Bacterial 2′-Deoxyguanosine Riboswitch Classes as Potential Targets for Antibiotics: A Structure and Dynamics Study
Abstract
:1. Introduction
2. Results
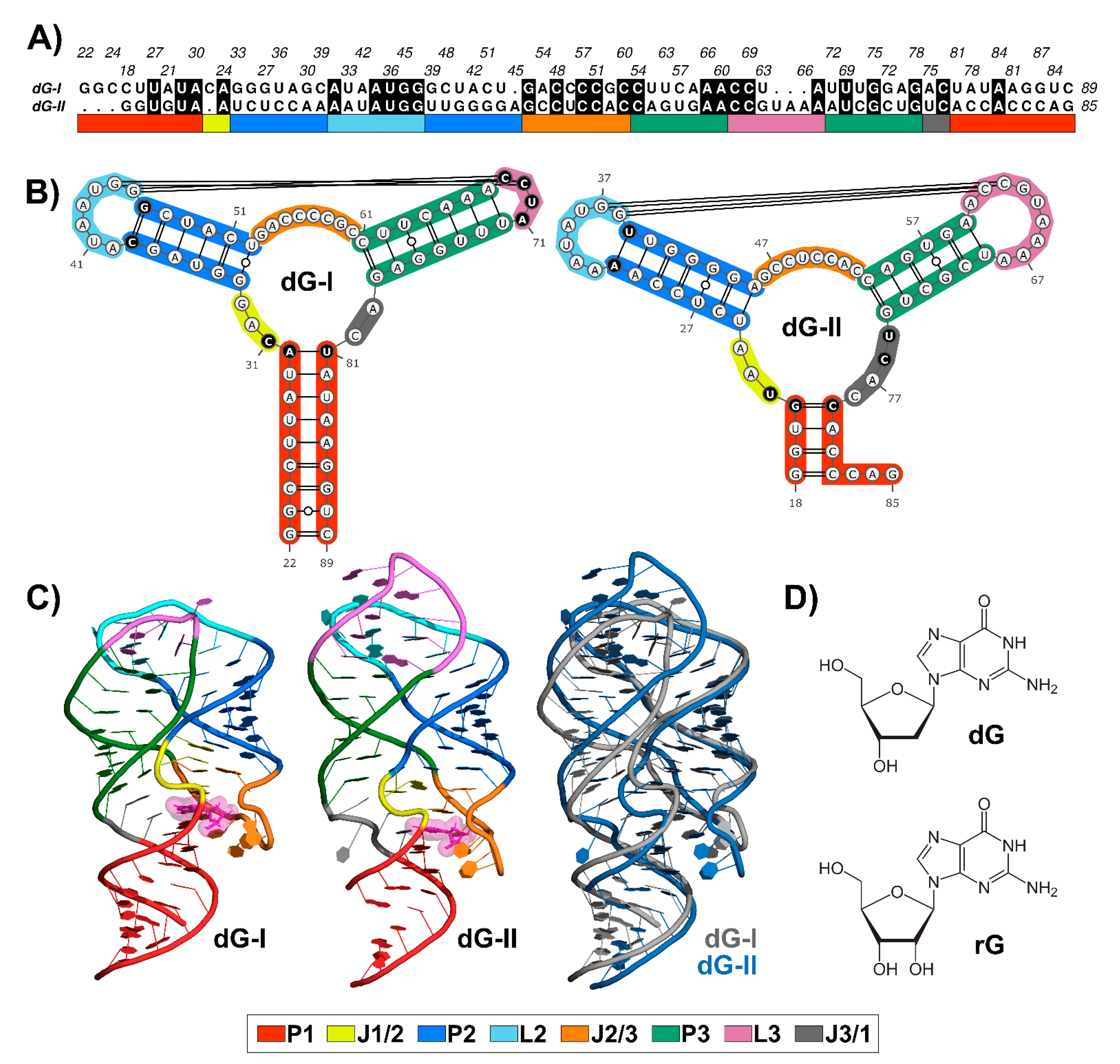
2.1. RNA Content Analysis
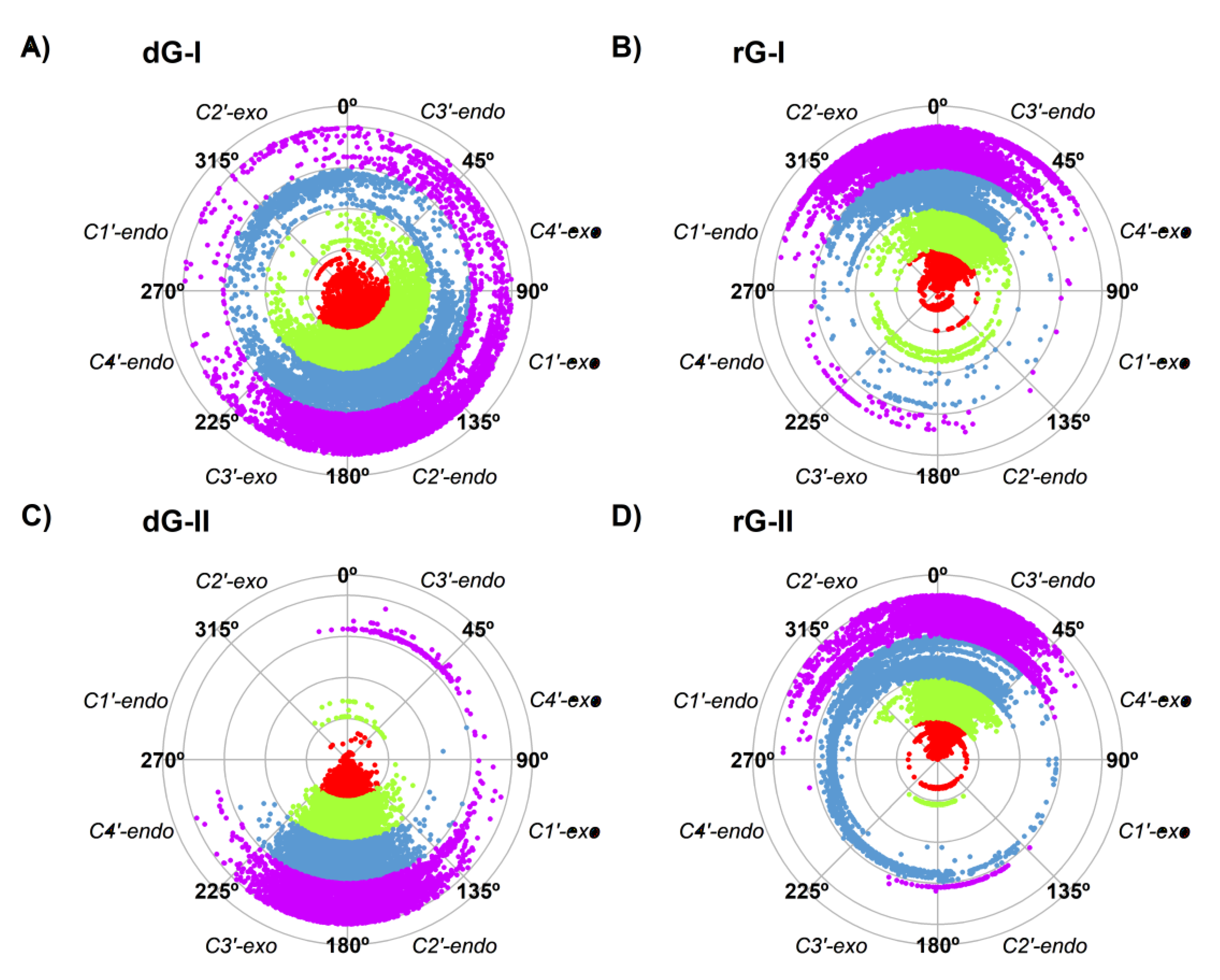
2.2. Influence of Ligands on Dynamics of 2′dG Riboswitches
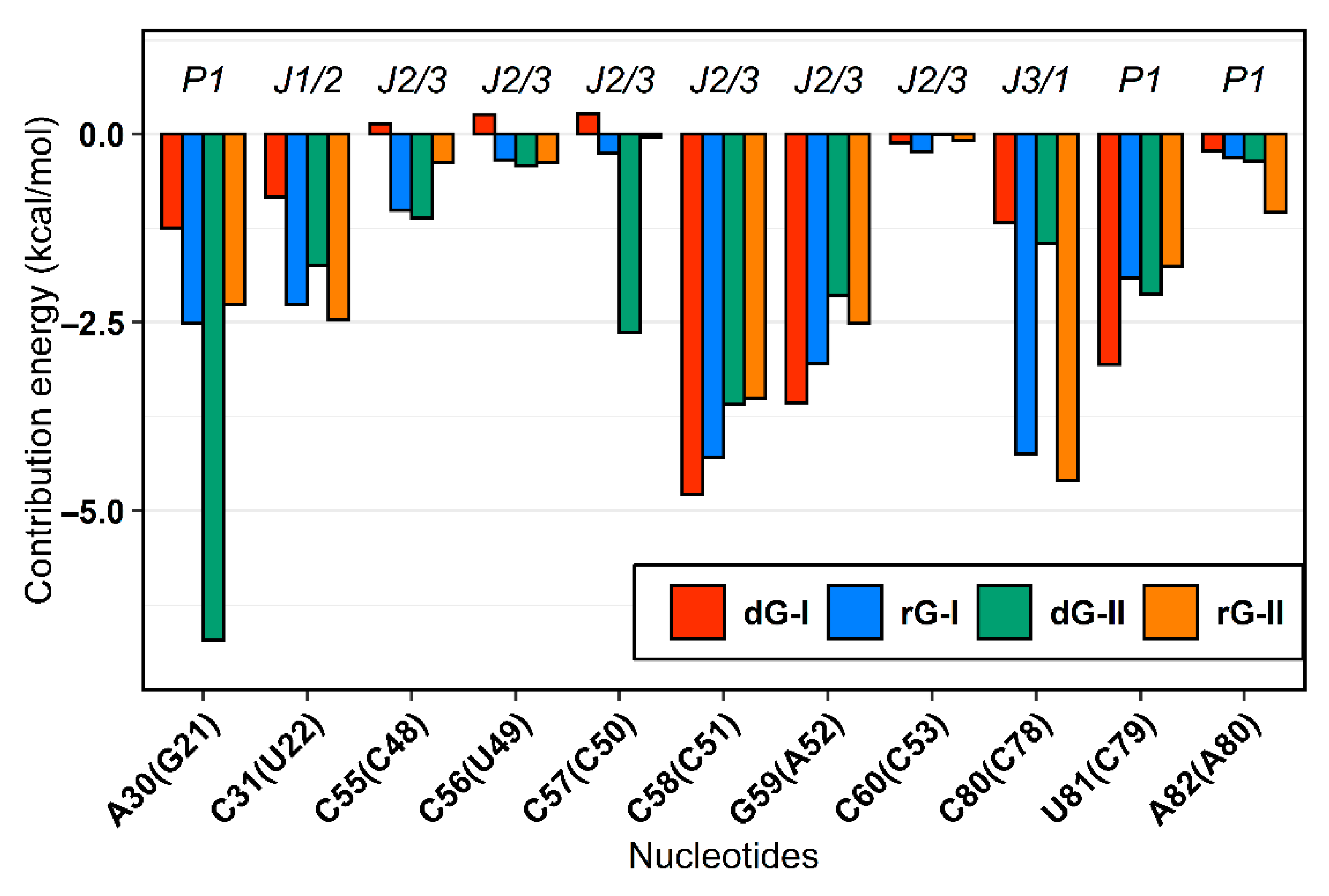
2.3. Analysis of Binding Free Energy
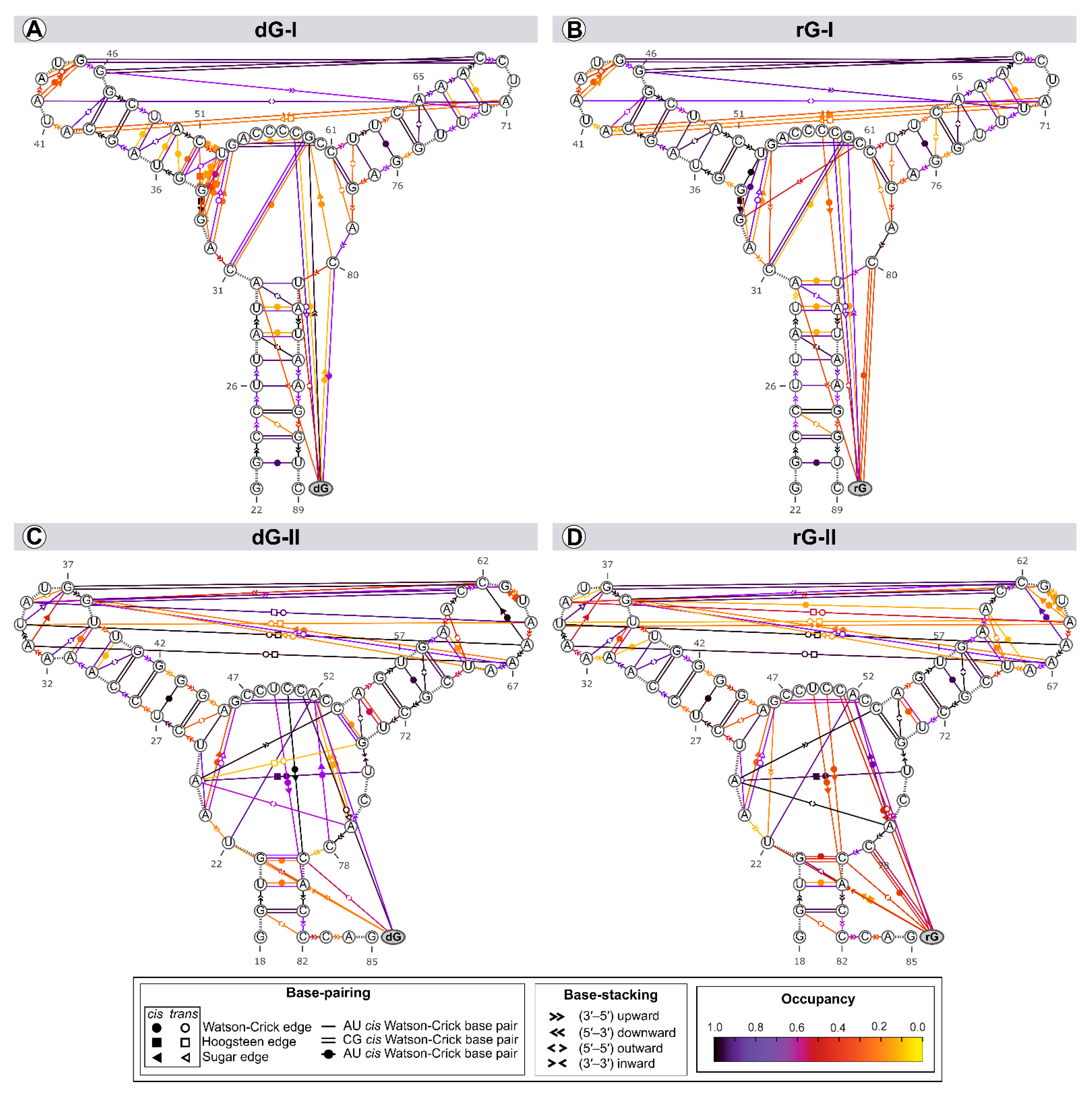
2.4. The Purine Riboswitch Family Share Identical Critical Nucleotides to Ligand Binding
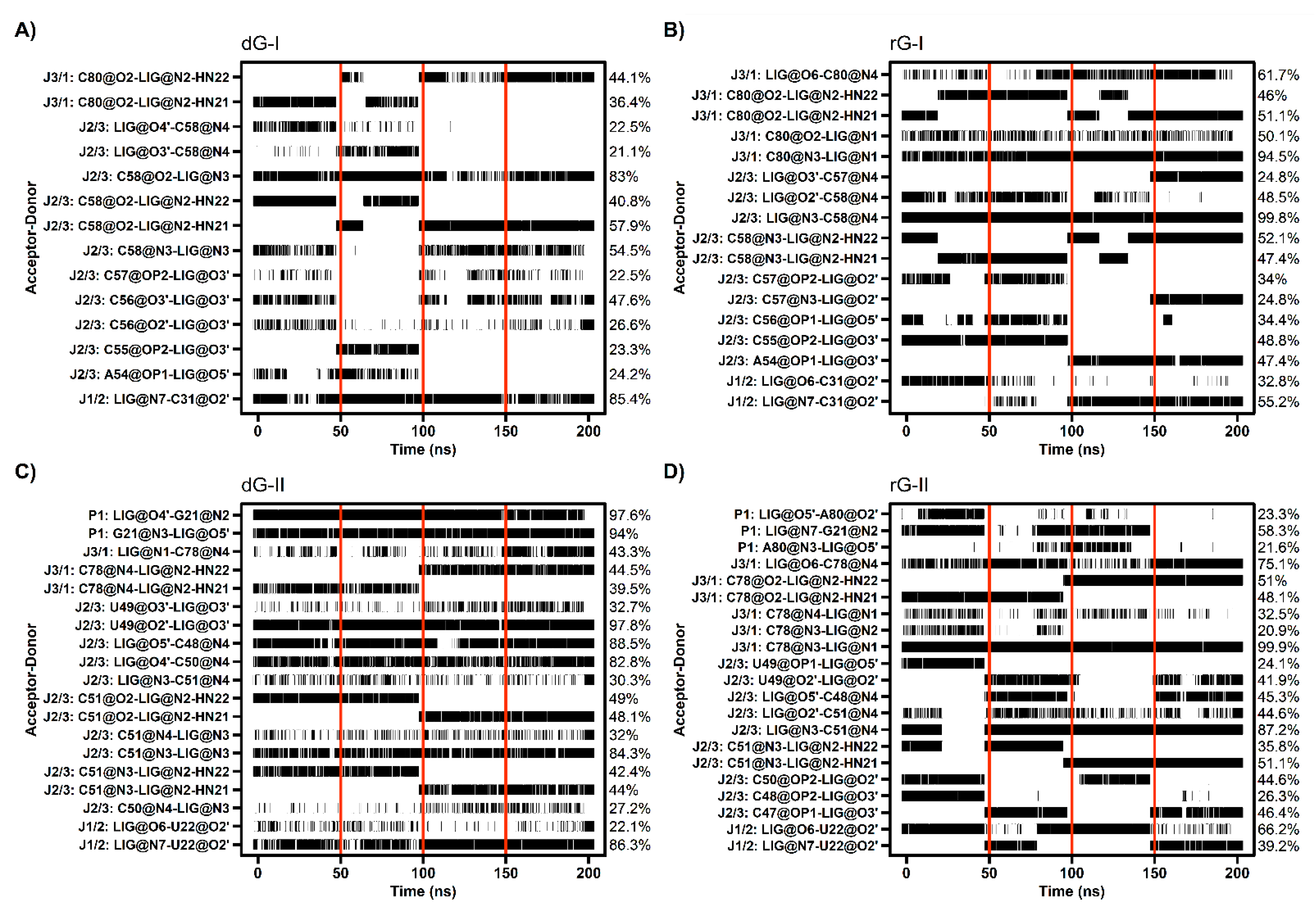
2.5. Cognate and Noncognate Ligands Alter the Dynamical Secondary Structure
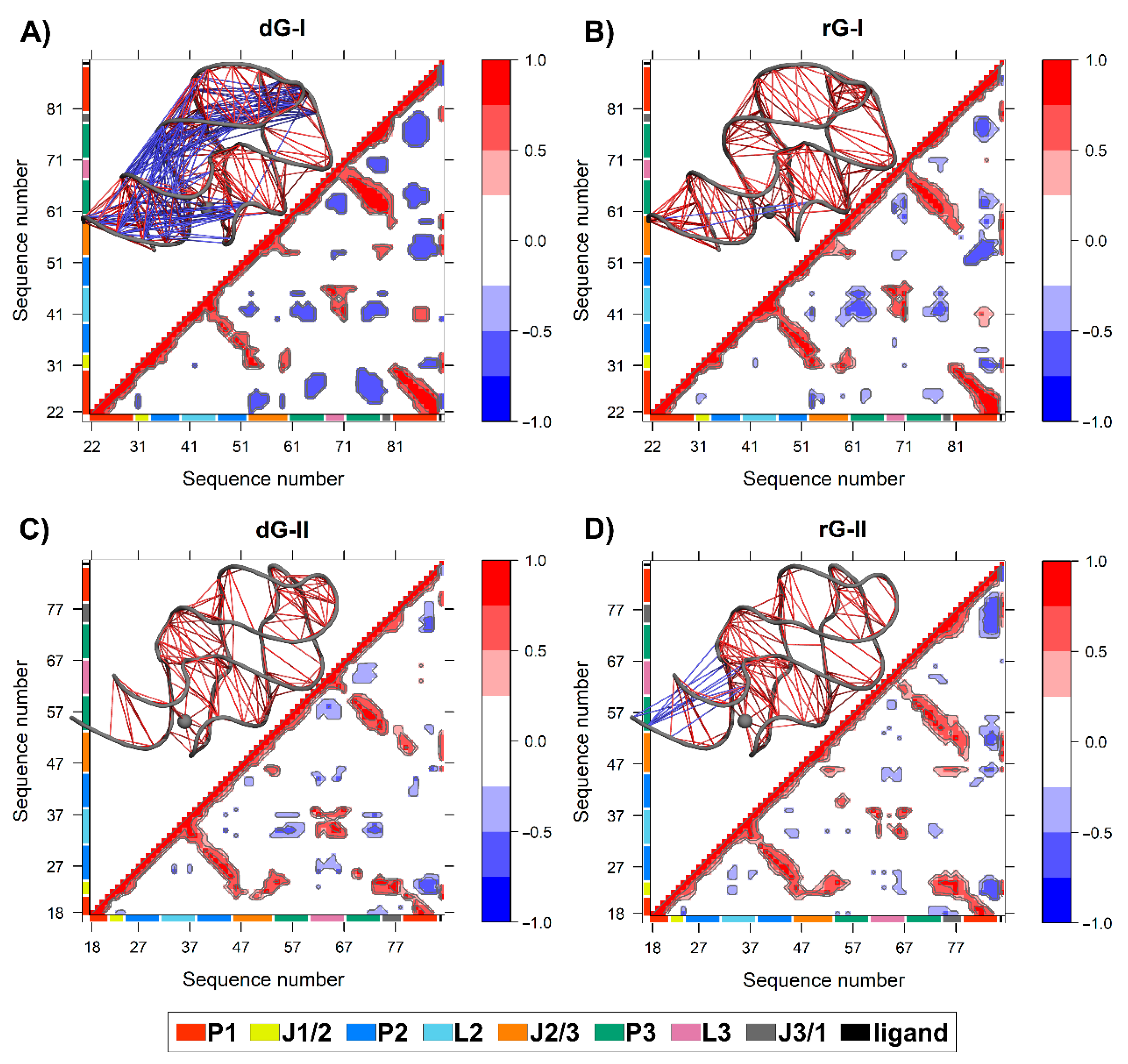
2.6. Anticorrelated Motions Are Changed by Cognate and Noncognate Ligands
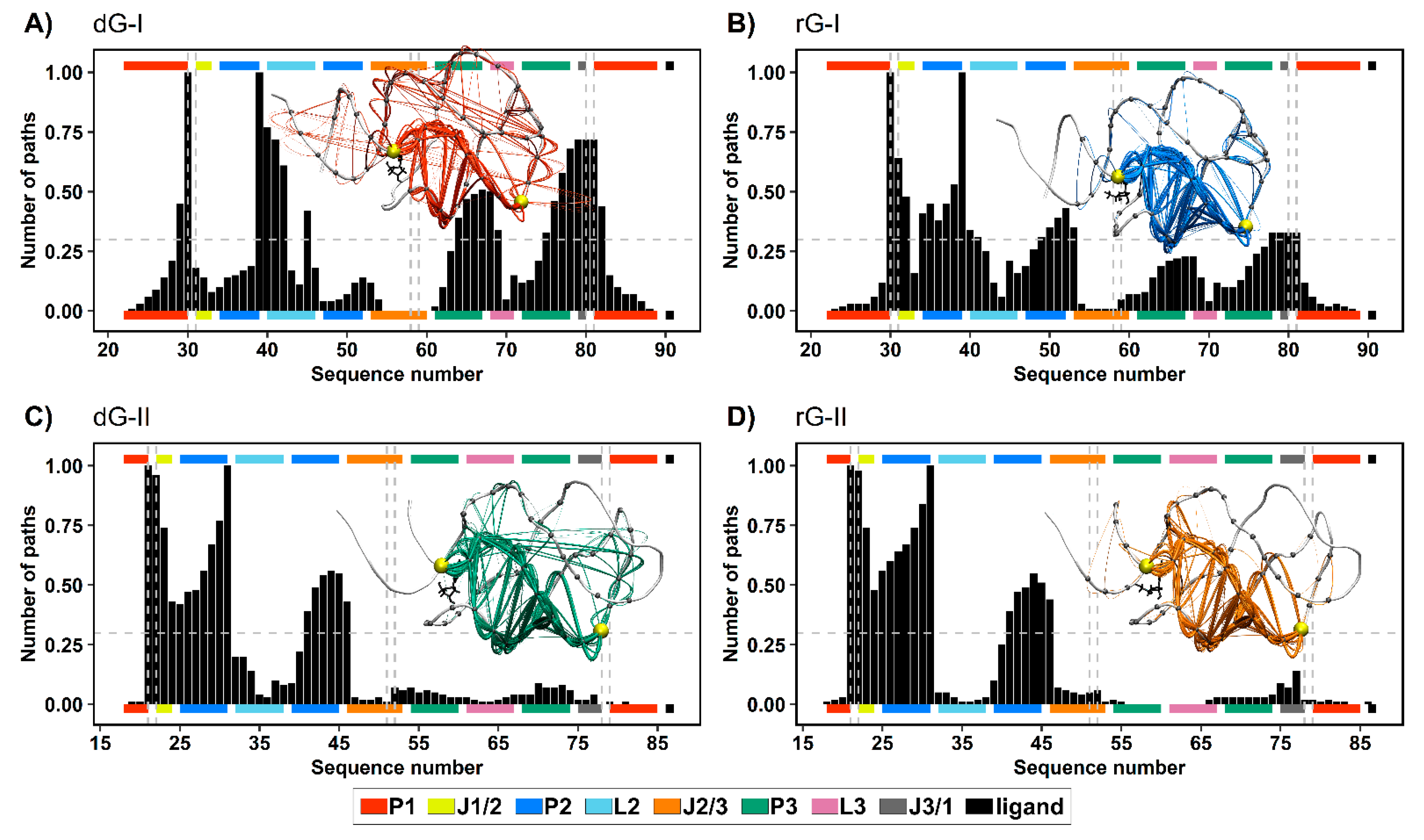
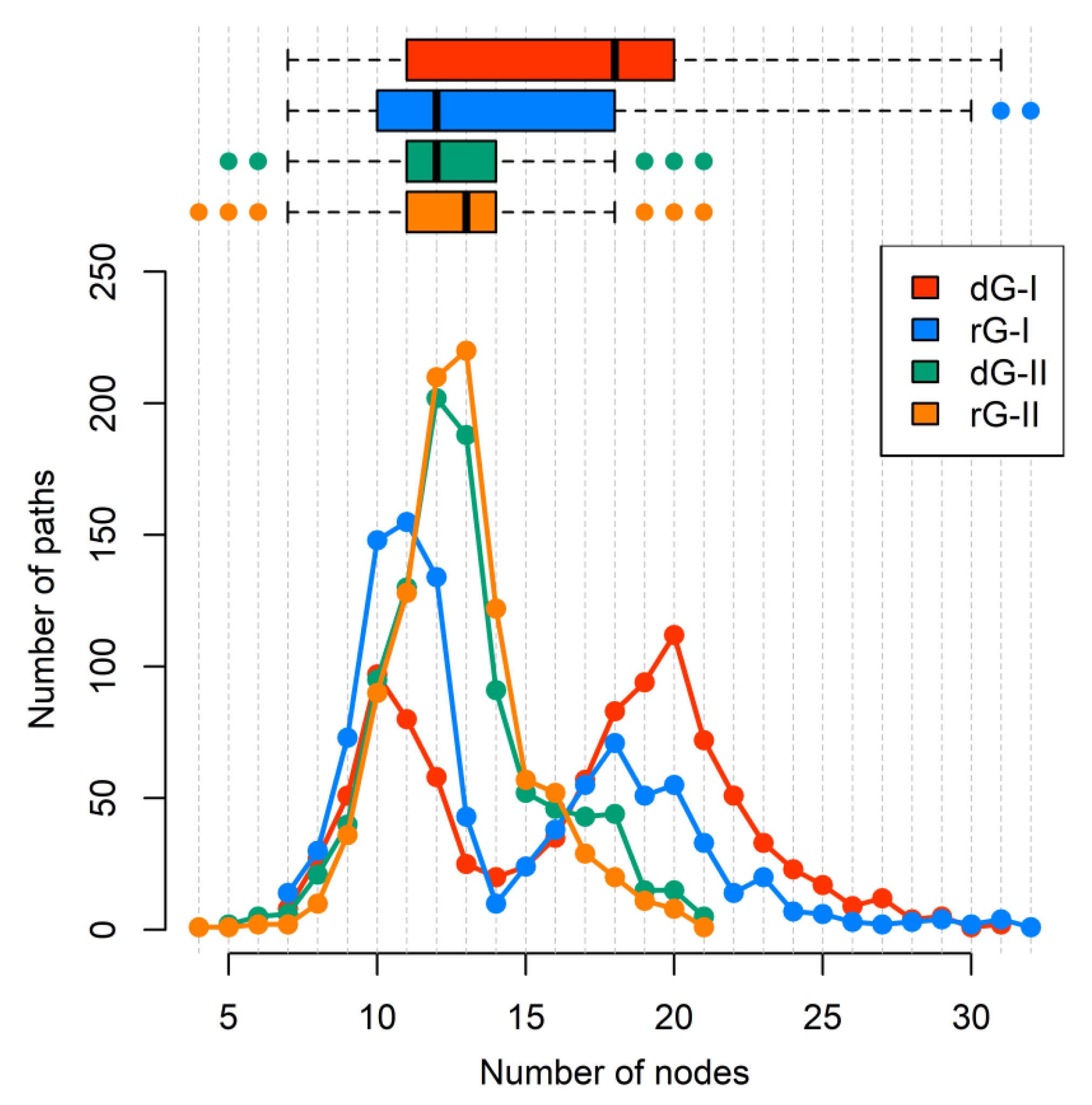
2.7. Communication Pathways between P1–P3
3. Discussion
4. Materials and Methods
4.1. Construction and Analysis of Molecular Systems
4.2. Molecular Dynamics Simulations
4.3. Trajectory Analysis
Author Contributions
Funding
Conflicts of Interest
References
