A Cross-Sectional Study on the Occurrence of the Intestinal Protist, Dientamoeba fragilis, in the Gut-Healthy Volunteers and Their Animals
Abstract
:1. Introduction
2. Results
2.1. Prevalence of Dientamoeba fragilis Based on Two Molecular Approaches
2.2. Comparison of the Sensitivity of cPCR and qPCR for Dientamoeba fragilis Detection
2.3. Influence of the Specific Factors on the Occurrence of Dientamoeba fragilis
2.3.1. Lifestyle
2.3.2. Contact with Animals
2.3.3. Age and Gender
2.4. Occurrence of Dientamoeba fragilis in the Family Environment
2.5. Coinfection of Dientamoeba fragilis with Blastocystis spp.
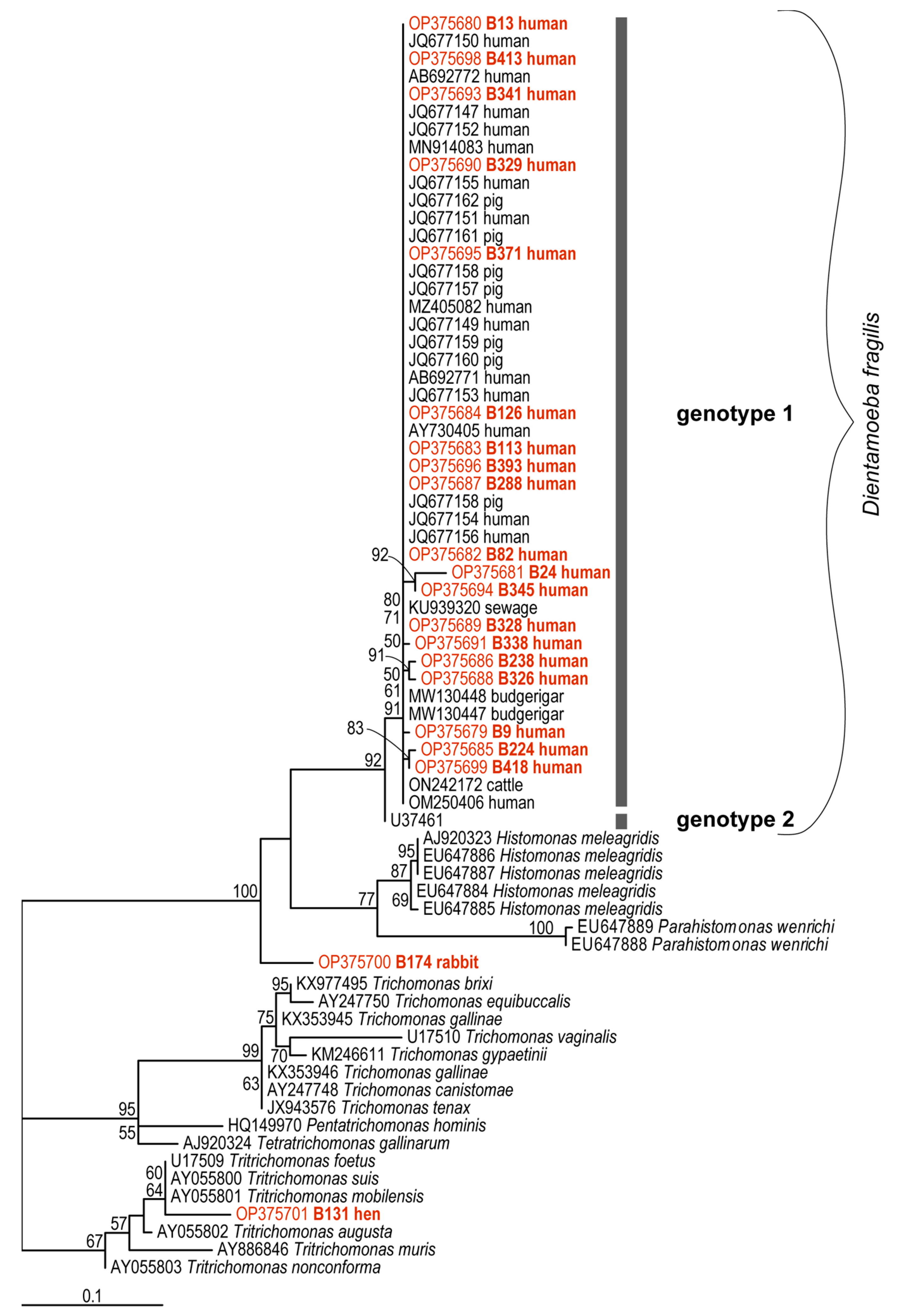
2.6. Genetic Diversity of Dientamoeba fragilis Based on Molecular Phylogenies
3. Discussion
4. Materials and Methods
4.1. Sample Collection and Ethical Approval
4.2. DNA Extraction and Samples Processing
4.3. Conventional PCR
4.4. Real-Time Diagnostics
4.5. Cloning and Sequencing of Positive Samples
4.6. Phylogenetic Analysis
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cacciò, S.M. Molecular Epidemiology of Dientamoeba fragilis. Acta Trop. 2018, 184, 73–77. [Google Scholar] [CrossRef] [PubMed]
- Stark, D.; Barratt, J.; Chan, D.; Ellis, J.T. Dientamoeba fragilis, the Neglected Trichomonad of the Human Bowel. Clin. Microbiol. Rev. 2016, 29, 553–580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Calderaro, A.; Buttrini, M.; Montecchini, S.; Rossi, S.; Farina, B.; Arcangeletti, M.C.; De Conto, F.; Chezzi, C. Prevalence of Intestinal Parasitoses in a Non-Endemic Setting during a 10-Year Period (2011–2020): A Focus on Dientamoeba fragilis. Microorganisms 2022, 10, 426. [Google Scholar] [CrossRef] [PubMed]
- Barratt, J.L.N.; Harkness, J.; Marriott, D.; Ellis, J.T.; Stark, D. A Review of Dientamoeba fragilis Carriage in Humans: Several Reasons Why This Organism Should Be Considered in the Diagnosis of Gastrointestinal Illness. Gut Microbes 2011, 2, 3–12. [Google Scholar] [CrossRef] [Green Version]
- Johnson, E.H.; Windsor, J.J.; Clark, C.G. Emerging from Obscurity: Biological, Clinical, and Diagnostic Aspects of Dientamoeba fragilis. Clin. Microbiol. Rev. 2004, 17, 553–570. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ögren, J.; Dienus, O.; Löfgren, S.; Iveroth, P.; Matussek, A. Dientamoeba fragilis DNA Detection in Enterobius Vermicularis Eggs. Pathog. Dis. 2013, 69, 157–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Röser, D.; Nejsum, P.; Carlsgart, A.J.; Nielsen, H.V.; Stensvold, C.R. DNA of Dientamoeba fragilis Detected within Surface-Sterilized Eggs of Enterobius vermicularis. Exp. Parasitol. 2013, 133, 57–61. [Google Scholar] [CrossRef] [PubMed]
- Ockert, G. Symptomatology, Pathology, Epidemiology, and Diagnosis of Dientamoeba fragilis. In Trichomonads Parasitic in Humans; Honigberg, B.M., Ed.; Springer: New York, NY, USA, 1990; pp. 394–410. ISBN 978-1-4612-3224-7. [Google Scholar]
