Smart Nucleic Acid Hydrogels with High Stimuli-Responsiveness in Biomedical Fields
Abstract
:1. Introduction
2. Assembly Mechanism of Stimuli-Responsive NAHs
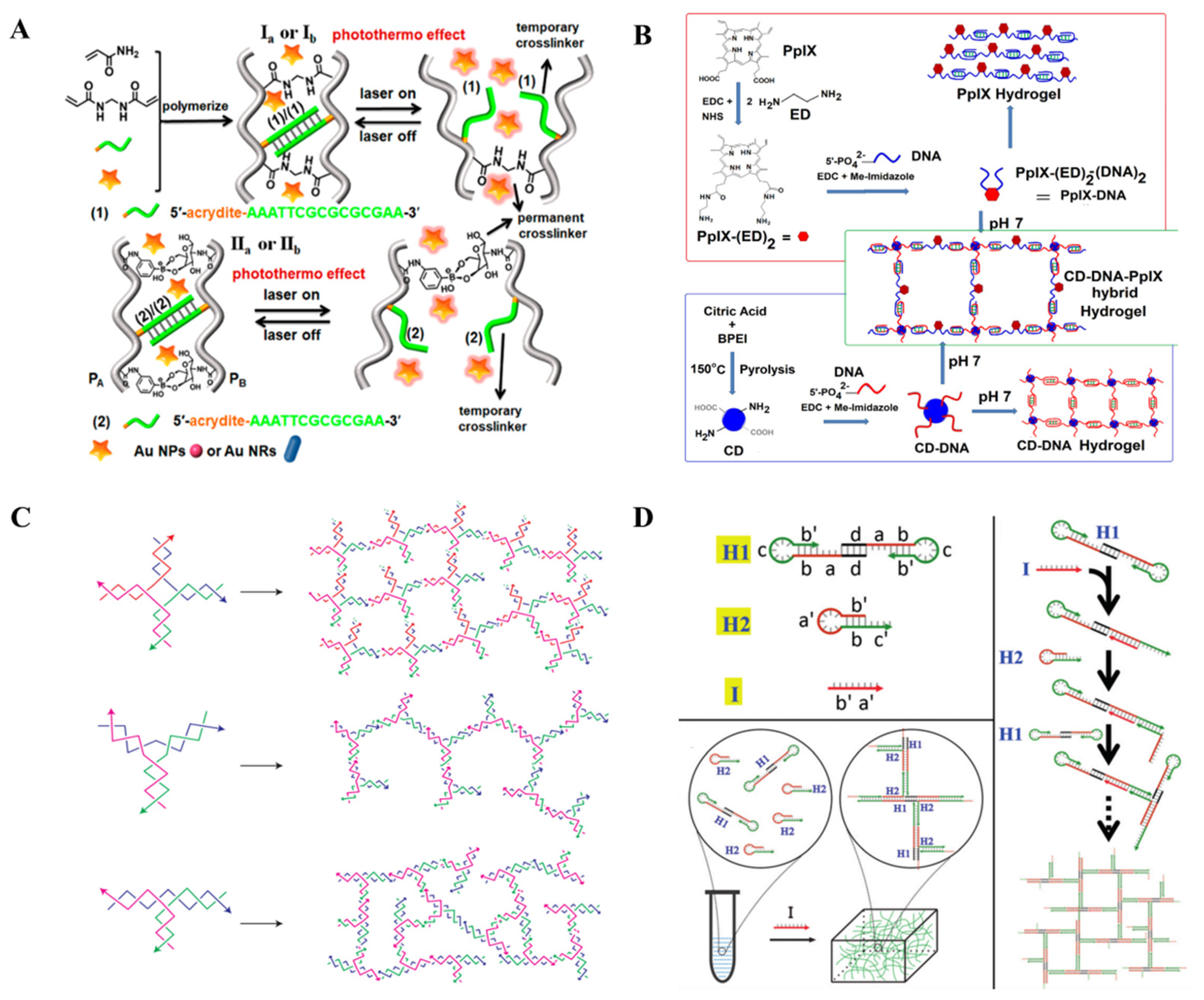
2.1. Hybrid NAHs
2.2. Pure NAHs
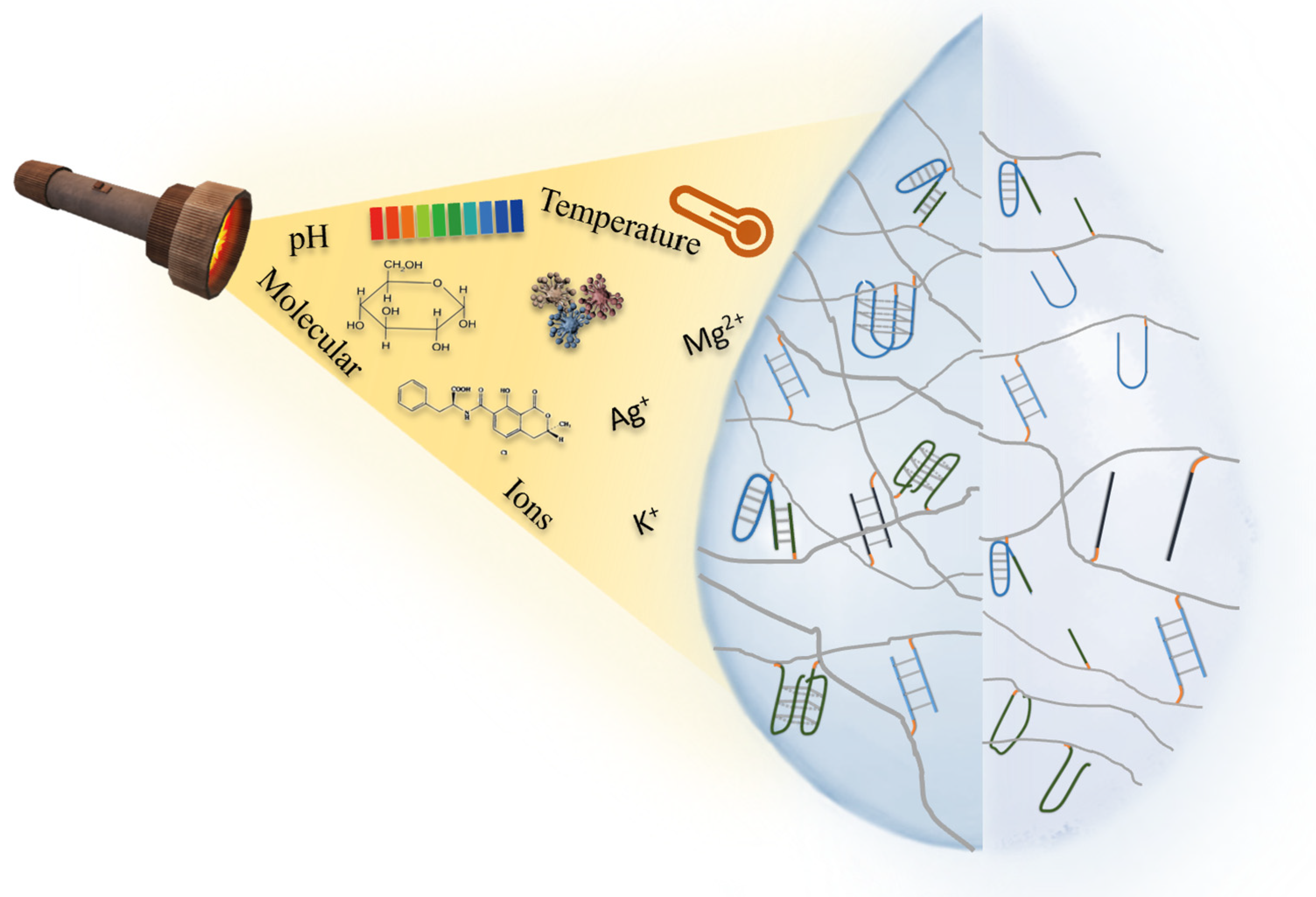
3. Stimuli-Responsive Mechanism of Stimuli-Responsive NAHs
3.1. pH-Responsive NAHs
3.2. Target-Responsive NAHs
3.3. Ions-Responsive NAHs
3.4. Temperature-Responsive NAHs

4. Biomedical Applications of Stimuli-Responsive NAHs
4.1. Biosensing
4.2. Drug Loading and Targeted Delivery
4.3. Tissue Engineering
5. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Veigl, S.J.; Harman, O.; Lamm, E. Friedrich Miescher’s Discovery in the Historiography of Genetics: From Contamination to Confusion, from Nuclein to DNA. J. Hist. Biol. 2020, 53, 451–484. [Google Scholar] [CrossRef] [PubMed]
- Watson, J.D.; Crick, F. Molecular structure of nucleic acids. Nature 1953, 171, 737–738. [Google Scholar] [CrossRef] [PubMed]
- Skopinska-Wisniewska, J.; De la Flor, S.; Kozlowska, J. From Supramolecular Hydrogels to Multifunctional Carriers for Biologically Active Substances. Int. J. Mol. Sci. 2021, 22, 7402. [Google Scholar] [CrossRef] [PubMed]
- Wichterle, O.; Lim, D. Hydrophilic Gels for Biological Use. Nature 1960, 185, 117–118. [Google Scholar] [CrossRef]
- Nagahara, S.; Matsuda, T. Hydrogel formation via hybridization of oligonucleotides derivatized in water-soluble vinyl polymers. Polym. Gels Netw. 1996, 4, 111–127. [Google Scholar] [CrossRef]
- Um, S.H.; Lee, J.B.; Park, N.; Kwon, S.Y.; Umbach, C.C.; Luo, D. Enzyme-catalysed assembly of DNA hydrogel. Nat. Mater. 2006, 5, 797–801. [Google Scholar] [CrossRef]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Joyce, G.F. RNA Cleavage by the 10–23 DNA Enzyme. In Ribonucleases—Part A; Academic Press: New York, NY, USA, 2001; pp. 503–517. [Google Scholar] [CrossRef]
- Mirkin, C.A.; Letsinger, R.L.; Mucic, R.C.; Storhoff, J.J. A DNA-based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [Google Scholar] [CrossRef]
- Egholm, M.; Nielsen, P.E.; Buchardt, O.; Berg, R.H. Recognition of guanine and adenine in DNA by cytosine and thymine containing peptide nucleic acids (PNA). J. Am. Chem. Soc. 1992, 114, 9677–9678. [Google Scholar] [CrossRef]
- Leroy, J.L.; Gueron, M.; Mergny, J.L.; Helene, C. Intramolecular folding of a fragment of the cytosine-rich strand of telomeric DNA into an i-motif. Nucleic Acids Res. 1994, 22, 1600–1606. [Google Scholar] [CrossRef] [Green Version]
- Travascio, P.; Bennet, A.J.; Wang, D.Y.; Sen, D. A ribozyme and a catalytic DNA with peroxidase activity: Active sites versus cofactor-binding sites. Chem. Biol. 1999, 6, 779–787. [Google Scholar] [CrossRef]
- Xu, W.; He, W.; Du, Z.; Zhu, L.; Huang, K.; Lu, Y.; Luo, Y. Functional Nucleic Acid Nanomaterials: Development, Properties, and Applications. Angew. Chem. Int. Ed. Engl. 2019, 60, 6890–6918. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhu, L.; Tian, J.; Zhu, L.; Ma, X.; He, X.; Huang, K.; Ren, F.; Xu, W. Smart and Functionalized Development of Nucleic Acid-Based Hydrogels: Assembly Strategies, Recent Advances, and Challenges. Adv. Sci. 2021, 8, 2100216. [Google Scholar] [CrossRef]
