Protein-Based Oncopanel as Addition to Target Sequencing in Head and Neck Squamous Cell Carcinoma to Individualize Treatment Decisions
Abstract
1. Introduction
2. Results
2.1. Patient Cohort
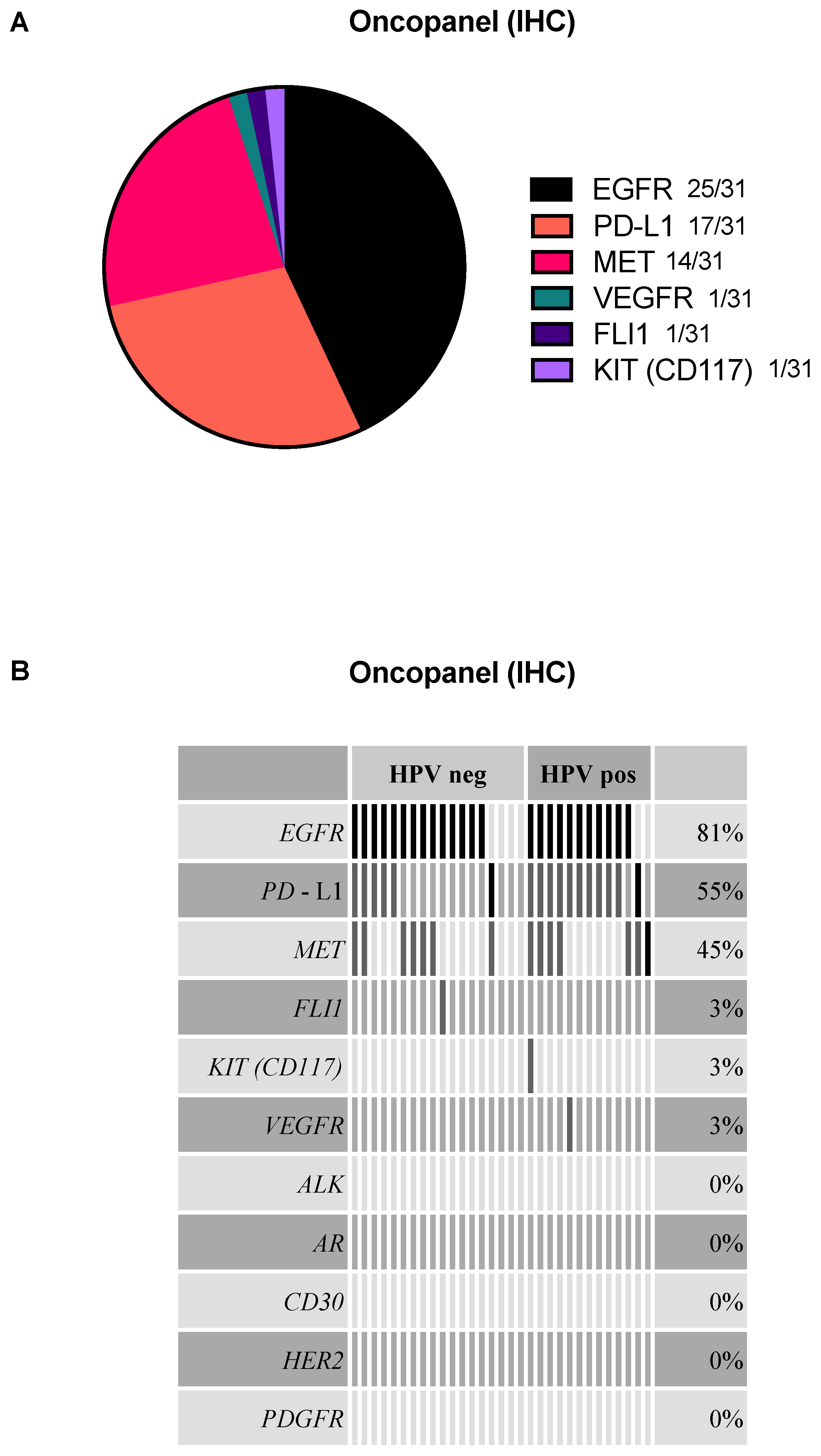
2.2. EGFR, PD-L1, and MET Are the Most Frequently Detected Targets in the Oncopanel
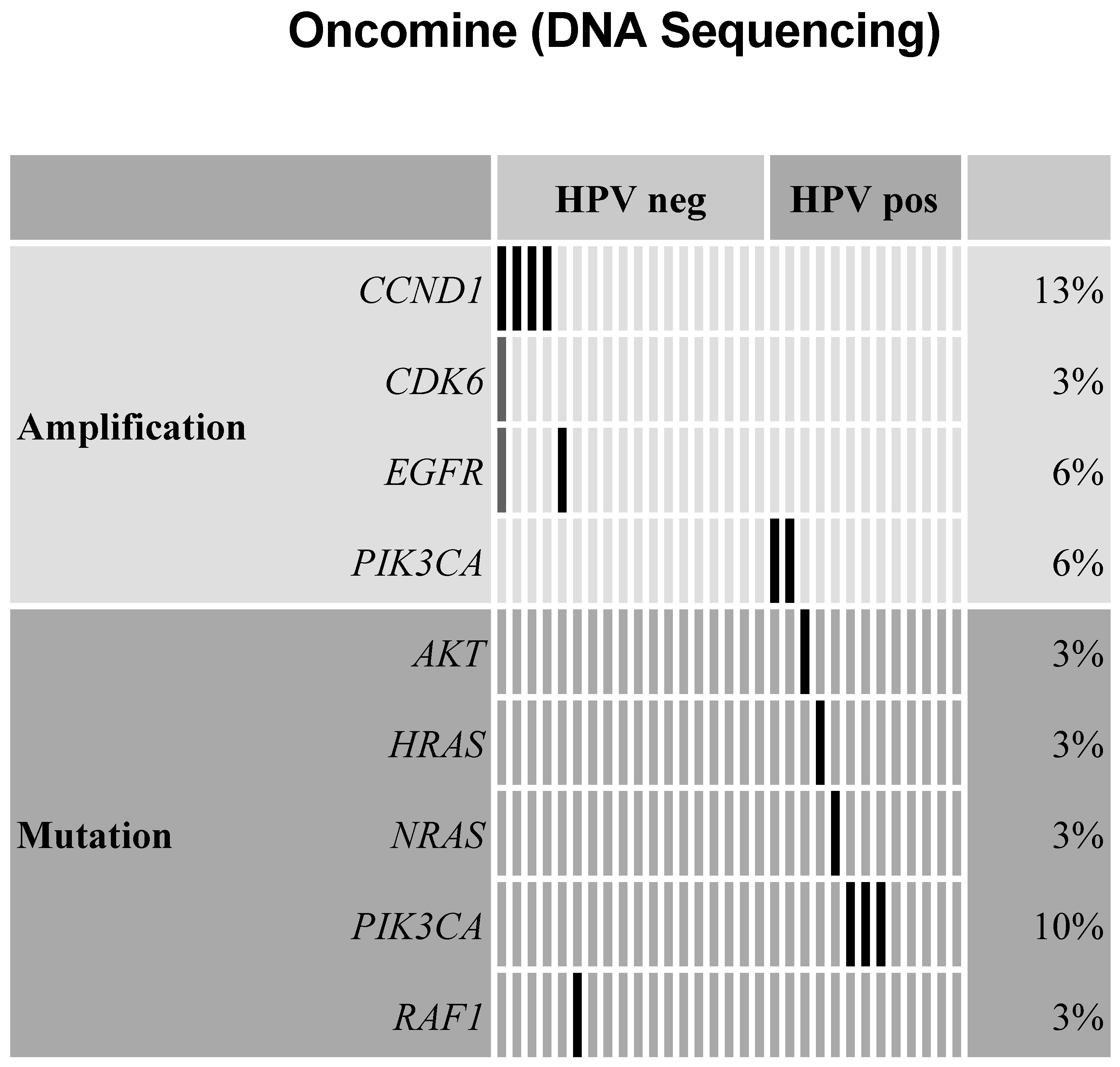
2.3. Amplifications Found for Cell Cycle Genes, EGFR, and PI3-Kinase in Oncomine Targeted Sequencing