- Girginkardeşler, N.; Kurt, Ö.; Kilimcioǧlu, A.A.; Ok, Ü.Z. Transmission of Dientamoeba fragilis: Evaluation of the Role of Enterobius Vermicularis. Parasitol. Int. 2008, 57, 72–75. [Google Scholar] [CrossRef] [PubMed]
- Ögren, J.; Dienus, O.; Löfgren, S.; Einemo, I.M.; Iveroth, P.; Matussek, A. Dientamoeba fragilis Prevalence Coincides with Gastrointestinal Symptoms in Children Less than 11 Years Old in Sweden. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1995–1998. [Google Scholar] [CrossRef] [Green Version]
- Yıldız, İ.; Ertuğ, S.; Tileklioğlu, E.; Malatyalı, E.; Güçlü, Ö.; Ertabaklar, H. Investigation of Dientamoeba fragilis Frequency in Faecal Samples of Patients with Enterobius vermicularis Infection by Polymerase Chain Reaction. Turk. Parazitolojii Derg. 2021, 45, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Munasinghe, V.S.; Vella, N.G.F.; Ellis, J.T.; Windsor, P.A.; Stark, D. Cyst Formation and Faecal-Oral Transmission of Dientamoeba fragilis—The Missing Link in the Life Cycle of an Emerging Pathogen. Int. J. Parasitol. 2013, 43, 879–883. [Google Scholar] [CrossRef] [PubMed]
- Stark, D.; Garcia, L.S.; Barratt, J.L.N.; Phillips, O.; Roberts, T.; Marriott, D.; Harkness, J.; Ellis, J.T. Description of Dientamoeba fragilis Cyst and Precystic Forms from Human Samples. J. Clin. Microbiol. 2014, 52, 2680–2683. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clark, C.G.; Röser, D.; Stensvold, C.R. Transmission of Dientamoeba fragilis: Pinworm or Cysts? Trends Parasitol. 2014, 30, 136–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Norberg, A.; Nord, C.E.; Evengård, B. Dientamoeba fragilis—A Protozoal Infection Which May Cause Severe Bowel Distress. Clin. Microbiol. Infect. 2003, 9, 65–68. [Google Scholar] [CrossRef] [Green Version]
- Stark, D.; Beebe, N.; Marriott, D.; Ellis, J.; Harkness, J. Prospective Study of the Prevalence, Genotyping, and Clinical Relevance of Dientamoeba fragilis Infections in an Australian Population. J. Clin. Microbiol. 2005, 43, 2718–2723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aykur, M.; Calıskan Kurt, C.; Dirim Erdogan, D.; Biray Avcı, C.; Vardar, R.; Aydemir, S.; Girginkardeşler, N.; Gündüz, C.; Dagci, H. Investigation of Dientamoeba fragilis Prevalence and Evaluation of Sociodemographic and Clinical Features in Patients with Gastrointestinal Symptoms. Acta Parasitol. 2019, 64, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. Prevalence, Incidence, and Risk Factors of Intestinal Parasites in Danish Primary Care Patients with Irritable Bowel Syndrome. Scand. J. Infect. Dis. 2014, 46, 204–209. [Google Scholar] [CrossRef] [PubMed]
- Stark, D.; van Hal, S.; Marriott, D.; Ellis, J.; Harkness, J. Irritable Bowel Syndrome: A Review on the Role of Intestinal Protozoa and the Importance of Their Detection and Diagnosis. Int. J. Parasitol. 2007, 37, 11–20. [Google Scholar] [CrossRef]
