Identification and Allele Combination Analysis of Rice Grain Shape-Related Genes by Genome-Wide Association Study
Abstract
:1. Introduction
2. Results
2.1. Distribution and Correlation of Phenotype and Heritability of Grain Shape
2.2. Population Structure, Kinship, and LD Decay
2.3. Identification of Significant Loci for Related Traits through GWAS
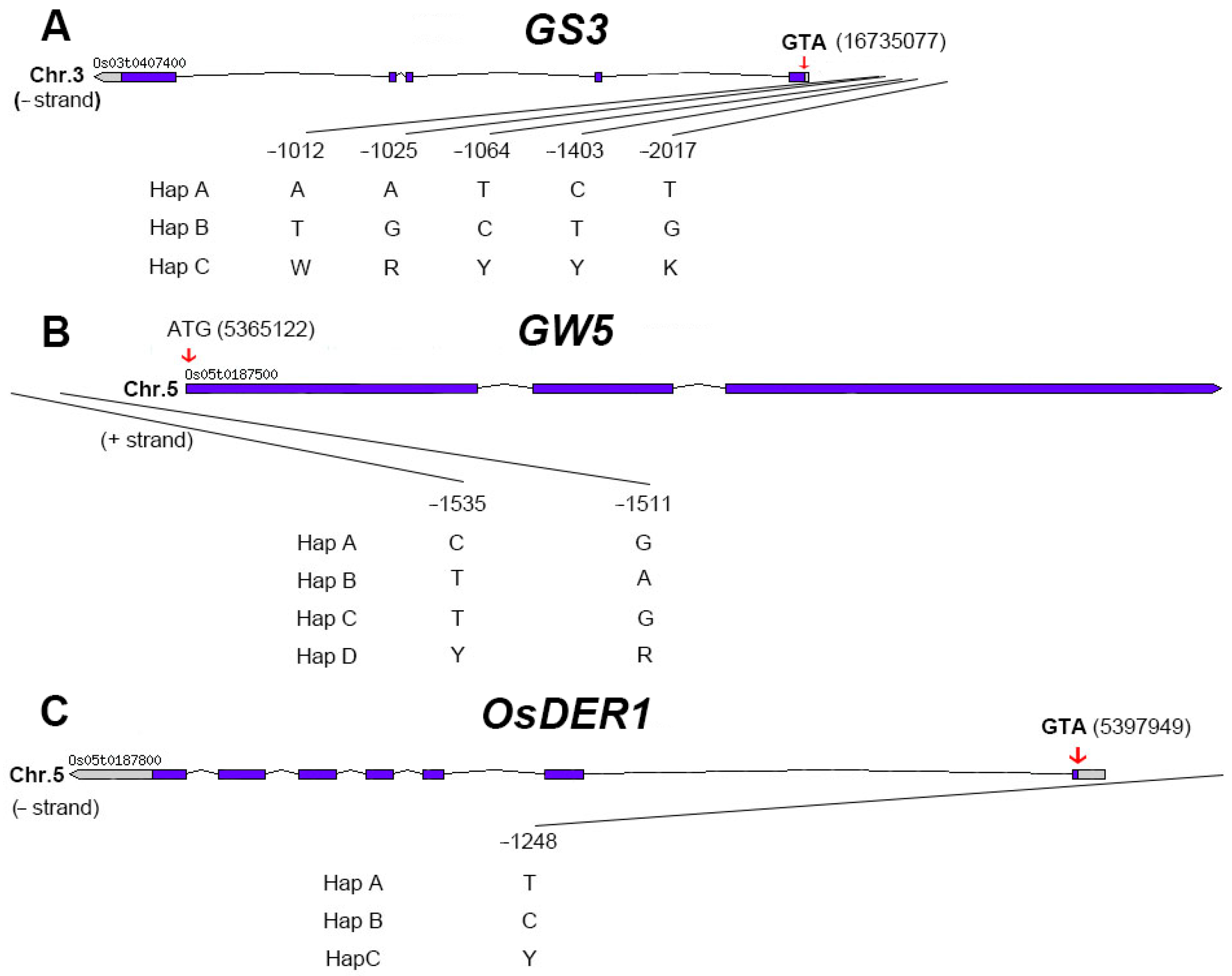
2.4. Candidate Genes Screen in Important QTL Regions
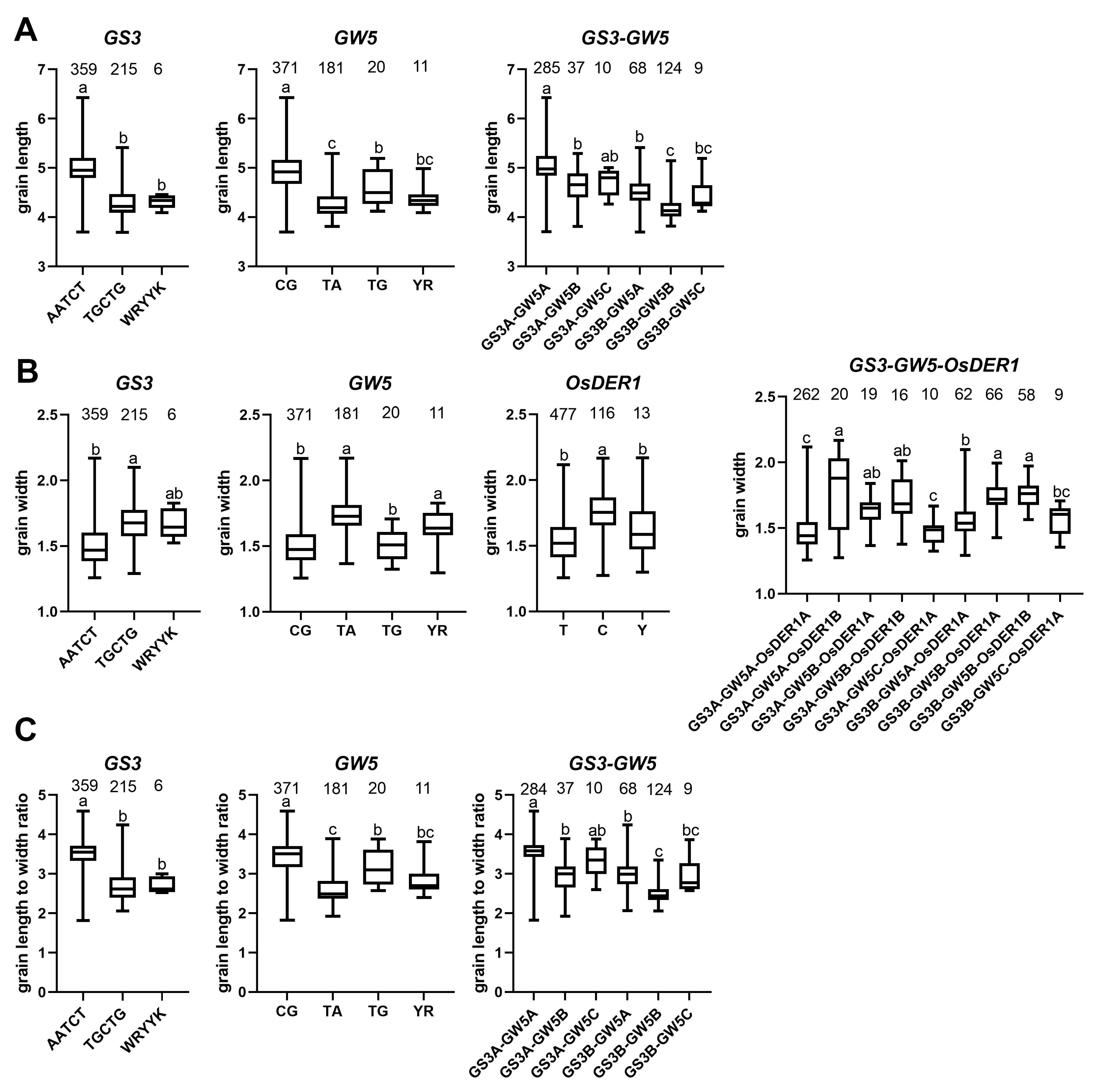
2.5. Extreme Combination of Alleles for Each Trait
3. Discussion
4. Materials and Methods
4.1. Materials
4.2. Field Trials and Trait Measurements
4.3. Statistical Analysis
4.4. Genome-Wide Association Study
4.4.1. Genotyping
4.4.2. Population Structure and Kinship Analysis
4.4.3. Linkage Disequilibrium Analysis
4.4.4. Genome-Wide Association Study and Candidate Genes Identification
4.5. Allele Combination Analysis
4.5.1. Allele Analysis of Different Genes
4.5.2. Extreme Combination of Alleles for Each Trait
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sakamoto, T.; Matsuoka, M. Identifying and exploiting grain yield genes in rice. Curr. Opin. Plant Biol. 2008, 11, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.F.; Xing, Y.Z.; Li, J.X.; Yu, S.B.; Xu, C.G.; Zhang, Q. Genetic bases of appearance quality of rice grains in Shanyou 63, an elite rice hybrid. Theor. Appl. Genet. 2000, 101, 823–829. [Google Scholar] [CrossRef]
- Redoña, E.D.; Mackill, D.J. Quantitative trait locus analysis for rice panicle and grain characteristics. Theor. Appl. Genet. 1998, 96, 957–963. [Google Scholar] [CrossRef]
- Li, Z.; Tang, L.; Qiu, J.; Zhang, W.; Wang, Y.; Tong, X.; Wei, X.; Hou, Y.; Zhang, J. Serine carboxypeptidase 46 Regulates Grain Filling and Seed Germination in Rice (Oryza sativa L.). PLoS ONE 2016, 11, e0159737. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Chen, J.; Zheng, X.; Wu, F.; Lin, Q.; Heng, Y.; Tian, P.; Cheng, Z.; Yu, X.; Zhou, K.; et al. GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 2017, 3, 17043. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, L.; Mao, H.; Shao, L.; Li, X.; Xiao, J.; Ouyang, Y.; Zhang, Q. A G-protein pathway determines grain size in rice. Nat. Commun. 2018, 9, 851. [Google Scholar] [CrossRef] [Green Version]
- Qian, D.; Chen, G.; Tian, L.; Qu, L.Q. OsDER1 Is an ER-Associated Protein Degradation Factor That Responds to ER Stress. Plant Physiol. 2018, 178, 402–412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Zhao, X.; Dai, Z.; Ma, F.; Miao, X.; Shi, Z. OsmiR396/growth regulating factor modulate rice grain size through direct regulation of embryo-specific miR408. Plant Physiol. 2021, 186, 519–533. [Google Scholar] [CrossRef]
- Li, S.; Gao, F.; Xie, K.; Zeng, X.; Cao, Y.; Zeng, J.; He, Z.; Ren, Y.; Li, W.; Deng, Q.; et al. The OsmiR396c-OsGRF4-OsGIF1 regulatory module determines grain size and yield in rice. Plant Biotechnol. J. 2016, 14, 2134–2146. [Google Scholar] [CrossRef]
- Cui, X.; Jin, P.; Cui, X.; Gu, L.; Lu, Z.; Xue, Y.; Wei, L.; Qi, J.; Song, X.; Luo, M.; et al. Control of transposon activity by a histone H3K4 demethylase in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 1953–1958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, J.; Cho, L.H.; Kim, S.R.; Tun, W.; Peng, X.; Pasriga, R.; Moon, S.; Hong, W.J.; Ji, H.; Jung, K.H.; et al. CTP synthase is essential for early endosperm development by regulating nuclei spacing. Plant Biotechnol. J. 2021, 19, 2177–2191. [Google Scholar] [CrossRef]