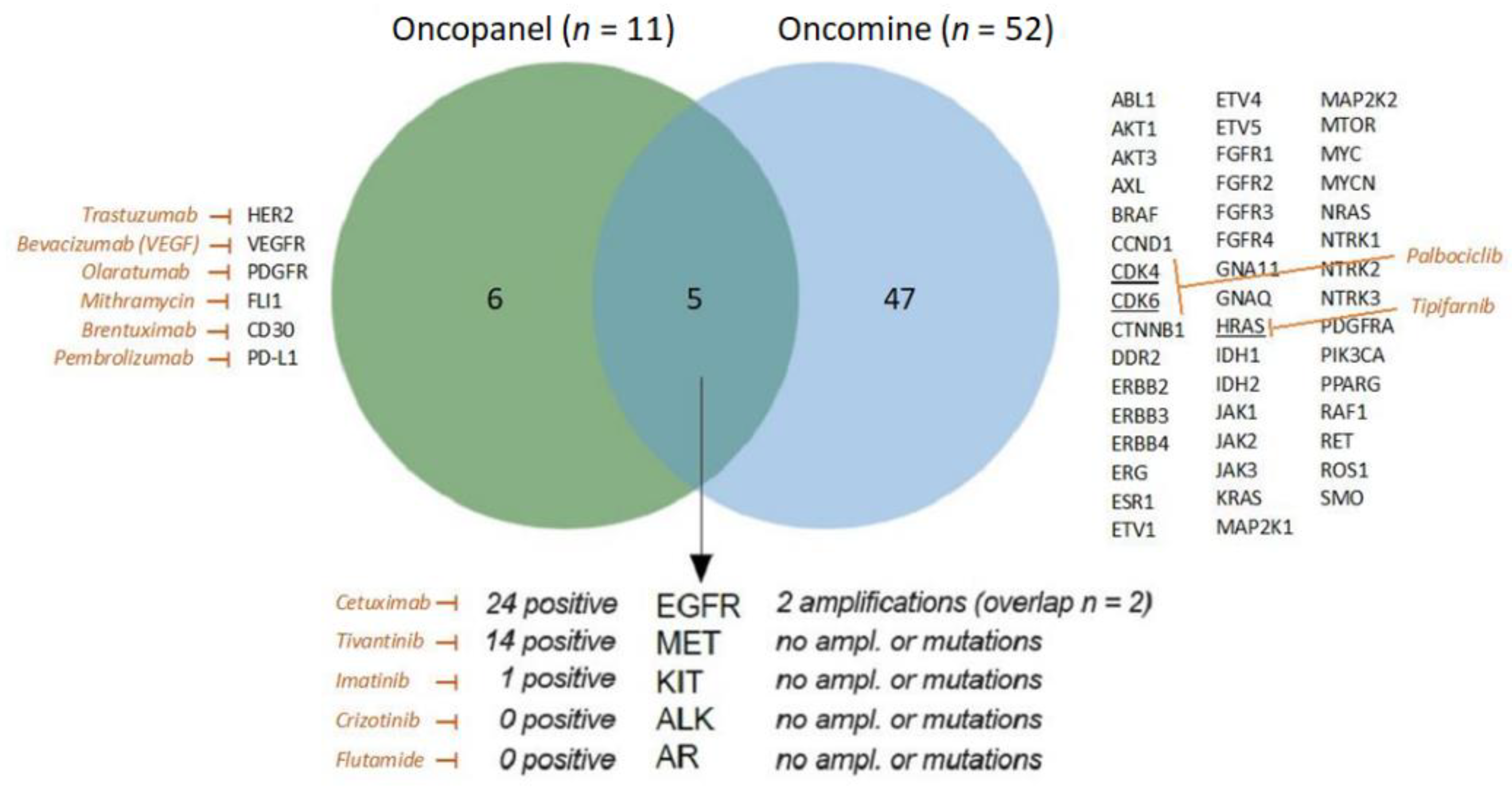
2.4. Overlap of Oncopanel and Oncomine
2.5. HPV-Positive and HPV-Negative HNSCC Analyzed by Oncopanel and Oncomine
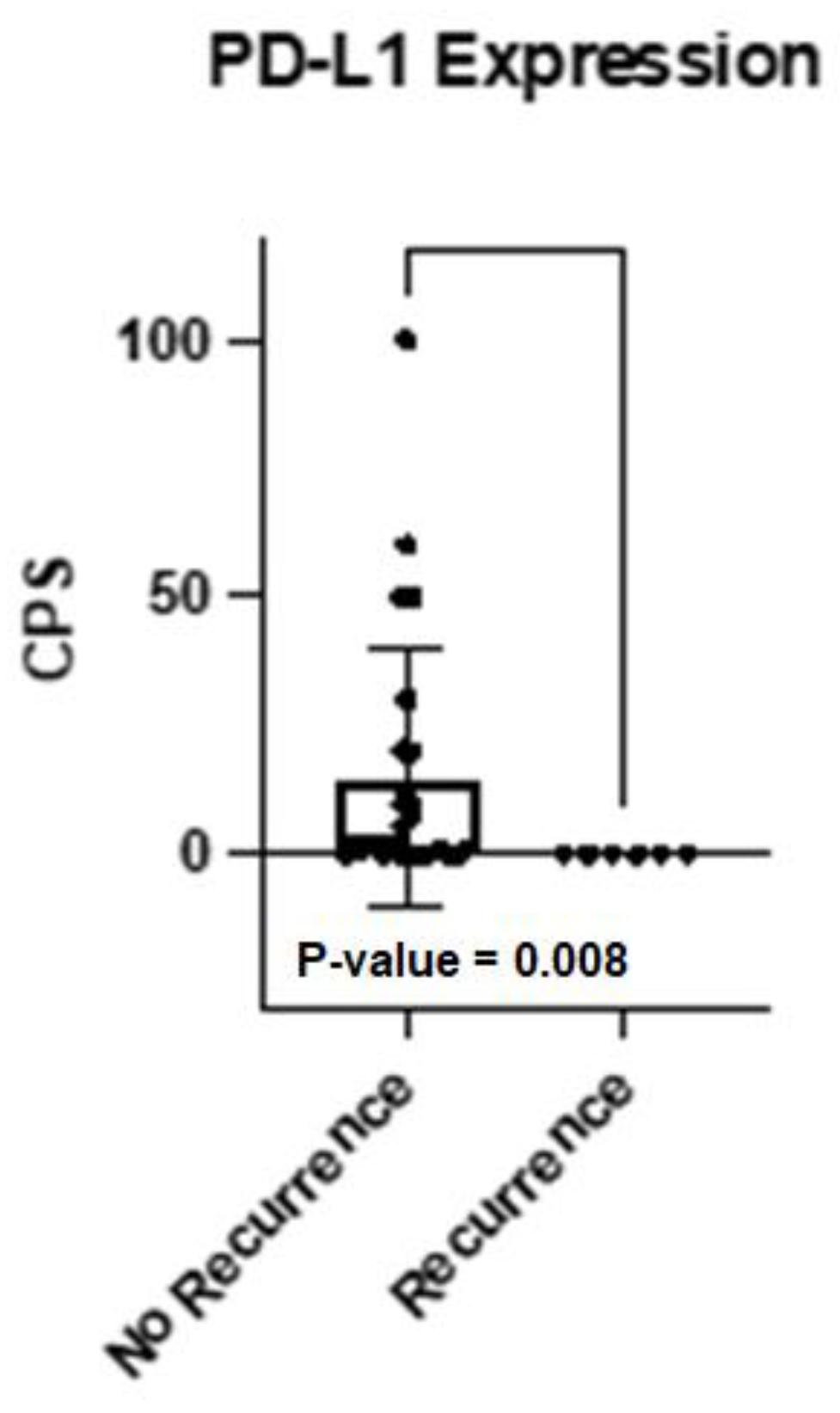
2.6. HNSCC Recurrences Analyzed by Oncopanel and Oncomine
2.7. Characteristics of Recurrent and Deceased Patients
3. Discussion
4. Materials and Methods
4.1. Study Design and Patient Cohort
4.2. Oncopanel
4.3. DNA Extraction
4.4. Next Generation Sequencing (NGS)
4.5. Data Analysis and Statistics
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- D’Souza, G.; Dempsey, A. The role of HPV in head and neck cancer and review of the HPV vaccine. Prev. Med. 2011, 53 (Suppl. 1), S5–S11. [Google Scholar] [CrossRef] [PubMed]
- Sethi, S.; Ali-Fehmi, R.; Franceschi, S.; Struijk, L.; van Doorn, L.J.; Quint, W.; Albashiti, B.; Ibrahim, M.; Kato, I. Characteristics and survival of head and neck cancer by HPV status: A cancer registry-based study. Int. J. Cancer 2012, 131, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- National Comprehensive Cancer Network (NCCN) Guidelines: Head and Neck Cancers. Version 1. Available online: https://www.nccn.org/professionals/physician_gls/pdf/head-and-neck.pdf (accessed on 1 February 2020).