- World Health Organization. Antimicrobial Resistance: Global Report on Surveillance; WHO: Geneva, Switzerland, 2014; ISBN 9789241564748. [Google Scholar]
- United Nations News Centre. At UN, global leaders commit to act on antimicrobial resistance. UN News Cent. 2016, 42, 21–22. [Google Scholar]
- Lünse, C.E.; Schüller, A.; Mayer, G. The promise of riboswitches as potential antibacterial drug targets. Int. J. Med. Microbiol. 2014, 304, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Edwards, A.L.; Batey, R.T. Riboswitches: A common RNA regulatory element. Nat. Educ. 2010, 3, 9. [Google Scholar]
- Breaker, R.R. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012, 4, a003566. [Google Scholar] [CrossRef] [Green Version]
- Tucker, B.J.; Breaker, R.R. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005, 15, 342–348. [Google Scholar] [CrossRef]
- Boström, J.; Hogner, A.; Schmitt, S. Do structurally similar ligands bind in a similar fashion? J. Med. Chem. 2006, 49, 6716–6725. [Google Scholar] [CrossRef]
- Barrick, J.E.; Breaker, R.R. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol. 2007, 8, R239. [Google Scholar] [CrossRef] [Green Version]
- Batey, R.T.; Gilbert, S.D.; Montange, R.K. Structure of a natural guanine-responsive riboswitch complexed with the metabolite hypoxanthine. Nature 2004, 432, 411–415. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Breaker, R.R. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004, 11, 29–35. [Google Scholar] [CrossRef]
- Pikovskaya, O.; Polonskaia, A.; Patel, D.J.; Serganov, A. Structural principles of nucleoside selectivity in a 2′-deoxyguanosine riboswitch. Nat. Chem. Biol. 2011, 7, 748–755. [Google Scholar] [CrossRef] [Green Version]
- Matyjasik, M.M.; Batey, R.T. Structural basis for 2′-deoxyguanosine recognition by the 2′-dG-II class of riboswitches. Nucleic Acids Res. 2019, 47, 10931–10941. [Google Scholar] [CrossRef]
- Porter, E.B.; Marcano-Velázquez, J.G.; Batey, R.T. The purine riboswitch as a model system for exploring RNA biology and chemistry. Biochim. Biophys. Acta-Gene Regul. Mech. 2014, 1839, 919–930. [Google Scholar] [CrossRef] [Green Version]