- Ma, X.; Cheng, Z.; Qin, R.; Qiu, Y.; Heng, Y.; Yang, H.; Ren, Y.; Wang, X.; Bi, J.; Ma, X.; et al. OsARG encodes an arginase that plays critical roles in panicle development and grain production in rice. Plant J. 2013, 73, 190–200. [Google Scholar] [CrossRef]
- Guo, Z.; Liu, X.; Zhang, B.; Yuan, X.; Xing, Y.; Liu, H.; Luo, L.; Chen, G.; Xiong, L. Genetic analyses of lodging resistance and yield provide insights into post-Green-Revolution breeding in rice. Plant Biotechnol. J. 2021, 19, 814–829. [Google Scholar] [CrossRef] [PubMed]
- Tenaillon, M.I.; Sawkins, M.C.; Long, A.D.; Gaut, R.L.; Doebley, J.F.; Gaut, B.S. Patterns of DNA sequence polymorphism along chromosome 1 of maize (Zea mays ssp. mays L.). Proc. Natl. Acad. Sci. USA 2001, 98, 9161–9166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Wei, X.; Sang, T.; Zhao, Q.; Feng, Q.; Zhao, Y.; Li, C.; Zhu, C.; Lu, T.; Zhang, Z.; et al. Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 2010, 42, 961–967. [Google Scholar] [CrossRef]
- Yoo, S.; Cho, S.; Sugimoto, H.; Li, J.; Kusumi, K.; Koh, H.; Iba, K.; Paek, N. Rice Virescent3 and Stripe1 Encoding the Large and Small Subunits of Ribonucleotide Reductase Are Required for Chloroplast Biogenesis during Early Leaf Development. Plant Physiol. 2009, 150, 388–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Wang, J.; Huang, J.; Lan, H.; Wang, C.; Yin, C.; Wu, Y.; Tang, H.; Qian, Q.; Li, J.; et al. Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. USA 2012, 109, 21534–21539. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Zhang, Y.; Hou, M.; Sun, F.; Shen, Y.; Xiu, Z.; Wang, X.; Chen, Z.; Sun, S.S.M.; Small, I.; et al. Small kernel 1 encodes a pentatricopeptide repeat protein required for mitochondrial nad7 transcript editing and seed development in maize (Zea mays) and rice (Oryza sativa). Plant J. 2014, 79, 797–809. [Google Scholar] [CrossRef]
- Lam, P.Y.; Zhu, F.; Chan, W.L.; Liu, H.; Lo, C. Cytochrome P450 93G1 Is a Flavone Synthase II That Channels Flavanones to the Biosynthesis of TricinO-Linked Conjugates in Rice. Plant Physiol. 2014, 165, 1315–1327. [Google Scholar] [CrossRef] [Green Version]
- Xu, F.; Fang, J.; Ou, S.; Gao, S.; Zhang, F.; Du, L.; Xiao, Y.; Wang, H.; Sun, X.; Chu, J.; et al. Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 2015, 38, 800–811. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Fu, Y.; Zhao, S.; Gu, P.; Zhu, Z.; Sun, C.; Tan, L. CLUSTERED PRIMARY BRANCH 1, a new allele of DWARF11, controls panicle architecture and seed size in rice. Plant Biotechnol. J. 2016, 14, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Ren, Z.; Bai, F.; Xu, J.; Wang, L.; Wang, X.; Zhang, Q.; Feng, C.; Niu, Q.; Zhang, L.; Song, J.; et al. A chloride efflux transporter, BIG RICE GRAIN 1, is involved in mediating grain size and salt tolerance in rice. J. Integr. Plant Biol. 2021. [Google Scholar] [CrossRef] [PubMed]
- Fan, C.; Xing, Y.; Mao, H.; Lu, T.; Han, B.; Xu, C.; Li, X.; Zhang, Q. GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 2006, 112, 1164–1171. [Google Scholar] [CrossRef] [PubMed]
- Tian, P.; Liu, J.; Mou, C.; Shi, C.; Zhang, H.; Zhao, Z.; Lin, Q.; Wang, J.; Wang, J.; Zhang, X.; et al. GW5-Like, a homolog of GW5, negatively regulates grain width, weight and salt resistance in rice. J. Integr. Plant Biol. 2019, 61, 1171–1185. [Google Scholar] [CrossRef] [PubMed]
- The 3000 rice genomes project. Gigascience 2014, 3, 7.