- Kim, M.-G.; Shon, Y.; Miao, W.; Lee, J.; Oh, Y.-K. Biodegradable graphene oxide and polyaptamer DNA hybrid hydrogels for implantable drug delivery. Carbon 2016, 105, 14–22. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Q.; Sun, Y.; Bai, H.; Shi, G. Three-dimensional self-assembly of graphene oxide and DNA into multifunctional hydrogels. Acs Nano 2010, 4, 7358–7362. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Hu, N.; Peng, J.; Chen, L.; Weng, J. Ultrasensitive Detection of Mitochondrial DNA Mutation by Graphene Oxide/DNA Hydrogel Electrode. Adv. Funct. Mater. 2014, 24, 6905–6913. [Google Scholar] [CrossRef]
- Singh, S.; Mishra, A.; Kumari, R.; Sinha, K.K.; Singh, M.K.; Das, P. Carbon dots assisted formation of DNA hydrogel for sustained release of drug. Carbon 2017, 114, 169–176. [Google Scholar] [CrossRef]
- Pandey, P.K.; Preeti; Rawat, K.; Prasad, T.; Bohidar, H.B. Multifunctional, fluorescent DNA-derived carbon dots for biomedical applications: Bioimaging, luminescent DNA hydrogels, and dopamine detection. J. Mater. Chem. B 2020, 8, 1277–1289. [Google Scholar] [CrossRef]
- Kumari, S.; Rajit Prasad, S.; Mandal, D.; Das, P. Carbon dot-DNA-protoporphyrin hybrid hydrogel for sustained photoinduced antimicrobial activity. J. Colloid Interface Sci. 2019, 553, 228–238. [Google Scholar] [CrossRef]
- Mansukhani, N.D.; Guiney, L.M.; Wei, Z.; Roth, E.W.; Putz, K.W.; Luijten, E.; Hersam, M.C. Optothermally Reversible Carbon Nanotube-DNA Supramolecular Hybrid Hydrogels. Macromol. Rapid Commun. 2018, 39, 1700587. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jean, S.R.; Ahmed, S.; Aldridge, P.M.; Li, X.; Fan, F.; Sargent, E.H.; Kelley, S.O. Multifunctional quantum dot DNA hydrogels. Nat. Commun. 2017, 8, 381. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.; Bian, P.; Kim, J.; McCarthy, T.J.; Hayward, R.C. Local switching of chemical patterns through light-triggered unfolding of creased hydrogel surfaces. Angew. Chem. Int. Ed. Engl. 2012, 51, 7146–7149. [Google Scholar] [CrossRef]
- Umeno, D.; Kano, T.; Maeda, M. Affinity adsorption separation of mutagenic molecules by polyacrylamide hydrogels comprising double-stranded DNA. Anal. Chim. Acta 1998, 365, 101–108. [Google Scholar] [CrossRef]
- Dave, N.; Chan, M.Y.; Huang, P.-J.J.; Smith, B.D.; Liu, J. Regenerable DNA-Functionalized Hydrogels for Ultrasensitive, Instrument-Free Mercury(II) Detection and Removal in Water. J. Am. Chem. Soc. 2010, 132, 12668–12673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.H.; Guo, W.; Hu, Y.; Qi, X.J.; Willner, I. Multitriggered Shape-Memory Acrylamide-DNA Hydrogels. J. Am. Chem. Soc. 2015, 137, 15723–15731. [Google Scholar] [CrossRef]
- Liao, W.C.; Lilienthal, S.; Kahn, J.S.; Riutin, M.; Sohn, Y.S.; Nechushtai, R.; Willner, I. pH- and ligand-induced release of loads from DNA-acrylamide hydrogel microcapsules. Chem. Sci. 2017, 8, 3362–3373. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.-H.; Liao, W.-C.; Sohn, Y.S.; Fadeev, M.; Cecconello, A.; Nechushtai, R.; Willner, I. Stimuli-Responsive Nucleic Acid-Based Polyacrylamide Hydrogel-Coated Metal-Organic Framework Nanoparticles for Controlled Drug Release. Adv. Funct. Mater. 2018, 28, 1705137. [Google Scholar] [CrossRef]