- Burtness, B.; Harrington, K.J.; Greil, R.; Soulieres, D.; Tahara, M.; de Castro, G., Jr.; Psyrri, A.; Basté, N.; Neupane, P.; Bratland, Å.; et al. Pembrolizumab alone or with chemotherapy versus cetuximab with chemotherapy for recurrent or metastatic squamous cell carcinoma of the head and neck (KEYNOTE-048): A randomised, open-label, phase 3 study. Lancet 2019, 394, 1915–1928. [Google Scholar] [CrossRef]
- Cohen, E.E.W.; Soulieres, D.; Le Tourneau, C.; Dinis, J.; Licitra, L.; Ahn, M.J.; Soria, A.; Machiels, J.P.; Mach, N.; Mehra, R.; et al. Pembrolizumab versus methotrexate, docetaxel, or cetuximab for recurrent or metastatic head-and-neck squamous cell carcinoma (KEYNOTE-040): A randomised, open-label, phase 3 study. Lancet 2019, 393, 156–167. [Google Scholar] [CrossRef] [PubMed]
- Ferris, R.L.; Blumenschein, G., Jr.; Fayette, J.; Guigay, J.; Colevas, A.D.; Licitra, L.; Harrington, K.; Kasper, S.; Vokes, E.E.; Even, C.; et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N. Engl. J. Med. 2016, 375, 1856–1867. [Google Scholar] [CrossRef] [PubMed]
- Doescher, J.; Busch, C.J.; Wollenberg, B.; Dietz, A.; Wurdemann, N.; Schuler, P.; Hoffmann, T.K.; Laban, S. Immunotherapy for head and neck cancer: Highlights of the 2019 ASCO Annual Meeting. HNO 2019, 67, 905–911. [Google Scholar] [CrossRef]
- Doescher, J.; Laban, S.; Schuler, P.J.; Brunner, C.; Hoffmann, T.K. Immunotherapy for head and neck cancers: An update and future perspectives. Immunotherapy 2019, 11, 561–564. [Google Scholar] [CrossRef]
- Manukian, G.; Bar-Ad, V.; Lu, B.; Argiris, A.; Johnson, J.M. Combining Radiation and Immune Checkpoint Blockade in the Treatment of Head and Neck Squamous Cell Carcinoma. Front. Oncol. 2019, 9, 122. [Google Scholar] [CrossRef]
- Guigay, J.; Auperin, A.; Fayette, J.; Saada-Bouzid, E.; Lafond, C.; Taberna, M.; Geoffrois, L.; Martin, L.; Capitain, O.; Cupissol, D.; et al. Cetuximab, docetaxel, and cisplatin versus platinum, fluorouracil, and cetuximab as first-line treatment in patients with recurrent or metastatic head and neck squamous-cell carcinoma (GORTEC 2014-01 TPExtreme): A multicentre, open-label, randomised, phase 2 trial. Lancet Oncol. 2021, 22, 463–475. [Google Scholar]
- Herbst, R.S.; Arquette, M.; Shin, D.M.; Dicke, K.; Vokes, E.E.; Azarnia, N.; Hong, W.K.; Kies, M.S. Phase II multicenter study of the epidermal growth factor receptor antibody cetuximab and cisplatin for recurrent and refractory squamous cell carcinoma of the head and neck. J. Clin. Oncol. 2005, 23, 5578–5587. [Google Scholar] [CrossRef]
- Chan, A.T.; Hsu, M.M.; Goh, B.C.; Hui, E.P.; Liu, T.W.; Millward, M.J.; Hong, R.L.; Whang-Peng, J.; Ma, B.B.; To, K.F.; et al. Multicenter, phase II study of cetuximab in combination with carboplatin in patients with recurrent or metastatic nasopharyngeal carcinoma. J. Clin. Oncol. 2005, 23, 3568–3576. [Google Scholar] [CrossRef] [PubMed]