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DePristo, M.A.; Banks, E.; Poplin, R.; Garimella, K.V.; Maguire, J.R.; Hartl, C.; Philippakis, A.A.; Del Angel, G.; Rivas, M.A.; Hanna, M.; et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 2011, 43, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Howie, B.N.; Donnelly, P.; Marchini, J. A Flexible and Accurate Genotype Imputation Method for the Next Generation of Genome-Wide Association Studies. PLoS Genet. 2009, 5, e1000529. [Google Scholar] [CrossRef] [Green Version]
- Yin, L.; Zhang, H.; Tang, Z.; Xu, J.; Yin, D.; Zhang, Z.; Yuan, X.; Zhu, M.; Zhao, S.; Li, X.; et al. rMVP: A Memory-efficient, Visualization-enhanced, and Parallel-accelerated tool for Genome-Wide Association Study. Genom. Proteom. Bioinform. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Patterson, N.J.; Plenge, R.M.; Weinblatt, M.E.; Shadick, N.A.; Reich, D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006, 38, 904–909. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.; Xu, J.; He, W.; Yang, T. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Gao, Y.; Xie, W.; Gong, L.; Lu, K.; Wang, W.; Li, Y.; Liu, X.; Zhang, H.; Dong, H.; et al. Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat. Genet. 2014, 46, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xie, W.; Xing, H.; Yan, J.; Meng, X.; Li, X.; Fu, X.; Xu, J.; Lian, X.; Yu, S.; et al. Genetic Architecture of Natural Variation in Rice Chlorophyll Content Revealed by a Genome-Wide Association Study. Mol. Plant 2015, 8, 946–957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Z.; Yang, W.; Chang, Y.; Ma, X.; Tu, H.; Xiong, F.; Jiang, N.; Feng, H.; Huang, C.; Yang, P.; et al. Genome-Wide Association Studies of Image Traits Reveal Genetic Architecture of Drought Resistance in Rice. Mol. Plant 2018, 11, 789–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Pang, Y.; Wang, C.; Chen, K.; Zhu, Y.; Shen, C.; Ali, J.; Xu, J.; Li, Z. New Candidate Genes Affecting Rice Grain Appearance and Milling Quality Detected by Genome-Wide and Gene-Based Association Analyses. Front. Plant Sci. 2017, 7, 1998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yano, K.; Yamamoto, E.; Aya, K.; Takeuchi, H.; Lo, P.; Hu, L.; Yamasaki, M.; Yoshida, S.; Kitano, H.; Hirano, K.; et al. Genome-wide association study using whole-genome sequencing rapidly identifies new genes influencing agronomic traits in rice. Nat. Genet. 2016, 48, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Lu, Y.; Feng, L.; Hao, W.; Li, C.; Yang, Y.; Fan, X.; Li, Q.; Zhang, C.; Liu, Q. Genetic Dissection and Functional Differentiation of ALKa and ALKb, Two Natural Alleles of the ALK/SSIIa Gene, Responding to Low Gelatinization Temperature in Rice. Rice 2020, 13, 39. [Google Scholar] [CrossRef]
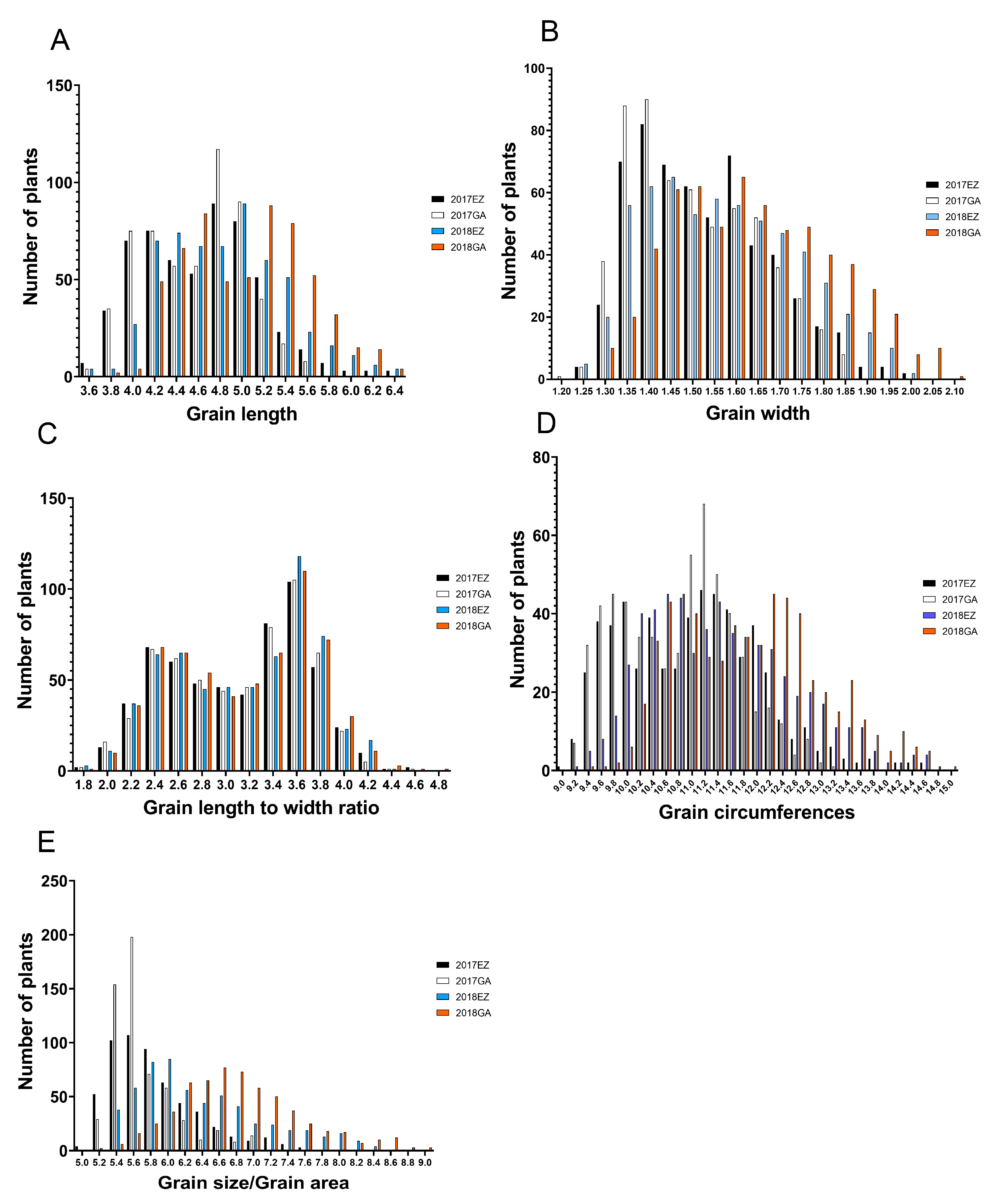

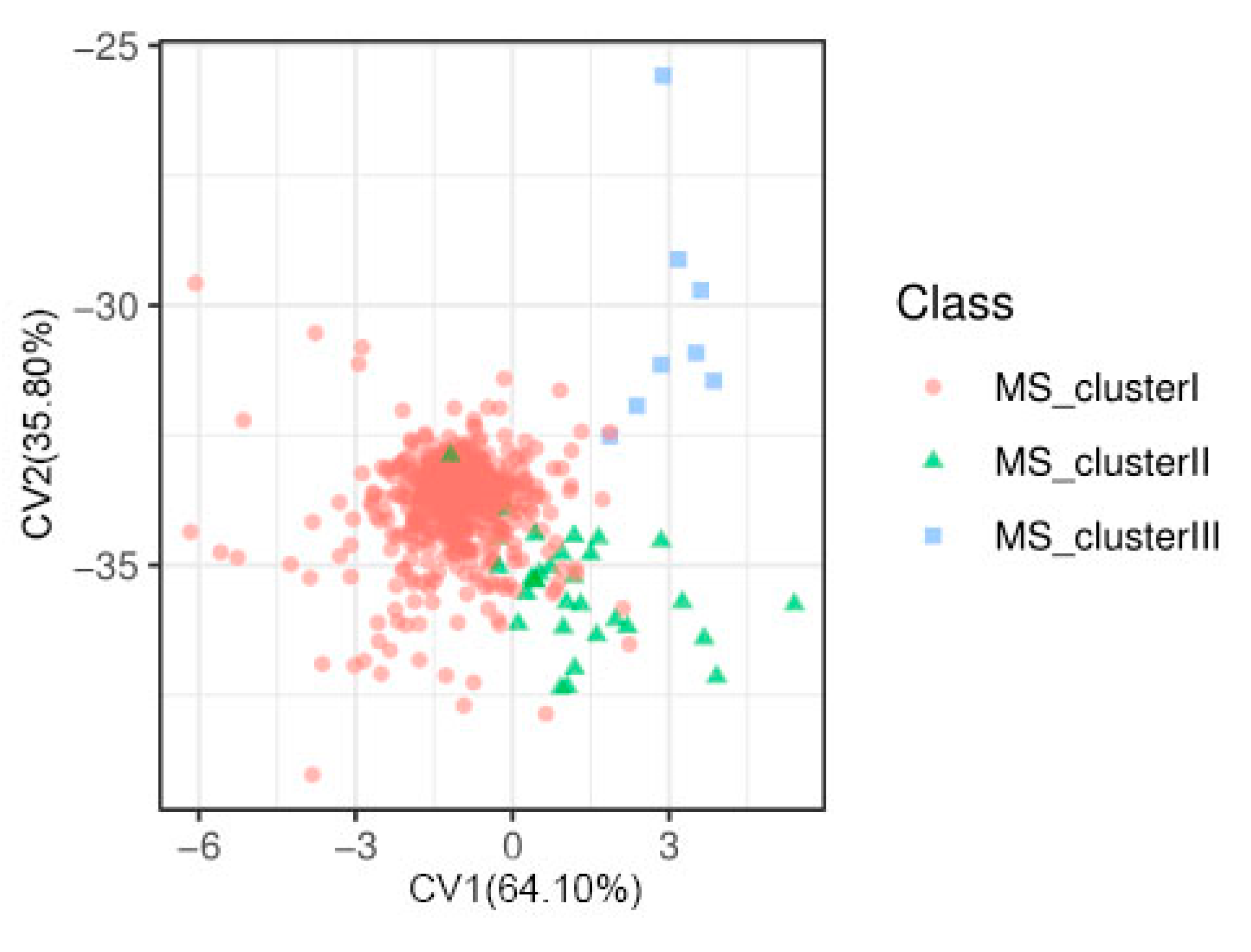