- Wang, C.; Fischer, A.; Ehrlich, A.; Nahmias, Y.; Willner, I. Biocatalytic reversible control of the stiffness of DNA-modified responsive hydrogels: Applications in shape-memory, self-healing and autonomous controlled release of insulin. Chem. Sci. 2020, 11, 4516–4524. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Liu, X.; Wulf, V.; Vazquez-Gonzalez, M.; Fadeev, M.; Willner, I. DNA-Based Hydrogels Loaded with Au Nanoparticles or Au Nanorods: Thermoresponsive Plasmonic Matrices for Shape-Memory, Self-Healing, Controlled Release, and Mechanical Applications. ACS Nano 2019, 13, 3424–3433. [Google Scholar] [CrossRef]
- Guo, W.; Lu, C.H.; Qi, X.J.; Orbach, R.; Fadeev, M.; Yang, H.H.; Willner, I. Switchable bifunctional stimuli-triggered poly-N-isopropylacrylamide/DNA hydrogels. Angew. Chem. Int. Ed. Engl. 2014, 53, 10134–10138. [Google Scholar] [CrossRef] [PubMed]
- Matthews, B.M.; Maley, A.M.; Kartub, K.M.; Corn, R.M. Characterizing the Incorporation of DNA into Single NIPAm Hydrogel Nanoparticles with Surface Plasmon Resonance Imaging Measurements. J. Phys. Chem. C 2019, 123, 6090–6096. [Google Scholar] [CrossRef]
- Abune, L.; Zhao, N.; Lai, J.; Peterson, B.; Szczesny, S.; Wang, Y. Macroporous Hydrogels for Stable Sequestration and Sustained Release of Vascular Endothelial Growth Factor and Basic Fibroblast Growth Factor Using Nucleic Acid Aptamers. ACS Biomater. Sci. Eng. 2019, 5, 2382–2390. [Google Scholar] [CrossRef]
- Chen, N.; Zhang, Z.; Soontornworajit, B.; Zhou, J.; Wang, Y. Cell adhesion on an artificial extracellular matrix using aptamer-functionalized PEG hydrogels. Biomaterials 2012, 33, 1353–1362. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Du, J.; Li, Y.; Wu, J.; Yu, F.; Chen, Y. An aptamer-patterned hydrogel for the controlled capture and release of proteins via biorthogonal click chemistry and DNA hybridization. J. Mater. Chem. B 2017, 5, 5974–5982. [Google Scholar] [CrossRef]
- Tanaka, S.; Yukami, S.; Fukushima, K.; Wakabayashi, K.; Ohya, Y.; Kuzuya, A. Bulk pH-Responsive DNA Quadruplex Hydrogels Prepared by Liquid-Phase, Large-Scale DNA Synthesis. ACS Macro Lett. 2018, 7, 295–299. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, Y.; Yu, F.; Niu, C.; Du, Z.; Chen, Y.; Du, J. Rapid and annealing-free self-assembly of DNA building blocks for 3D hydrogel chaperoned by cationic comb-type copolymers. J. Biomater. Sci. Polym. Ed. 2017, 28, 1511–1524. [Google Scholar] [CrossRef] [PubMed]
- Ranjbar-Navazi, Z.; Fathi, M.; Abdolahinia, E.D.; Omidi, Y.; Davaran, S. MUC-1 aptamer conjugated InP/ZnS quantum dots/nanohydrogel fluorescent composite for mitochondria-mediated apoptosis in MCF-7 cells. Mater. Sci. Eng. C Mater. Biol. Appl. 2021, 118, 111469. [Google Scholar] [CrossRef]
- Li, C.; Faulkner-Jones, A.; Dun, A.R.; Jin, J.; Chen, P.; Xing, Y.; Yang, Z.; Li, Z.; Shu, W.; Liu, D.; et al. Rapid formation of a supramolecular polypeptide-DNA hydrogel for in situ three-dimensional multilayer bioprinting. Angew. Chem. Int. Ed. Engl. 2015, 54, 3957–3961. [Google Scholar] [CrossRef]