- Yakoob, J.; Jafri, W.; Beg, A.; Abbas, Z.; Naz, S.; Islam, M.; Khan, R. Blastocystis hominis and Dientamoeba fragilis in Patients Fulfilling Irritable Bowel Syndrome Criteria. Parasitol. Res. 2010, 107, 679–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Piubelli, C.; Soleymanpoor, H.; Giorli, G.; Formenti, F.; Buonfrate, D.; Bisoffi, Z.; Perandin, F. Blastocystis Prevalence and Subtypes in Autochthonous and Immigrant Patients in a Referral Centre for Parasitic Infections in Italy. PLoS ONE 2019, 14, e0210171. [Google Scholar] [CrossRef] [PubMed]
- Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. Treatment of Dientamoeba fragilis in Patients with Irritable Bowel Syndrome. Am. J. Trop. Med. Hyg. 2012, 87, 1046–1052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Röser, D.; Simonsen, J.; Stensvold, C.R.; Olsen, K.E.P.; Bytzer, P.; Nielsen, H.V.; Mølbak, K. Metronidazole Therapy for Treating Dientamoebiasis in Children Is Not Associated with Better Clinical Outcomes: A Randomized, Double-Blinded and Placebo-Controlled Clinical Trial. Clin. Infect. Dis. Off. Publ. Infect. Dis. Soc. Am. 2014, 58, 1692–1699. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unalan-Altintop, T.; Vahabov, C.; Ergunay, K.; Kurt, O.; Kav, T.; Akyon, Y.; Erguven, S. Investigation of Dientamoeba fragilis and Blastocystis in Patients from Turkey with Ulcerative Colitis and Irritable Bowel Syndrome: Any Relation with Genotypes? Acta Trop. 2022, 231, 106451. [Google Scholar] [CrossRef] [PubMed]
- Brands, M.R.; Van de Vijver, E.; Haisma, S.M.; Heida, A.; van Rheenen, P.F. No Association between Abdominal Pain and Dientamoeba in Dutch and Belgian Children. Arch. Dis. Child. 2019, 104, 686–689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Boer, M.D.; Schuurs, T.A.; Vermeer, M.; Ruijs, G.J.H.M.; van der Zanden, A.G.M.; Weel, J.F.; Bruijnesteijn van Coppenraet, L.E.S. Distribution and Relevance of Dientamoeba fragilis and Blastocystis Species in Gastroenteritis: Results from a Case-Control Study. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 197–203. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.J.; Korterink, J.J.; Benninga, M.A.; Hilbink, M.; Widdershoven, J.; Deckers-Kocken, J.M. Dientamoeba fragilis and Chronic Abdominal Pain in Children: A Case-Control Study. Archives of Disease in Childhood: Educ. Pract. Ed. 2014, 99, 1109–1113. [Google Scholar] [CrossRef] [PubMed]
- Holtman, G.A.; Kranenberg, J.J.; Blanker, M.H.; Ott, A.; Lisman-van Leeuwen, Y.; Berger, M.Y. Dientamoeba fragilis Colonization Is Not Associated with Gastrointestinal Symptoms in Children at Primary Care Level. Fam. Pract. 2017, 34, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jokelainen, P.; Jensen, B.H.; Andreassen, B.U.; Petersen, A.M.; Röser, D.; Krogfelt, K.A.; Nielsen, H.V.; Stensvold, C.R. Dientamoeba fragilis, a Commensal in Children in Danish Day Care Centers. J. Clin. Microbiol. 2017, 55, 1707–1713. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krogsgaard, L.R.; Engsbro, A.L.; Stensvold, C.R.; Nielsen, H.V.; Bytzer, P. The Prevalence of Intestinal Parasites Is Not Greater among Individuals with Irritable Bowel Syndrome: A Population-Based Case-Control Study. Clin. Gastroenterol. Hepatol. 2015, 13, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Stensvold, C.R.; van der Giezen, M. Associations between Gut Microbiota and Common Luminal Intestinal Parasites. Trends Parasitol. 2018, 34, 369–377. [Google Scholar] [CrossRef]