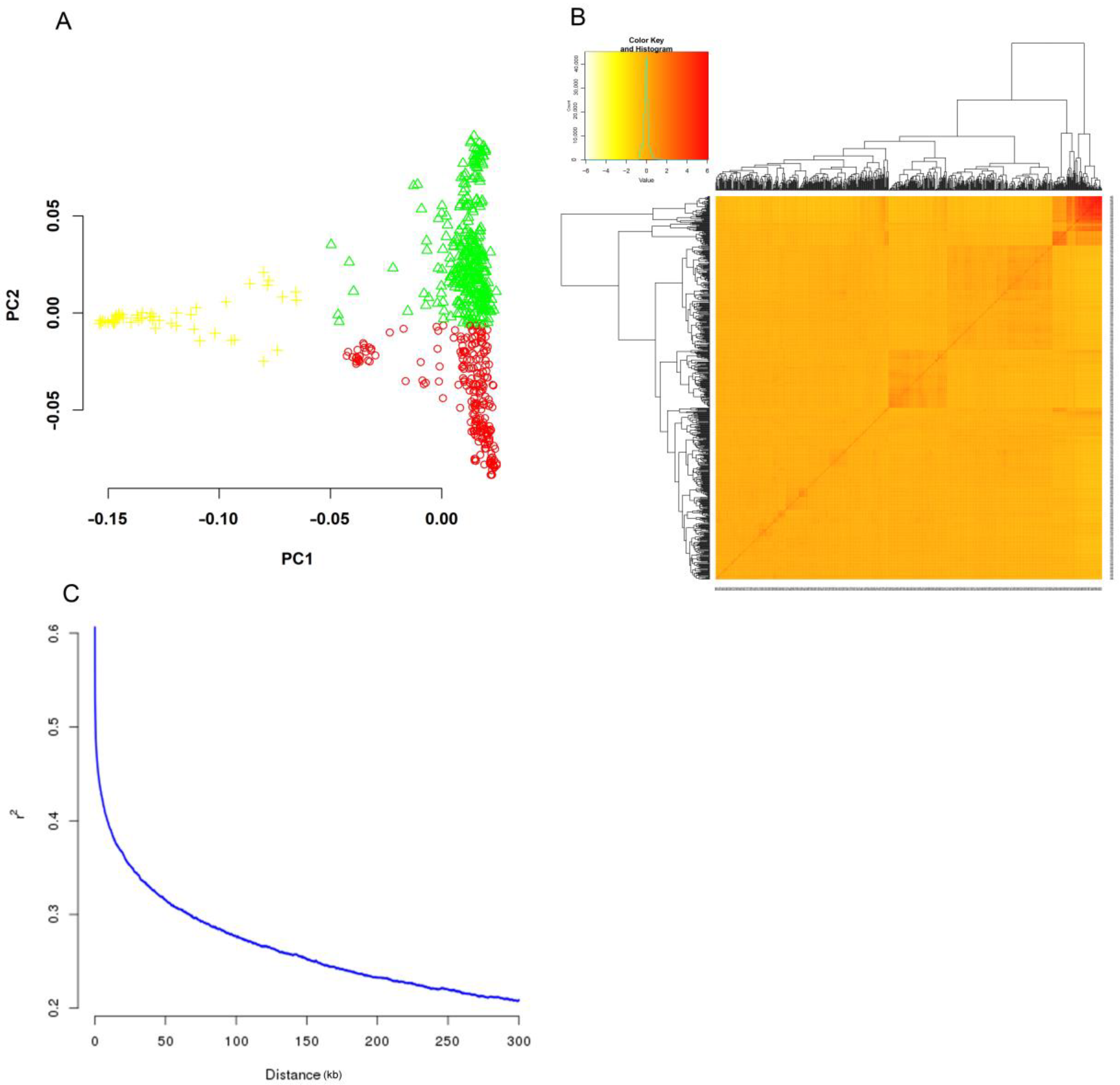
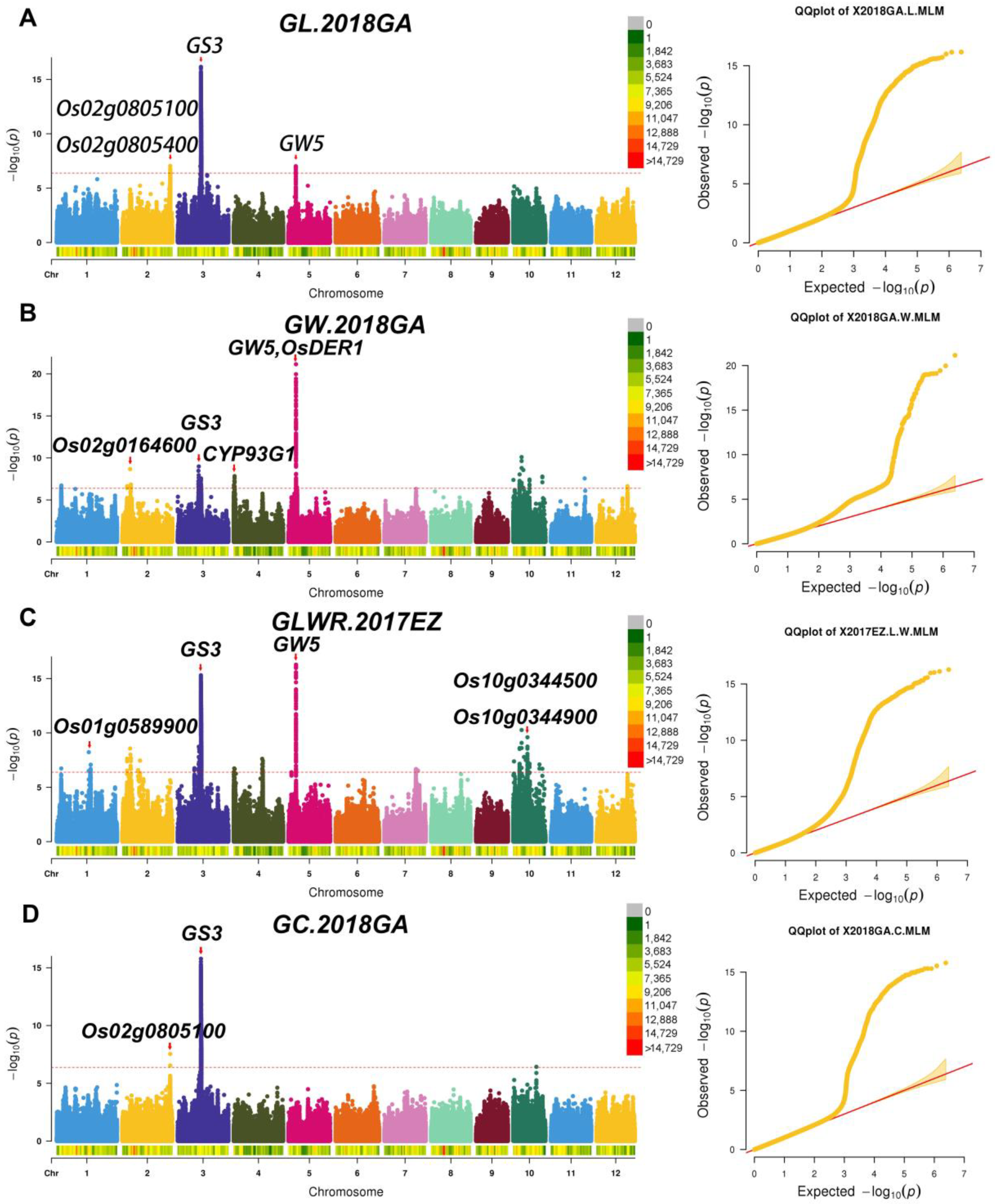

| GL | GW | GLWR | GC | GS | |
|---|---|---|---|---|---|
| GL | − | ||||
| GW | −111,507.80 | − | |||
| GLWR | 6039.21 | −19,306.47 | − | ||
| GC | 49,771.01 | −588,205.40 | 78,805.02 | − | |
| GS | 7704.96 | 754,904.50 | 81,783.15 | 40,385.27 | − |
| Env\Trait | GL | GW | GLWR | GC | GS |
|---|---|---|---|---|---|
| 2017EZ | 0.561198 | 0.587232 | 0.816784 | 0.435194 | 0.102882 |
| 2017GA | 0.790264 | 0.842533 | 0.863557 | 0.750632 | 0.403186 |
| 2018EZ | 0.417942 | 0.593973 | 0.780337 | 0.439132 | 0.220128 |
| 2018GA | 0.682506 | 0.755747 | 0.802726 | 0.651486 | 0.519685 |
| mean | 0.6129775 | 0.69487125 | 0.815851 | 0.569111 | 0.31147025 |
| QTL | Env | Trait | CHRO | Position | Peak-SNP | Ref/Alt | Effect | SE | P | PVE(%) | Cloned Gene |
|---|---|---|---|---|---|---|---|---|---|---|---|
| qGC2 | 2018GA | GC | 2 | 34214439-34472981 | chr02_34347981 | G/A | −0.555 | 0.099 | 2.88316 × 10−8 | 2.96 | OsmiR396a; OsmiR396c |
| qGC3 | 2017GA | GC | 3 | 16538239-17145970 | chr03_16707611 | T/C | −0.392 | 0.047 | 4.618 × 1016 | 42.07 | GS3 |
| 2017EZ | GC | 3 | 16538239-17145970 | chr03_16707611 | T/C | −0.429 | 0.055 | 1.98333 × 10−14 | 41.50 | GS3 | |
| 2018GA | GC | 3 | 16538239-17145970 | chr03_16692834 | A/G | −0.488 | 0.057 | 1.64375 × 10−16 | 38.46 | GS3 | |
| 2018EZ | GC | 3 | 16538239-17145970 | chr03_16669223 | T/C | −0.518 | 0.060 | 4.16047 × 10−17 | 36.18 | GS3 | |
| qGC5 | 2017GA | GC | 5 | 5233756-5486894 | chr05_5361894 | G/A | −0.215 | 0.039 | 4.55499 × 10−8 | 33.45 | GW5; OsDER1 |
| qGL2 | 2018GA | GL | 2 | 34202268-34492940 | chr02_34339439 | G/A | −0.239 | 0.044 | 9.10759 × 10−8 | 4.34 | OsmiR396a; OsmiR396c |
| qGL3.2 | 2018GA | GL | 3 | 16121544-16371544 | chr03_16246544 | G/A | 0.110 | 0.020 | 9.6657 × 10−8 | 20.37 | |
| qGL3.3 | 2017EZ | GL | 3 | 16538239-17145970 | chr03_16692706 | C/T | −0.215 | 0.028 | 3.9741 × 10−14 | 41.52 | GS3 |
| 2018GA | GL | 3 | 16538239-17145970 | chr03_16692834 | A/G | −0.236 | 0.027 | 7.18544 × 10−14 | 40.03 | GS3 | |
| 2017GA | GL | 3 | 16538239-17145970 | chr03_16727804 | G/A | −0.187 | 0.023 | 1.00333 × 10−15 | 39.81 | GS3 | |
| 2018EZ | GL | 3 | 16538239-17145970 | chr03_16669223 | T/C | −0.242 | 0.029 | 3.25938 × 10−16 | 35.72 | GS3 | |
| qGL5 | 2017GA | GL | 5 | 5233629-5496042 | chr05_5361894 | G/A | −0.132 | 0.020 | 3.72538 × 10−11 | 35.53 | GW5; OsDER1 |
| 2017EZ | GL | 5 | 5233629-5496042 | chr05_5361894 | G/A | −0.145 | 0.024 | 1.49047 × 10−9 | 34.90 | GW5; OsDER1 | |
| 2018EZ | GL | 5 | 5233629-5488611 | chr05_5359598 | G/A | −0.138 | 0.026 | 9.59975 × 10−8 | 24.92 | GW5; OsDER1 | |
| 2018GA | GL | 5 | 5234598-5484598 | chr05_5359598 | G/A | −0.121 | 0.024 | 4.07439 × 10−7 | 27.56 | GW5; OsDER1 | |
| qGLWR1.1 | 2017GA | GLWR | 1 | 3182916-3432916 | chr01_3307916 | A/G | 0.222 | 0.039 | 1.93411 × 10−8 | 0.57 | |