- Grunmuller, L.; Thierauf, J.; Weissinger, S.E.; Bergmann, C.; Bankfalvi, A.; Veit, J.; Hoffmann, T.K.; Möller, P.; Lennerz, J.K. Biopanel identifies expression status of targetable proteins in sinonasal melanoma. Pers. Med. 2016, 13, 291–301. [Google Scholar] [CrossRef]
- Doescher, J.; Weissinger, S.E.; Schonsteiner, S.S.; Lisson, C.; Bullinger, L.; Barth, T.F.; Leithäuser, F.; Mueller-Richter, U.; Laban, S.; Hoffmann, T.K.; et al. Clinical utility of a protein-based oncopanel in patients with end-stage head and neck cancer. Immunotherapy 2019, 11, 1193–1203. [Google Scholar] [CrossRef]
- Guo, Y.; Xiao, P.; Lei, S.; Deng, F.; Xiao, G.G.; Liu, Y.; Chen, X.; Li, L.; Wu, S.; Chen, Y.; et al. How is mRNA expression predictive for protein expression? A correlation study on human circulating monocytes. Acta Biochim. Biophys. Sin. 2008, 40, 426–436. [Google Scholar] [CrossRef]
- de Abreu, R.S.; Penalva, L.O.; Marcotte, E.M.; Vogel, C. Global signatures of protein and mRNA expression levels. Mol. BioSyst. 2009, 5, 1512–1526. [Google Scholar] [CrossRef]
- Sacco, A.G.; Chen, R.; Worden, F.P.; Wong, D.J.L.; Adkins, D.; Swiecicki, P.; Chai-Ho, W.; Oppelt, P.; Ghosh, D.; Bykowski, J.; et al. Pembrolizumab plus cetuximab in patients with recurrent or metastatic head and neck squamous cell carcinoma: An open-label, multi-arm, non-randomised, multicentre, phase 2 trial. Lancet Oncol. 2021, 22, 883–892. [Google Scholar] [CrossRef]
- Doescher, J.; Minkenberg, P.; Laban, S.; Kostezka, U.; von Witzleben, A.; Hoffmann, T.K.; Schuler, P.J.; Weissinger, S.E. Immune checkpoint expression in HNSCC patients before and after definitive chemoradiotherapy. Head Neck 2021, 43, 778–787. [Google Scholar] [CrossRef]
- Baldin, V.; Lukas, J.; Marcote, M.J.; Pagano, M.; Draetta, G. Cyclin D1 is a nuclear protein required for cell cycle progression in G1. Genes Dev. 1993, 7, 812–821. [Google Scholar] [CrossRef]
- Ku, B.M.; Yi, S.Y.; Koh, J.; Bae, Y.H.; Sun, J.M.; Lee, S.H.; Ahn, J.S.; Park, K.; Ahn, M.J. The CDK4/6 inhibitor LY2835219 has potent activity in combination with mTOR inhibitor in head and neck squamous cell carcinoma. Oncotarget 2016, 7, 14803–14813. [Google Scholar] [CrossRef]
- Michel, L.; Ley, J.; Wildes, T.M.; Schaffer, A.; Robinson, A.; Chun, S.E.; Lee, W.; Lewis, J., Jr.; Trinkaus, K.; Adkins, D. Phase I trial of palbociclib, a selective cyclin dependent kinase 4/6 inhibitor, in combination with cetuximab in patients with recurrent/metastatic head and neck squamous cell carcinoma. Oral Oncol. 2016, 58, 41–48. [Google Scholar] [CrossRef]
- Ho, A.L.; Brana, I.; Haddad, R.; Bauman, J.; Bible, K.; Oosting, S.; Wong, D.J.; Ahn, M.J.; Boni, V.; Even, C.; et al. Tipifarnib in Head and Neck Squamous Cell Carcinoma with HRAS Mutations. J. Clin. Oncol. 2021, 39, 1856–1864. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef]