- Weinberg, Z.; Nelson, J.W.; Lünse, C.E.; Sherlock, M.E.; Breaker, R.R. Bioinformatic analysis of riboswitch structures uncovers variant classes with altered ligand specificity. Proc. Natl. Acad. Sci. USA 2017, 114, E2077–E2085. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.N.; Roth, A.; Breaker, R.R. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc. Natl. Acad. Sci. USA 2007, 104, 16092–16097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kolberg, M.; Strand, K.R.; Graff, P.; Kristoffer Andersson, K. Structure, function, and mechanism of ribonucleotide reductases. Biochim. Biophys. Acta-Proteins Proteom. 2004, 1699, 1–34. [Google Scholar] [CrossRef]
- Wereszczynski, J.; McCammon, J.A. Statistical mechanics and molecular dynamics in evaluating thermodynamic properties of biomolecular recognition. Q. Rev. Biophys. 2012, 45, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Genheden, S.; Ryde, U. The MM/PBSA and MM/GBSA methods to estimate ligand-binding affinities. Expert Opin. Drug Discov. 2015, 10, 449–461. [Google Scholar] [CrossRef]
- Chen, J.; Wang, X.; Pang, L.; Zhang, J.Z.H.; Zhu, T. Effect of mutations on binding of ligands to guanine riboswitch probed by free energy perturbation and molecular dynamics simulations. Nucleic Acids Res. 2019, 47, 6618–6631. [Google Scholar] [CrossRef]