| 2018EZ | GLWR | 1 | 3182916-3432916 | chr01_3307916 | A/G | 0.214 | 0.041 | 2.85427 × 10−7 | 0.38 | ||
| 2017EZ | GLWR | 1 | 3182916-3432916 | chr01_3307916 | A/G | 0.213 | 0.040 | 1.86665 × 10−7 | 0.32 | ||
| qGLWR1.2 | 2018EZ | GLWR | 1 | 22885450-23135450 | chr01_23010450 | A/G | −0.287 | 0.052 | 4.5957 × 10−8 | 9.95 | |
| 2018GA | GLWR | 1 | 22885450-23135450 | chr01_23010450 | A/G | −0.276 | 0.052 | 1.61289 × 10−7 | 9.36 | ||
| 2017EZ | GLWR | 1 | 22885450-23137896 | chr01_23010450 | A/G | −0.301 | 0.051 | 5.90519 × 10−9 | 9.03 | ||
| qGLWR2.1 | 2018EZ | GLWR | 2 | 3328503-3578511 | chr02_3453511 | C/A | −0.180 | 0.035 | 2.76336 × 10−7 | 7.68 | |
| 2017EZ | GLWR | 2 | 3328503-3578581 | chr02_3453511 | C/A | −0.192 | 0.034 | 2.33924 × 10−8 | 7.39 | ||
| 2018GA | GLWR | 2 | 3328511-3578511 | chr02_3453511 | C/A | −0.179 | 0.035 | 3.7586 × 10−7 | 7.16 | ||
| qGLWR2.2 | 2017GA | GLWR | 2 | 5535710-5785710 | chr02_5660710 | T/A | −0.330 | 0.056 | 5.91233 × 10−9 | 6.60 | |
| 2018GA | GLWR | 2 | 5535710-5999873 | chr02_5660710 | T/A | −0.324 | 0.060 | 8.1989 × 10−8 | 6.34 | ||
| 2018EZ | GLWR | 2 | 5535710-5999873 | chr02_5874873 | C/T | −0.373 | 0.071 | 2.23701 × 10−7 | 6.08 | ||
| 2017EZ | GLWR | 2 | 5535710-6323430 | chr02_5660710 | T/A | −0.347 | 0.057 | 2.70846 × 10−9 | 5.92 | ||
| 2018EZ | GLWR | 2 | 6073121-6323430 | chr02_6198419 | G/T | −0.338 | 0.064 | 1.90726 × 10−7 | 6.22 | ||
| 2018GA | GLWR | 2 | 6073121-6323430 | chr02_6198419 | G/T | −0.358 | 0.065 | 4.58012 × 10−8 | 5.76 | ||
| qGLWR2.4 | 2017GA | GLWR | 2 | 12281126-12531126 | chr02_12406126 | C/T | −0.313 | 0.058 | 1.20528 × 10−7 | 11.47 | |
| qGLWR2.5 | 2018EZ | GLWR | 2 | 12581170-12854381 | chr02_12706170 | G/A | −0.375 | 0.067 | 3.83638 × 10−8 | 12.35 | |
| 2017GA | GLWR | 2 | 12581170-12854381 | chr02_12706170 | G/A | −0.356 | 0.065 | 5.90344 × 10−8 | 12.21 | ||
| 2018GA | GLWR | 2 | 12581170-12831170 | chr02_12706170 | G/A | −0.367 | 0.068 | 1.02856 × 10−7 | 11.60 | ||
| 2017EZ | GLWR | 2 | 12581170-12854381 | chr02_12729381 | C/T | −0.377 | 0.067 | 3.44126 × 10−8 | 10.28 | ||
| qGLWR2.6 | 2018EZ | GLWR | 2 | 13775664-14025664 | chr02_13900664 | A/G | −0.341 | 0.066 | 3.74571 × 10−7 | 11.84 | |
| qGLWR2.7 | 2017GA | GLWR | 2 | 15065202-15315202 | chr02_15190202 | C/T | −0.253 | 0.049 | 2.83929 × 10−7 | 10.74 | |
| qGLWR3.1 | 2017GA | GLWR | 3 | 11862123-12195220 | chr03_12040893 | G/A | −0.238 | 0.042 | 2.0198 × 10−8 | 10.29 | |
| qGLWR3.2 | 2017GA | GLWR | 3 | 14887077-15259083 | chr03_15097804 | T/C | −0.379 | 0.066 | 1.73085 × 10−8 | 11.94 | |
| 2018EZ | GLWR | 3 | 14972804-15262319 | chr03_15137319 | G/C | −0.370 | 0.063 | 8.16489 × 10−9 | 10.84 | ||
| 2017EZ | GLWR | 3 | 14972804-15775796 | chr03_15137319 | G/C | −0.377 | 0.062 | 1.86381 × 10−9 | 9.87 | ||
| 2018GA | GLWR | 3 | 15007020-15499312 | chr03_15137319 | G/C | −0.377 | 0.064 | 6.37505 × 10−9 | 10.27 | ||