- Volkel, G.; Laban, S.; Furstberger, A.; Kuhlwein, S.D.; Ikonomi, N.; Hoffman, T.K.; Brunner, C.; Neuberg, D.S.; Gaidzik, V.; Döhner, H.; et al. Analysis, identification and visualization of subgroups in genomics. Brief. Bioinform. 2021, 22, bbaa217. [Google Scholar] [CrossRef]
- Bardou, P.; Mariette, J.; Escudie, F.; Djemiel, C.; Klopp, C. Jvenn: An interactive Venn diagram viewer. BMC Bioinform. 2014, 15, 293. [Google Scholar] [CrossRef]
| PatID | Gen-der | Age | Life Status | OS | Recurrence | PFS | HPV Status | Localization | T-Status | N-Status | M-Status | Therapy |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | m | 52 | alive | 35 | no | negative | Oropharynx | 3 | 1 | 0 | SRG+RT | |
| 2 | m | 47 | alive | 35 | no | negative | Oropharynx | 3 | 1 | 0 | SRG+RT | |
| 3 | m | 65 | alive | 35 | no | positive | Oropharynx | 1 | 1 | 0 | SRG+CRT | |
| 4 | m | 61 | alive | 26 | no | positive | Oropharynx | 2 | 2b | 0 | SRG+CRT | |
| 5 | m | 71 | alive | 32 | yes | 16 | positive | Oropharynx | 4 | 2c | 0 | CRT |
| 6 | m | 50 | alive | 35 | no | negative | Oropharynx | 2 | 0 | 0 | SRG | |
| 7 | f | 59 | alive | 33 | yes | 34 | negative | Oropharynx | 2 | 0 | 0 | SRG |
| 8 | m | 69 | alive | 34 | no | negative | Oropharynx | 2 | 3b | 0 | SRG+CRT | |
| 9 | m | 71 | alive | 22 | no | negative | Oropharynx | 2 | 2b | 0 | SRG+RT | |
| 10 | f | 54 | dead | 10 | yes | 8 | negative | Oropharynx | 2 | 0 | 0 | SRG+RT |
| 11 | m | 68 | alive | 32 | no | positive | Oropharynx | 2 | 2c | 0 | SRG+RT | |
| 12 | m | 64 | alive | 29 | no | positive | Oropharynx | 2 | 0 | 0 | SRG | |
| 13 | m | 54 | alive | 29 | no | negative | Oropharynx | 4 | 2c | 0 | SRG+CRT | |
| 14 | m | 58 | alive | 26 | yes | 23 | negative | Oropharynx | 3 | 0 | 0 | CRT |
| 15 | m | 56 | alive | 28 | no | positive | Oropharynx | 2 | 1 | 0 | SRG+CRT | |
| 16 | f | 56 | alive | 27 | yes | 5 | negative | Oropharynx | 4 | 2a | 0 | SRG |
| 17 | f | 38 | alive | 25 | no | na | Oral Cavity | 2 | 0 | 0 | SRG+RT | |
| 18 | m | 69 | alive | 19 | no | na | Oral Cavity | 3 | 1 | 0 | SRG | |
| 19 | f | 66 | alive | 21 | no | na | Oral Cavity | 3 | 1 | 0 | SRG+RT | |
| 20 | f | 69 | alive | 19 | no | na | Oral Cavity | 2 | 0 | 0 | SRG | |
| 21 | m | 64 | alive | 15 | no | positive | Oropharynx | 3 | 2c | 0 | CRT | |
| 22 | m | 64 | alive | 14 | no | positive | Oropharynx | 2 | 1 | 0 | SRG+RT | |
| 23 | m | 83 | alive | 14 | no | positive | Oropharynx | 2 | 1 | 0 | SRG+RT | |
| 24 | m | 76 | alive | 16 | no | positive | Oropharynx | 2 | 0 | 0 | SRG+RT | |
| 25 | m | 68 | alive | 13 | no | negative | Oropharynx | 2 | 2b | 0 | SRG | |
| 26 | f | 64 | alive | 1 | no | positive | Oropharynx | 4 | 2a | 0 | SRG+CRT | |
| 27 | f | 57 | alive | 12 | no | positive | Oropharynx | 2 | 0 | 0 | SRG | |
| 28 | m | 76 | alive | 13 | yes | 6 | negative | Oropharynx | 4 | 2b | 0 | SRG+RT |