- Li, Y.; Tseng, Y.D.; Kwon, S.Y.; D’Espaux, L.; Bunch, J.S.; McEuen, P.L.; Luo, D. Controlled assembly of dendrimer-like DNA. Nat. Mater. 2004, 3, 38–42. [Google Scholar] [CrossRef]
- Li, Y.; Cu, Y.T.; Luo, D. Multiplexed detection of pathogen DNA with DNA-based fluorescence nanobarcodes. Nat. Biotechnol. 2005, 23, 885–889. [Google Scholar] [CrossRef]
- Nishida, Y.; Ohtsuki, S.; Araie, Y.; Umeki, Y.; Endo, M.; Emura, T.; Hidaka, K.; Sugiyama, H.; Takahashi, Y.; Takakura, Y.; et al. Self-assembling DNA hydrogel-based delivery of immunoinhibitory nucleic acids to immune cells. Nanomedicine 2016, 12, 123–130. [Google Scholar] [CrossRef]
- Hartman, M.R.; Yang, D.; Tran, T.N.; Lee, K.; Kahn, J.S.; Kiatwuthinon, P.; Yancey, K.G.; Trotsenko, O.; Minko, S.; Luo, D. Thermostable branched DNA nanostructures as modular primers for polymerase chain reaction. Angew. Chem. Int. Ed. Engl. 2013, 52, 8699–8702. [Google Scholar] [CrossRef] [PubMed]
- Cheng, E.; Xing, Y.; Chen, P.; Yang, Y.; Sun, Y.; Zhou, D.; Xu, L.; Fan, Q.; Liu, D. A pH-triggered, fast-responding DNA hydrogel. Angew. Chem. Int. Ed. Engl. 2009, 48, 7660–7663. [Google Scholar] [CrossRef]
- Lu, S.; Wang, S.; Zhao, J.; Sun, J.; Yang, X. A pH-controlled bidirectionally pure DNA hydrogel: Reversible self-assembly and fluorescence monitoring. Chem. Commun. 2018, 54, 4621–4624. [Google Scholar] [CrossRef]
- Wang, J.; Chao, J.; Liu, H.; Su, S.; Wang, L.; Huang, W.; Willner, I.; Fan, C. Clamped Hybridization Chain Reactions for the Self-Assembly of Patterned DNA Hydrogels. Angew. Chem. Int. Ed. Engl. 2017, 56, 2171–2175. [Google Scholar] [CrossRef]
- Lee, J.B.; Peng, S.; Yang, D.; Roh, Y.H.; Funabashi, H.; Park, N.; Rice, E.J.; Chen, L.; Long, R.; Wu, M.; et al. A mechanical metamaterial made from a DNA hydrogel. Nat. Nanotechnol. 2012, 7, 816–820. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Xu, W.; Liu, G.; Tian, L. A pure DNA hydrogel with stable catalytic ability produced by one-step rolling circle amplification. Chem. Commun. 2017, 53, 3038–3041. [Google Scholar] [CrossRef] [PubMed]
- Langridge, R.; Rich, A. Molecular structure of helical polycytidylic acid. Nature 1963, 198, 725–728. [Google Scholar] [CrossRef]
- Brown, D.M.; Gray, D.M.; Patrick, M.H.; Ratliff, R.L. Photochemical demonstration of stacked C.C+ base pairs in a novel DNA secondary structure. Biochemistry 1985, 24, 1676–1683. [Google Scholar] [CrossRef]
- Rajendran, A.; Nakano, S.; Sugimoto, N. Molecular crowding of the cosolutes induces an intramolecular i-motif structure of triplet repeat DNA oligomers at neutral pH. Chem. Commun. 2010, 46, 1299–1301. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Yang, Z.; Liu, D. DNA nanotechnology based on i-motif structures. Acc. Chem. Res. 2014, 47, 1853–1860. [Google Scholar] [CrossRef]
- Guo, W.; Lu, C.H.; Orbach, R.; Wang, F.; Qi, X.J.; Cecconello, A.; Seliktar, D.; Willner, I. pH-stimulated DNA hydrogels exhibiting shape-memory properties. Adv. Mater. 2015, 27, 73–78. [Google Scholar] [CrossRef]
- Hu, Y.; Lu, C.-H.; Guo, W.; Aleman-Garcia, M.A.; Ren, J.; Willner, I. A Shape Memory Acrylamide/DNA Hydrogel Exhibiting Switchable Dual pH-Responsiveness. Adv. Funct. Mater. 2015, 25, 6867–6874. [Google Scholar] [CrossRef]