- Hu, G.; Ma, A.; Wang, J. Ligand Selectivity Mechanism and Conformational Changes in Guanine Riboswitch by Molecular Dynamics Simulations and Free Energy Calculations. J. Chem. Inf. Model. 2017, 57, 918–928. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Li, H.; Xu, S.; Wang, J. Ligand binding mechanism and its relationship with conformational changes in adenine riboswitch. Int. J. Mol. Sci. 2020, 21, 1926. [Google Scholar] [CrossRef] [Green Version]
- Mignon, P.; Loverix, S.; Steyaert, J.; Geerlings, P. Influence of the pi-pi interaction on the hydrogen bonding capacity of stacked DNA/RNA bases. Nucleic Acids Res. 2005, 33, 1779–1789. [Google Scholar] [CrossRef] [Green Version]
- Skjærven, L.; Yao, X.-Q.; Scarabelli, G.; Grant, B.J. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinform. 2014, 15, 399. [Google Scholar] [CrossRef] [Green Version]
- Edwards, A.L.; Batey, R.T. A Structural Basis for the Recognition of 2′-Deoxyguanosine by the Purine Riboswitch. J. Mol. Biol. 2009, 385, 938–948. [Google Scholar] [CrossRef] [Green Version]
- Mandal, M.; Boese, B.; Barrick, J.E.; Winkler, W.C.; Breaker, R.R. Riboswitches Control Fundamental Biochemical Pathways in Bacillus subtilis and Other Bacteria. Cell 2003, 113, 577–586. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.H.; Le Roux, A.; Boyapelly, K.; Lamontagne, A.M.; Archambault, M.A.; Picard-Jean, F.; Lalonde-Seguin, D.; St-Pierre, E.; Najmanovich, R.J.; Fortier, L.C.; et al. Purine analogs targeting the guanine riboswitch as potential antibiotics against Clostridioides difficile. Eur. J. Med. Chem. 2018, 143, 755–768. [Google Scholar] [CrossRef]
- Dixon, N.; Duncan, J.N.; Geerlings, T.; Dunstan, M.S.; McCarthy, J.E.G.; Leys, D.; Micklefield, J. Reengineering orthogonally selective riboswitches. Proc. Natl. Acad. Sci. USA 2010, 107, 2830–2835. [Google Scholar] [CrossRef] [Green Version]
- Robinson, C.J.; Vincent, H.A.; Wu, M.C.; Lowe, P.T.; Dunstan, M.S.; Leys, D.; Micklefield, J. Modular riboswitch toolsets for synthetic genetic control in diverse bacterial species. J. Am. Chem. Soc. 2014, 136, 10615–10624. [Google Scholar] [CrossRef] [PubMed]
