
| GSU2507 | HisK | 0.73 | 0.79 | 0.94 | 0.98 | 0.91 | 0.92 |
| GSU3356 | GGDEF | 0.76 | 0.8 | 0.99 | 0.98 | 0.89 | 0.97 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tan, Z.; Chan, C.H.; Maleska, M.; Banuelos Jara, B.; Lohman, B.K.; Ricks, N.J.; Bond, D.R.; Hammond, M.C. The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens. Int. J. Mol. Sci. 2022, 23, 1183. https://doi.org/10.3390/ijms23031183
Tan Z, Chan CH, Maleska M, Banuelos Jara B, Lohman BK, Ricks NJ, Bond DR, Hammond MC. The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens. International Journal of Molecular Sciences. 2022; 23(3):1183. https://doi.org/10.3390/ijms23031183
Chicago/Turabian StyleTan, Zhesen, Chi Ho Chan, Michael Maleska, Bryan Banuelos Jara, Brian K. Lohman, Nathan J. Ricks, Daniel R. Bond, and Ming C. Hammond. 2022. "The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens" International Journal of Molecular Sciences 23, no. 3: 1183. https://doi.org/10.3390/ijms23031183
APA StyleTan, Z., Chan, C. H., Maleska, M., Banuelos Jara, B., Lohman, B. K., Ricks, N. J., Bond, D. R., & Hammond, M. C. (2022). The Signaling Pathway That cGAMP Riboswitches Found: Analysis and Application of Riboswitches to Study cGAMP Signaling in Geobacter sulfurreducens. International Journal of Molecular Sciences, 23(3), 1183. https://doi.org/10.3390/ijms23031183