- Ren, J.; Hu, Y.; Lu, C.H.; Guo, W.; Aleman-Garcia, M.A.; Ricci, F.; Willner, I. pH-responsive and switchable triplex-based DNA hydrogels. Chem. Sci. 2015, 6, 4190–4195. [Google Scholar] [CrossRef] [Green Version]
- Yang, M.; Chen, X.; Zhu, L.; Lin, S.; Li, C.; Li, X.; Huang, K.; Xu, W. Aptamer-Functionalized DNA-Silver Nanocluster Nanofilm for Visual Detection and Elimination of Bacteria. ACS Appl. Mater. Interfaces 2021, 13, 38647–38655. [Google Scholar] [CrossRef]
- Liu, D.; Jia, S.; Zhang, H.; Ma, Y.; Guan, Z.; Li, J.; Zhu, Z.; Ji, T.; Yang, C.J. Integrating Target-Responsive Hydrogel with Pressuremeter Readout Enables Simple, Sensitive, User-Friendly, Quantitative Point-of-Care Testing. ACS Appl. Mater. Interfaces 2017, 9, 22252–22258. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Wang, X.; Sun, Y.; Dai, Y.; Sun, W.; Zhu, X.; Liu, H.; Han, R.; Gao, D.; Luo, C. A chemiluminescent biosensor for ultrasensitive detection of adenosine based on target-responsive DNA hydrogel with Au@HKUST-1 encapsulation. Sens. Actuators B Chem. 2019, 289, 56–64. [Google Scholar] [CrossRef]
- Battig, M.R.; Huang, Y.; Chen, N.; Wang, Y. Aptamer-functionalized superporous hydrogels for sequestration and release of growth factors regulated via molecular recognition. Biomaterials 2014, 35, 8040–8048. [Google Scholar] [CrossRef] [PubMed]
- Zhao, N.; Suzuki, A.; Zhang, X.; Shi, P.; Abune, L.; Coyne, J.; Jia, H.; Xiong, N.; Zhang, G.; Wang, Y. Dual Aptamer-Functionalized in Situ Injectable Fibrin Hydrogel for Promotion of Angiogenesis via Codelivery of Vascular Endothelial Growth Factor and Platelet-Derived Growth Factor-BB. ACS Appl. Mater. Interfaces 2019, 11, 18123–18132. [Google Scholar] [CrossRef]
- Lilienthal, S.; Shpilt, Z.; Wang, F.; Orbach, R.; Willner, I. Programmed DNAzyme-Triggered Dissolution of DNA-Based Hydrogels: Means for Controlled Release of Biocatalysts and for the Activation of Enzyme Cascades. ACS Appl. Mater. Interfaces 2015, 7, 8923–8931. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Bedrat, A.; Lacroix, L.; Mergny, J.L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016, 44, 1746–1759. [Google Scholar] [CrossRef]
- Tanaka, S.; Wakabayashi, K.; Fukushima, K.; Yukami, S.; Maezawa, R.; Takeda, Y.; Tatsumi, K.; Ohya, Y.; Kuzuya, A. Intelligent, Biodegradable, and Self-Healing Hydrogels Utilizing DNA Quadruplexes. Chem. Asian J. 2017, 12, 2388–2392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, Y.; Cheng, E.; Yang, Y.; Chen, P.; Zhang, T.; Sun, Y.; Yang, Z.; Liu, D. Self-assembled DNA hydrogels with designable thermal and enzymatic responsiveness. Adv. Mater. 2011, 23, 1117–1121. [Google Scholar] [CrossRef] [PubMed]
- Yata, T.; Takahashi, Y.; Tan, M.; Nakatsuji, H.; Ohtsuki, S.; Murakami, T.; Imahori, H.; Umeki, Y.; Shiomi, T.; Takakura, Y.; et al. DNA nanotechnology-based composite-type gold nanoparticle-immunostimulatory DNA hydrogel for tumor photothermal immunotherapy. Biomaterials 2017, 146, 136–145. [Google Scholar] [CrossRef]
- Zhang, X.; Battig, M.R.; Chen, N.; Gaddes, E.R.; Duncan, K.L.; Wang, Y. Chimeric Aptamer-Gelatin Hydrogels as an Extracellular Matrix Mimic for Loading Cells and Growth Factors. Biomacromolecules 2016, 17, 778–787. [Google Scholar] [CrossRef] [Green Version]