- Antczak, M.; Zok, T.; Popenda, M.; Lukasiak, P.; Adamiak, R.W.; Blazewicz, J.; Szachniuk, M. RNApdbee—A webserver to derive secondary structures from pdb files of knotted and unknotted RNAs. Nucleic Acids Res. 2014, 42, W368–W372. [Google Scholar] [CrossRef]
- Zok, T.; Antczak, M.; Zurkowski, M.; Popenda, M.; Blazewicz, J.; Adamiak, R.W.; Szachniuk, M. RNApdbee 2.0: Multifunctional tool for RNA structure annotation. Nucleic Acids Res. 2018, 46, W30–W35. [Google Scholar] [CrossRef] [Green Version]
- Will, S.; Joshi, T.; Hofacker, I.L.; Stadler, P.F.; Backofen, R. LocARNA-P: Accurate boundary prediction and improved detection of structural RNAs. RNA 2012, 18, 900–914. [Google Scholar] [CrossRef] [Green Version]
- Bond, C.S.; Schüttelkopf, A.W. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009, 65, 510–512. [Google Scholar] [CrossRef] [Green Version]
- Blin, G.; Denise, A.; Dulucq, S.; Herrbach, C.; Touzet, H. VARNA: Interactive drawing and editing of the RNA secondary structure. IEEE/ACM Trans. Comput. Biol. Bioinform. 2010, 7, 309–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLano, W.L. The PyMOL Molecular Graphics System. Available online: https://pymol.org/2/ (accessed on 9 December 2021).