| 29 | m | 67 | alive | 13 | no | positive | Oropharynx | 1 | 2a | 0 | SRG+RT | |
| 30 | m | 63 | alive | 6 | no | na | Oral Cavity | 1 | 0 | 0 | SRG+RT | |
| 31 | m | 64 | alive | 6 | no | na | Oral Cavity | 2 | 0 | 0 | SRG |
| n | % | ||
|---|---|---|---|
| Primary Site | Oropharynx | 25 | 81% |
| Oral Cavity | 6 | 19% | |
| Total | 31 | ||
| Sex | Male | 23 | 74% |
| Female | 8 | 26% | |
| Age | mean (range) | 62.6 (37.9–83.0) | |
| Tumor Status | T1 | 3 | 10% |
| T2 | 17 | 55% | |
| T3 | 6 | 19% | |
| T4 | 5 | 16% | |
| Nodal Status | N- | 11 | 35% |
| N+ | 20 | 65% | |
| Distant Metastases | M0 | 31 | 100% |
| HPV-status (DNA) | negative | 12 | |
| positive | 13 | ||
| missing (not OPSCC) | 6 | ||
| Treatment | CRT only | 3 (2 *) | |
| Surgery only | 9 | ||
| Surgery+RT | 6 | ||
| Surgery+CRT | 13 | ||
| Primary | Recurrence | |
|---|---|---|
| Oncopanel (IHC) | ||
| Patient 7 | no target | no target |
| Patient 10 | EGFR+ | EGFR+/PD-L1 |
| Patient 16 | EGFR+ | EGFR+/PD-L1 |
| Oncomine (amplification) | ||
| Patient 7 | no target | no target |
| Patient 10 | no target | CCND1 |
| Patient 16 | CCND1 | CCND1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
von Witzleben, A.; Müller-Richter, U.; Maurus, K.; Brändlein, S.; Theodoraki, M.-N.; Brunner, C.; Laban, S.; Lennerz, J.; Möller, P.; Hoffmann, T.K.; et al. Protein-Based Oncopanel as Addition to Target Sequencing in Head and Neck Squamous Cell Carcinoma to Individualize Treatment Decisions. Int. J. Mol. Sci. 2022, 23, 15835. https://doi.org/10.3390/ijms232415835
von Witzleben A, Müller-Richter U, Maurus K, Brändlein S, Theodoraki M-N, Brunner C, Laban S, Lennerz J, Möller P, Hoffmann TK, et al. Protein-Based Oncopanel as Addition to Target Sequencing in Head and Neck Squamous Cell Carcinoma to Individualize Treatment Decisions. International Journal of Molecular Sciences. 2022; 23(24):15835. https://doi.org/10.3390/ijms232415835
Chicago/Turabian Stylevon Witzleben, Adrian, Urs Müller-Richter, Katja Maurus, Stephanie Brändlein, Marie-Nicole Theodoraki, Cornelia Brunner, Simon Laban, Jochen Lennerz, Peter Möller, Thomas K. Hoffmann, and et al. 2022. "Protein-Based Oncopanel as Addition to Target Sequencing in Head and Neck Squamous Cell Carcinoma to Individualize Treatment Decisions" International Journal of Molecular Sciences 23, no. 24: 15835. https://doi.org/10.3390/ijms232415835
APA Stylevon Witzleben, A., Müller-Richter, U., Maurus, K., Brändlein, S., Theodoraki, M.-N., Brunner, C., Laban, S., Lennerz, J., Möller, P., Hoffmann, T. K., Doescher, J., & Schuler, P. J. (2022). Protein-Based Oncopanel as Addition to Target Sequencing in Head and Neck Squamous Cell Carcinoma to Individualize Treatment Decisions. International Journal of Molecular Sciences, 23(24), 15835. https://doi.org/10.3390/ijms232415835