| 2018GA | GLWR | 3 | 15525398-15798206 | chr03_15650795 | C/T | −0.261 | 0.048 | 8.10411 × 10−8 | 9.26 | ||
| 2018EZ | GLWR | 3 | 15525795-15798182 | chr03_15673182 | T/A | −0.253 | 0.049 | 2.89611 × 10−7 | 9.09 | ||
| qGLWR3.3 | 2018EZ | GLWR | 3 | 16384313-17145970 | chr03_16665078 | G/A | −0.253 | 0.030 | 5.54904 × 10−16 | 38.74 | GS3 |
| 2017EZ | GLWR | 3 | 16538239-17145970 | chr03_16667236 | A/C | −0.247 | 0.030 | 4.99029 × 10−16 | 40.92 | GS3 | |
| 2017GA | GLWR | 3 | 16538239-17144509 | chr03_16665078 | G/A | −0.242 | 0.030 | 1.84033 × 10−15 | 38.89 | GS3 | |
| 2018GA | GLWR | 3 | 16538239-17145970 | chr03_16665078 | G/A | −0.249 | 0.031 | 2.87413 × 10−15 | 38.73 | GS3 | |
| qGLWR4.4 | 2018EZ | GLWR | 4 | 20288335-20541326 | chr04_20416326 | G/A | −0.229 | 0.041 | 3.2422 × 10−8 | 7.48 | |
| 2017EZ | GLWR | 4 | 20288335-21006646 | chr04_20416326 | G/A | −0.226 | 0.040 | 2.42506 × 10−8 | 6.86 | ||
| 2018GA | GLWR | 4 | 20291326-20541326 | chr04_20416326 | G/A | −0.220 | 0.041 | 1.22254 × 10−7 | 6.94 | ||
| 2018EZ | GLWR | 4 | 20593177-21006646 | chr04_20718177 | G/A | −0.215 | 0.041 | 2.54712 × 10−7 | 5.45 | ||
| 2017GA | GLWR | 4 | 20638861-21006958 | chr04_20881646 | G/A | −0.254 | 0.042 | 3.01979 × 10−9 | 5.45 | ||
| qGLWR5.2 | 2017EZ | GLWR | 5 | 5231448-5561924 | chr05_5361894 | G/A | −0.214 | 0.025 | 5.32542 × 10−17 | 42.53 | GW5; OsDER1 |
| 2018GA | GLWR | 5 | 5231448-5561924 | chr05_5361894 | G/A | −0.231 | 0.025 | 1.41282 × 10−18 | 41.88 | GW5; OsDER1 | |
| 2017GA | GLWR | 5 | 5231448-5503981 | chr05_5359598 | G/A | −0.210 | 0.024 | 5.38154 × 10−17 | 41.49 | GW5; OsDER1 | |
| 2018EZ | GLWR | 5 | 5231448-5561924 | chr05_5359598 | G/A | −0.232 | 0.025 | 1.19804 × 10−18 | 40.73 | GW5; OsDER1 | |
| qGLWR7.2 | 2017GA | GLWR | 7 | 23874546-24124546 | chr07_23999546 | G/T | −0.273 | 0.051 | 1.32149 × 10−7 | 10.23 | |
| qGLWR10.1 | 2018EZ | GLWR | 10 | 461202-711202 | chr10_586202 | A/T | −0.241 | 0.045 | 1.11028 × 10−7 | 12.01 | |
| 2018GA | GLWR | 10 | 461202-711202 | chr10_586202 | A/T | −0.237 | 0.045 | 1.8842 × 10−7 | 11.28 | ||
| 2017EZ | GLWR | 10 | 461202-711202 | chr10_586202 | A/T | −0.253 | 0.045 | 4.17056 × 10−8 | 10.75 | ||
| qGLWR10.2 | 2017GA | GLWR | 10 | 981239-1231239 | chr10_1106239 | G/T | −0.251 | 0.049 | 3.35516 × 10−7 | 10.74 | |
| qGLWR10.4 | 2017GA | GLWR | 10 | 3451996-3701996 | chr10_3576996 | G/A | −0.256 | 0.048 | 1.5324 × 10−7 | 10.03 | |
| qGLWR10.5 | 2017EZ | GLWR | 10 | 4480128-4738419 | chr10_4612011 | A/G | −0.276 | 0.046 | 2.50401 × 10−9 | 11.20 | |
| 2017GA | GLWR | 10 | 4487011-4737058 | chr10_4612058 | T/G | −0.283 | 0.046 | 1.62479 × 10−9 | 11.26 | ||
| 2018EZ | GLWR | 10 | 4487011-4737058 | chr10_4612058 | T/G | −0.254 | 0.048 | 2.08188 × 10−7 | 10.79 | ||
| 2018GA | GLWR | 10 | 4487011-4737058 | chr10_4612058 | T/G | −0.263 | 0.049 | 9.96257 × 10−8 | 10.62 | ||