- Case, D.A.; Cheatham, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 11, 3696–3713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez, A.; Marchán, I.; Svozil, D.; Sponer, J.; Cheatham, T.E.; Laughton, C.A.; Orozco, M. Refinement of the AMBER force field for nucleic acids: Improving the description of α/γ conformers. Biophys. J. 2007, 92, 3817–3829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wolf, R.M.; Caldwell, J.W.; Kollman, P.A.; Case, D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004, 25, 1157–1174. [Google Scholar] [CrossRef] [PubMed]
- Jakalian, A.; Jack, D.B.; Bayly, C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002, 23, 1623–1641. [Google Scholar] [CrossRef]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006, 25, 247–260. [Google Scholar] [CrossRef]
- Jorgensen, W.L.; Chandrasekhar, J.; Madura, J.D.; Impey, R.W.; Klein, M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983, 79, 926–935. [Google Scholar] [CrossRef]
- Ryckaert, J.P.; Ciccotti, G.; Berendsen, H.J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comput. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Abraham, M.J.; Murtola, T.; Schulz, R.; Páll, S.; Smith, J.C.; Hess, B.; Lindahl, E. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar] [CrossRef] [Green Version]
- Bottaro, S.; Bussi, G.; Pinamonti, G.; Reiber, S.; Boomsma, W.; Lindorff-Larsen, K. Barnaba: Software for analysis of nucleic acid structures and trajectories. RNA 2019, 25, 219–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for Processing and Analysis of Molecular Dynamics Trajectory Data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Miller, B.R.; McGee, T.D.; Swails, J.M.; Homeyer, N.; Gohlke, H.; Roitberg, A.E. MMPBSA.py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput. 2012, 8, 3314–3321. [Google Scholar] [CrossRef] [PubMed]
- Csárdi, G.; Nepusz, T. The igraph software package for complex network research. InterJournal Complex Syst. 2006, 1695, 1–9. [Google Scholar]
- Antunes, D.; Jorge, N.A.N.; de Souza Costa, M.G.; Passetti, F.; Caffarena, E.R. Unraveling RNA dynamical behavior of TPP riboswitches: A comparison between Escherichia coli and Arabidopsis thaliana. Sci. Rep. 2019, 9, 4197. [Google Scholar] [CrossRef]
- Chen, J.; Zhang, S.; Wang, W.; Pang, L.; Zhang, Q.; Liu, X. Mutation-Induced Impacts on the Switch Transformations of the GDP-and GTP-Bound K-Ras: Insights from Multiple Replica Gaussian Accelerated Molecular Dynamics and Free Energy Analysis. J. Chem. Inf. Model. 2021, 61, 1954–1969. [Google Scholar] [CrossRef] [PubMed]
- Yen, J.Y. Finding the K Shortest Loopless Paths in a Network. Manag. Sci. 1971, 17, 712–716. [Google Scholar] [CrossRef]

| 2′dG-I | 2′dG-II | |||
|---|---|---|---|---|
| dG | rG | dG | rG | |
| RNA | 3.09 ± 0.65 | 2.62 ± 0.61 | 2.98 ± 0.65 | 2.53 ± 0.40 |
| P1 | 3.17 ± 0.75 | 2.43 ± 0.75 | 3.35 ± 1.11 | 1.30 ± 0.31 |
| J1/2 | 4.39 ± 0.80 | 2.62 ± 0.70 | 1.57 ± 0.47 | 1.37 ± 0.32 |
| P2 | 2.26 ± 0.45 | 2.49 ± 0.73 | 1.34 ± 0.25 | 1.30 ± 0.31 |
| L2 | 2.53 ± 0.64 | 2.42 ± 0.62 | 1.86 ± 0.52 | 1.91 ± 0.41 |
| J2/3 | 3.87 ± 0.94 | 3.38 ± 0.86 | 1.76 ± 0.46 | 2.77 ± 1.07 |
| P3 | 2.73 ± 1.02 | 2.26 ± 0.73 | 1.93 ± 0.28 | 1.85 ± 0.26 |
| L3 | 3.55 ± 0.77 | 3.19 ± 0.59 | 2.08 ± 0.39 | 2.95 ± 0.70 |
| J3/1 | 2.96 ± 1.13 | 1.97 ± 0.66 | 2.30 ± 0.50 | 1.57 ± 0.35 |
| Ligand | 3.03 ± 1.00 | 2.12 ± 0.58 | 1.68 ± 1.00 | 2.23 ± 1.13 |
| Items | 2′dG-I | 2′dG-II | ||
|---|---|---|---|---|
| dG | rG | dG | rG | |
| ΔEvdw | −41.82 ± 0.08 | −42.38 ± 0.10 | −46.73 ± 0.07 | −40.89 ± 0.09 |
| ΔEele | −22.52 ± 0.27 | −61.31 ± 0.21 | −30.00 ± 0.20 | −76.67 ± 0.32 |
| ΔGegb | 38.54 ± 0.25 | 70.23 ± 0.19 | 40.08 ± 0.18 | 86.45 ± 0.29 |
| ΔGesurf | −3.72 ± 0.004 | −4.26 ± 0.003 | −4.14 ± 0.004 | −4.34 ± 0.004 |
| b ΔGele+egb | 16.01 ± 0.26 | 8.92 ± 0.20 | 10.08 ± 0.19 | 9.78 ± 0.30 |
| c ΔH | −29.53 ± 0.08 | −37.72 ± 0.13 | −40.79 ± 0.09 | −35.44 ± 0.10 |
| d −TΔS | 23.04 ± 0.44 | 21.64 ± 1.03 | 23.47 ± 0.85 | 23.54 ± 1.44 |
| ΔGbind | −6.49 | −16.08 | −17.32 | −11.90 |
| e ΔGexp | −10.97 | −8.64 | −11.62 | −11.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Antunes, D.; Santos, L.H.S.; Caffarena, E.R.; Guimarães, A.C.R. Bacterial 2′-Deoxyguanosine Riboswitch Classes as Potential Targets for Antibiotics: A Structure and Dynamics Study. Int. J. Mol. Sci. 2022, 23, 1925. https://doi.org/10.3390/ijms23041925
Antunes D, Santos LHS, Caffarena ER, Guimarães ACR. Bacterial 2′-Deoxyguanosine Riboswitch Classes as Potential Targets for Antibiotics: A Structure and Dynamics Study. International Journal of Molecular Sciences. 2022; 23(4):1925. https://doi.org/10.3390/ijms23041925
Chicago/Turabian StyleAntunes, Deborah, Lucianna H. S. Santos, Ernesto Raul Caffarena, and Ana Carolina Ramos Guimarães. 2022. "Bacterial 2′-Deoxyguanosine Riboswitch Classes as Potential Targets for Antibiotics: A Structure and Dynamics Study" International Journal of Molecular Sciences 23, no. 4: 1925. https://doi.org/10.3390/ijms23041925
APA StyleAntunes, D., Santos, L. H. S., Caffarena, E. R., & Guimarães, A. C. R. (2022). Bacterial 2′-Deoxyguanosine Riboswitch Classes as Potential Targets for Antibiotics: A Structure and Dynamics Study. International Journal of Molecular Sciences, 23(4), 1925. https://doi.org/10.3390/ijms23041925