- Shahbazi, M.-A.; Bauleth-Ramos, T.; Santos, H.A. DNA Hydrogel Assemblies: Bridging Synthesis Principles to Biomedical Applications. Adv. Ther. 2018, 1, 1800042. [Google Scholar] [CrossRef]
- Hu, Y.; Guo, W.; Kahn, J.S.; Aleman-Garcia, M.A.; Willner, I. A Shape-Memory DNA-Based Hydrogel Exhibiting Two Internal Memories. Angew. Chem. Int. Ed. Engl. 2016, 55, 4210–4214. [Google Scholar] [CrossRef] [PubMed]
- Xiang, B.; He, K.; Zhu, R.; Liu, Z.; Zeng, S.; Huang, Y.; Nie, Z.; Yao, S. Self-Assembled DNA Hydrogel Based on Enzymatically Polymerized DNA for Protein Encapsulation and Enzyme/DNAzyme Hybrid Cascade Reaction. ACS Appl. Mater. Interfaces 2016, 8, 22801–22807. [Google Scholar] [CrossRef]
- Zhou, L.; Sun, N.; Xu, L.; Chen, X.; Cheng, H.; Wang, J.; Pei, R. Dual signal amplification by an “on-command” pure DNA hydrogel encapsulating HRP for colorimetric detection of ochratoxin A. RSC Adv. 2016, 6, 114500–114504. [Google Scholar] [CrossRef]
- Sun, Y.; Li, S.; Chen, R.; Wu, P.; Liang, J. Ultrasensitive and rapid detection of T-2 toxin using a target-responsive DNA hydrogel. Sens. Actuators B Chem. 2020, 311, 127912. [Google Scholar] [CrossRef]
- Wei, X.; Tian, T.; Jia, S.; Zhu, Z.; Ma, Y.; Sun, J.; Lin, Z.; Yang, C.J. Target-responsive DNA hydrogel mediated “stop-flow” microfluidic paper-based analytic device for rapid, portable and visual detection of multiple targets. Anal. Chem. 2015, 87, 4275–4282. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Huang, Y.; Ma, Y.; Jia, S.; Gao, M.; Li, J.; Zhang, H.; Xu, D.; Wu, M.; Chen, Y.; et al. Design and synthesis of target-responsive aptamer-cross-linked hydrogel for visual quantitative detection of ochratoxin A. ACS Appl. Mater. Interfaces 2015, 7, 6982–6990. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, Z.; Zou, Y.; Huang, Y.; Liu, D.; Jia, S.; Xu, D.; Wu, M.; Zhou, Y.; Zhou, S.; et al. Target-responsive “sweet” hydrogel with glucometer readout for portable and quantitative detection of non-glucose targets. J. Am. Chem. Soc. 2013, 135, 3748–3751. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, N.; Zhang, Z.; Wang, Y. Endonuclease-responsive aptamer-functionalized hydrogel coating for sequential catch and release of cancer cells. Biomaterials 2013, 34, 460–469. [Google Scholar] [CrossRef]
- Li, N.; Wang, X.Y.; Xiang, M.H.; Liu, J.W.; Yu, R.Q.; Jiang, J.H. Programmable Self-Assembly of Protein-Scaffolded DNA Nanohydrogels for Tumor-Targeted Imaging and Therapy. Anal. Chem. 2019, 91, 2610–2614. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Ma, Y.; Chen, Y.; Wu, X.; Fang, L.; Zhu, Z.; Yang, C.J. Target-responsive DNAzyme cross-linked hydrogel for visual quantitative detection of lead. Anal. Chem. 2014, 86, 11434–11439. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, J.; Fadeev, M.; Li, Z.; Wulf, V.; Tian, H.; Willner, I. Chemical and photochemical DNA “gears” reversibly control stiffness, shape-memory, self-healing and controlled release properties of polyacrylamide hydrogels. Chem. Sci. 2019, 10, 1008–1016. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, Y.; Ma, R.; Xu, Y.; Zhang, Y.; Li, B.; An, Y.; Shi, L. A G-Quadruplex Hydrogel via Multicomponent Self-Assembly: Formation and Zero-Order Controlled Release. ACS Appl. Mater. Interfaces 2017, 9, 13056–13067. [Google Scholar] [CrossRef]