| qGLWR10.6 | 2018EZ | GLWR | 10 | 5900422-6150495 | chr10_6025464 | G/A | −0.304 | 0.053 | 1.57353 × 10−8 | 9.73 | |
| 2017EZ | GLWR | 10 | 5900422-6150524 | chr10_6025464 | G/A | −0.346 | 0.052 | 5.39359 × 10−11 | 9.46 | ||
| 2018GA | GLWR | 10 | 5900422-6150495 | chr10_6025464 | G/A | −0.315 | 0.053 | 5.45461 × 10−9 | 9.25 | ||
| qGLWR10.7 | 2018EZ | GLWR | 10 | 7593442-7843442 | chr10_7718442 | T/C | −0.313 | 0.058 | 8.71587 × 10−8 | 9.06 | |
| 2018GA | GLWR | 10 | 7593442-7843442 | chr10_7718442 | T/C | −0.311 | 0.058 | 1.33752 × 10−7 | 8.50 | ||
| 2017EZ | GLWR | 10 | 7593442-7843442 | chr10_7718442 | T/C | −0.321 | 0.056 | 1.63274 × 10−8 | 8.11 | ||
| qGLWR10.8 | 2018EZ | GLWR | 10 | 8856195-9106195 | chr10_8981195 | G/T | −0.332 | 0.062 | 1.05397 × 10−7 | 10.16 | |
| 2018GA | GLWR | 10 | 8856195-9106195 | chr10_8981195 | G/T | −0.332 | 0.062 | 1.42668 × 10−7 | 9.51 | ||
| 2017EZ | GLWR | 10 | 8856195-9106195 | chr10_8981195 | G/T | −0.336 | 0.060 | 3.12832 × 10−8 | 9.06 | ||
| qGLWR10.9 | 2017EZ | GLWR | 10 | 10025822-10458283 | chr10_10223273 | C/T | −0.381 | 0.059 | 2.51359 × 10−10 | 8.23 | |
| 2017GA | GLWR | 10 | 10042380-10458283 | chr10_10272029 | G/A | −0.363 | 0.061 | 4.73434 × 10−9 | 9.43 | ||
| 2018EZ | GLWR | 10 | 10042380-10397029 | chr10_10223273 | C/T | −0.363 | 0.060 | 2.98474 × 10−9 | 9.16 | ||
| 2018GA | GLWR | 10 | 10042380-10397029 | chr10_10223273 | C/T | −0.372 | 0.061 | 1.97669 × 10−9 | 8.79 | ||
| qGW1.3 | 2018EZ | GW | 1 | 23933241-24512660 | chr01_24304654 | A/G | 0.113 | 0.022 | 2.42535 × 10−7 | 6.23 | OsCTPS1 |
| qGW2.1 | 2017EZ | GW | 2 | 3328503-3578511 | chr02_3453511 | C/A | 0.069 | 0.012 | 1.13817 × 10−8 | 6.07 | |
| 2018EZ | GW | 2 | 3328503-3578511 | chr02_3453511 | C/A | 0.065 | 0.012 | 2.15744 × 10−7 | 4.81 | ||
| 2018GA | GW | 2 | 3328503-3578511 | chr02_3453511 | C/A | 0.067 | 0.013 | 2.35387 × 10−7 | 4.12 | ||
| qGW3.4 | 2017GA | GW | 3 | 16538324-17028634 | chr03_16706516 | T/C | 0.053 | 0.008 | 5.96404 × 10−11 | 41.78 | GS3 |
| 2018EZ | GW | 3 | 16540072-17040318 | chr03_16706516 | T/C | 0.062 | 0.010 | 5.44229 × 10−10 | 30.86 | GS3 | |
| 2017EZ | GW | 3 | 16540424-16926472 | chr03_16706516 | T/C | 0.056 | 0.009 | 3.95865 × 10−9 | 30.65 | GS3 | |
| 2018GA | GW | 3 | 16570517-16998965 | chr03_16746142 | A/G | 0.059 | 0.011 | 2.73991 × 10−8 | 25.80 | GS3 | |
| qGW4.1 | 2018GA | GW | 4 | 11108-800893 | chr04_628540 | C/A | 0.111 | 0.019 | 1.47095 × 10−8 | 8.52 | OsARG |
| 2017EZ | GW | 4 | 11113-261113 | chr04_136113 | G/A | 0.091 | 0.017 | 1.77411 × 10−7 | 8.98 | ||
| 2018EZ | GW | 4 | 263820-769400 | chr04_644395 | A/G | 0.108 | 0.019 | 2.30805 × 10−8 | 7.48 | OsARG | |
| 2017EZ | GW | 4 | 546523-796523 | chr04_671523 | C/T | 0.099 | 0.019 | 3.60601 × 10−7 | 9.04 | OsARG | |
| qGW5.1 | 2017GA | GW | 5 | 5231448-5561924 | chr05_5359246 | G/A | 0.065 | 0.007 | 2.13646 × 10−20 | 42.12 | GW5; OsDER1 |