- Jiang, Y.; Wu, Y.; Huo, Y. Thermo-responsive hydrogels with N-isopropylacrylamide/acrylamide interpenetrating networks for controlled drug release. J. Biomater. Sci. Polym. Ed. 2015, 26, 917–930. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Liu, H.; Dai, W.; Lei, Y. Responsive principles and applications of smart materials in biosensing. Smart Mater. Med. 2020, 1, 54–65. [Google Scholar] [CrossRef] [PubMed]
- Cho, E.J.; Lee, J.W.; Ellington, A.D. Applications of aptamers as sensors. Annu. Rev. Anal. Chem. 2009, 2, 241–264. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, H.; Kang, H.; Donovan, M.; Zhu, Z.; Tan, W. Aptamer-incorporated hydrogels for visual detection, controlled drug release, and targeted cancer therapy. Anal. Bioanal. Chem. 2012, 402, 187–194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, W.; Wu, H.; Liu, X.; Fu, M.; Jiang, J.; Du, Y.; Yang, L.; Huang, Y. Simultaneous fluorescent detection of multiple metal ions based on the DNAzymes and graphene oxide. Anal. Chim. Acta 2017, 986, 115–121. [Google Scholar] [CrossRef]
- Huang, Y.; Fang, L.; Zhu, Z.; Ma, Y.; Zhou, L.; Chen, X.; Xu, D.; Yang, C. Design and synthesis of target-responsive hydrogel for portable visual quantitative detection of uranium with a microfluidic distance-based readout device. Biosens. Bioelectron. 2016, 85, 496–502. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manzari, M.T.; Shamay, Y.; Kiguchi, H.; Rosen, N.; Scaltriti, M.; Heller, D.A. Targeted drug delivery strategies for precision medicines. Nat. Rev. Mater. 2021, 6, 351–370. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Nandu, N.; Uyar, T.B.; Royzen, M.; Yigit, M.V. Small molecule-induced DNA hydrogel with encapsulation and release properties. Chem Commun 2020, 56, 7313–7316. [Google Scholar] [CrossRef]
- Chyzy, A.; Tomczykowa, M.; Plonska-Brzezinska, M.E. Hydrogels as Potential Nano-, Micro- and Macro-Scale Systems for Controlled Drug Delivery. Materials 2020, 13, 188. [Google Scholar] [CrossRef] [Green Version]
- Song, P.; Ye, D.; Zuo, X.; Li, J.; Wang, J.; Liu, H.; Hwang, M.T.; Chao, J.; Su, S.; Wang, L.; et al. DNA Hydrogel with Aptamer-Toehold-Based Recognition, Cloaking, and Decloaking of Circulating Tumor Cells for Live Cell Analysis. Nano Lett. 2017, 17, 5193–5198. [Google Scholar] [CrossRef]
- Biswal, T. Biopolymers for tissue engineering applications: A review. Mater. Today Proc. 2021, 41, 397–402. [Google Scholar] [CrossRef]
- Morya, V.; Walia, S.; Mandal, B.B.; Ghoroi, C.; Bhatia, D. Functional DNA Based Hydrogels: Development, Properties and Biological Applications. ACS Biomater. Sci. Eng. 2020, 6, 6021–6035. [Google Scholar] [CrossRef] [PubMed]
- Mironov, V.; Boland, T.; Trusk, T.; Forgacs, G.; Markwald, R.R. Organ printing: Computer-aided jet-based 3D tissue engineering. Trends Biotechnol. 2003, 21, 157–161. [Google Scholar] [CrossRef]