| 2018GA | GW | 5 | 5231448-5585712 | chr05_5359246 | G/A | 0.091 | 0.009 | 7.09101 × 10−22 | 36.43 | GW5; OsDER1 | |
| 2018EZ | GW | 5 | 5231448-5574689 | chr05_5359681 | C/T | 0.082 | 0.009 | 5.765 × 10−19 | 36.22 | GW5; OsDER1 | |
| 2017EZ | GW | 5 | 5231448-5581997 | chr05_5359246 | G/A | 0.071 | 0.008 | 7.27903 × 10−17 | 31.13 | GW5; OsDER1 | |
| qGW5.2 | 2018GA | GW | 5 | 5917480-6167480 | chr05_6042480 | C/T | 0.043 | 0.008 | 3.08835 × 10−7 | 12.62 | JMJ703 |
| qGW10.1 | 2018GA | GW | 10 | 52079-302079 | chr10_177079 | T/A | 0.124 | 0.022 | 4.17434 × 10−8 | 8.97 | OsSCP46 |
| 2018EZ | GW | 10 | 52079-302079 | chr10_177079 | T/A | 0.120 | 0.022 | 5.35845 × 10−8 | 8.72 | OsSCP46 | |
| qGW10.7 | 2017EZ | GW | 10 | 5900422-6150495 | chr10_6025464 | G/A | 0.126 | 0.019 | 1.49782 × 10−10 | 10.32 | |
| 2018GA | GW | 10 | 5900422-6150524 | chr10_6025464 | G/A | 0.142 | 0.021 | 7.97234 × 10−11 | 8.94 | ||
| 2018EZ | GW | 10 | 5900464-6150464 | chr10_6025464 | G/A | 0.112 | 0.022 | 3.22894 × 10−7 | 7.82 | ||
| qGW10.8 | 2017EZ | GW | 10 | 6986882-7439539 | chr10_7297872 | A/C | 0.121 | 0.020 | 4.16284 × 10−9 | 10.05 | |
| 2018GA | GW | 10 | 7154870-7422872 | chr10_7279870 | C/G | 0.108 | 0.018 | 2.7638 × 10−9 | 11.54 | ||
| qGW10.12 | 2017EZ | GW | 10 | 10078190-10459267 | chr10_10223273 | C/T | 0.122 | 0.021 | 9.81178 × 10−9 | 8.84 | |
| 2018EZ | GW | 10 | 10078190-10405532 | chr10_10203190 | A/G | 0.127 | 0.022 | 1.04852 × 10−8 | 7.51 | ||
| 2017GA | GW | 10 | 10147029-10418094 | chr10_10272029 | G/A | 0.095 | 0.018 | 1.02675 × 10−7 | 7.89 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, B.; Wang, T.; Luo, Y.; Guo, Y.; Xu, D.; Liu, C.; Zou, J.; Li, L.; Diao, Y.; Gao, Z.; et al. Identification and Allele Combination Analysis of Rice Grain Shape-Related Genes by Genome-Wide Association Study. Int. J. Mol. Sci. 2022, 23, 1065. https://doi.org/10.3390/ijms23031065
Meng B, Wang T, Luo Y, Guo Y, Xu D, Liu C, Zou J, Li L, Diao Y, Gao Z, et al. Identification and Allele Combination Analysis of Rice Grain Shape-Related Genes by Genome-Wide Association Study. International Journal of Molecular Sciences. 2022; 23(3):1065. https://doi.org/10.3390/ijms23031065
Chicago/Turabian StyleMeng, Bingxin, Tao Wang, Yi Luo, Ying Guo, Deze Xu, Chunhai Liu, Juan Zou, Lanzhi Li, Ying Diao, Zhiyong Gao, and et al. 2022. "Identification and Allele Combination Analysis of Rice Grain Shape-Related Genes by Genome-Wide Association Study" International Journal of Molecular Sciences 23, no. 3: 1065. https://doi.org/10.3390/ijms23031065
APA StyleMeng, B., Wang, T., Luo, Y., Guo, Y., Xu, D., Liu, C., Zou, J., Li, L., Diao, Y., Gao, Z., Hu, Z., & Zheng, X. (2022). Identification and Allele Combination Analysis of Rice Grain Shape-Related Genes by Genome-Wide Association Study. International Journal of Molecular Sciences, 23(3), 1065. https://doi.org/10.3390/ijms23031065