- Chen, X.; He, X.; Gao, R.; Lan, X.; Zhu, L.; Chen, K.; Hu, Y.; Huang, K.; Xu, W. Aptamer-Functionalized Binary-Drug Delivery System for Synergetic Obesity Therapy. ACS Nano 2021. [Google Scholar] [CrossRef] [PubMed]

| Stimuli | Responsive Units | Applications | References |
|---|---|---|---|
| pH | i-motif, CD | drug loading and targeted delivery | [18] |
| i-motif, pNIPAM | / | [31] | |
| i-motif, Y-DNA | / | [52] | |
| i-motif, acrylamide | / | [53] | |
| i-motif, PEG | / | [36] | |
| i-motif, G-quadruplex | biosensing | [26] | |
| triplex DNA | / | [45] | |
| triplex DNA, i-motif, acrylamide | drug loading and targeted delivery | [29] | |
| triplex DNA, acrylamide | drug loading and targeted delivery | [54,55,69] | |
| aptamer, i-motif, acrylamide | drug loading and targeted delivery | [27] | |
| Molecular | X-DNA, DNAzyme | biosensing | [70] |
| Y-DNA, aptamer | biosensing | [71] | |
| Y-DNA, aptamer, QDs | drug loading and targeted delivery | [22] | |
| GO, aptamer | drug loading and targeted delivery | [15] | |
| aptamer, acrylamide | biosensing | [57,72,73,74,75] | |
| aptamer, acrylamide | drug loading and targeted delivery | [28,76] | |
| aptamer, PEG | tissue engineering | [34,35] | |
| aptamer, PEG | drug loading and targeted delivery | [33,59] | |
| aptamer, nanomaterials | drug loading and targeted delivery | [38,77] | |
| aptamer, i-motif, acrylamide | drug loading and targeted delivery | [27] | |
| aptamer, G-quadruplex | biosensing | [58] | |
| Ions | DNAzyme, acrylamide | biosensing | [61,78] |
| G-quadruplex, acrylamide, dsDNA | drug loading and targeted delivery | [79] | |
| G-quadruplex, PEG | / | [64] | |
| G-quadruplex, FPBA | drug loading and targeted delivery | [80] | |
| i-motif, pNIPAM | / | [31] | |
| G-quadruplex, i-motif | biosensing | [26] | |
| rich-T DNA, acrylamide | biosensing | [25] | |
| Temperature | dsDNA, carbon nanotube | / | [21] |
| ds-DNA, acrylamide | drug loading and targeted delivery | [30] | |
| i-motif, p-NIPAM | / | [31] | |
| PNA, p-NIPAM | drug loading and targeted delivery | [81] | |
| Y-DNA, dsDNA | / | [65] | |
| AuNS, AuNP, hexapodna | drug loading and targeted delivery | [66] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, J.; Zhang, Y.; Zhu, L.; Chen, K.; Li, X.; Xu, W. Smart Nucleic Acid Hydrogels with High Stimuli-Responsiveness in Biomedical Fields. Int. J. Mol. Sci. 2022, 23, 1068. https://doi.org/10.3390/ijms23031068
Li J, Zhang Y, Zhu L, Chen K, Li X, Xu W. Smart Nucleic Acid Hydrogels with High Stimuli-Responsiveness in Biomedical Fields. International Journal of Molecular Sciences. 2022; 23(3):1068. https://doi.org/10.3390/ijms23031068
Chicago/Turabian StyleLi, Jie, Yangzi Zhang, Longjiao Zhu, Keren Chen, Xiangyang Li, and Wentao Xu. 2022. "Smart Nucleic Acid Hydrogels with High Stimuli-Responsiveness in Biomedical Fields" International Journal of Molecular Sciences 23, no. 3: 1068. https://doi.org/10.3390/ijms23031068
APA StyleLi, J., Zhang, Y., Zhu, L., Chen, K., Li, X., & Xu, W. (2022). Smart Nucleic Acid Hydrogels with High Stimuli-Responsiveness in Biomedical Fields. International Journal of Molecular Sciences, 23(3), 1068. https://doi.org/10.3390/ijms23031068