- Cacciò, S.M.; Sannella, A.R.; Manuali, E.; Tosini, F.; Sensi, M.; Crotti, D.; Pozio, E. Pigs as Natural Hosts of Dientamoeba fragilis Genotypes Found in Humans. Emerg. Infect. Dis. 2012, 18, 838–841. [Google Scholar] [CrossRef] [PubMed]
- Crotti, D.; Sensi, M.; Crotti, S.; Grelloni, V.; Manuali, E. Dientamoeba fragilis in Swine Population: A Preliminary Investigation. Vet. Parasitol. 2007, 145, 349–351. [Google Scholar] [CrossRef] [PubMed]
- Stark, D.; Phillips, O.; Peckett, D.; Munro, U.; Marriott, D.; Harkness, J.; Ellis, J. Gorillas Are a Host for Dientamoeba fragilis: An Update on the Life Cycle and Host Distribution. Vet. Parasitol. 2008, 151, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Lankester, F.; Kiyang, J.A.; Bailey, W.; Unwin, S. Dientamoeba fragilis: Initial Evidence of Pathogenicity in the Western Lowland Gorilla (Gorilla Gorilla Gorilla). J. Zoo Wildl. Med. 2010, 41, 350–352. [Google Scholar] [CrossRef] [PubMed]
- Helenbrook, W.D.; Wade, S.E.; Shields, W.M.; Stehman, S.V.; Whipps, C.M. Gastrointestinal Parasites of Ecuadorian Mantled Howler Monkeys (Alouatta Palliata Aequatorialis) Based on Fecal Analysis. J. Parasitol. 2015, 101, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Menu, E.; Davoust, B.; Mediannikov, O.; Akiana, J.; Mulot, B.; Diatta, G.; Levasseur, A.; Ranque, S.; Raoult, D.; Bittar, F. Occurrence of Ten Protozoan Enteric Pathogens in Three Non-Human Primate Populations. Pathogens 2021, 10, 280. [Google Scholar] [CrossRef] [PubMed]
- Chan, D.; Barratt, J.; Roberts, T.; Phillips, O.; Šlapeta, J.; Ryan, U.; Marriott, D.; Harkness, J.; Ellis, J.; Stark, D. Detection of Dientamoeba fragilis in Animal Faeces Using Species-Specific Real-Time PCR Assay. Vet. Parasitol. 2016, 227, 42–47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogunniyi, T.; Balogun, H.; Shasanya, B. Ectoparasites and Endoparasites of Peridomestic House-Rats in Ile-Ife, Nigeria and Implication on Human Health. Iran. J. Parasitol. 2014, 9, 134–140. [Google Scholar] [PubMed]
- Yildiz, I.; Aynur, Z.E. First Detection and Molecular Characterization of Dientamoeba fragilis in Cattle. Zoonoses Public Health 2022, 69, 897–903. [Google Scholar] [CrossRef]
- Yetismis, G.; Yildrim, A.; Pekmezci, D.; Duzlu, O.; Ciloglu, A.; Onder, Z.; Simsek, E.; Ercan, N.; Pekmezci, G.Z.; Inci, A. First Report and Genotyping of Dientamoeba fragilis in Pet Budgerigars (Melopsattacus undulatus), with Zoonotic Importance. Zoonoses Public Health 2022, 69, 572–578. [Google Scholar] [CrossRef] [PubMed]
- El-Gayar, E.K.; Mokhtar, A.B.; Hassan, W.A. Study of the Pathogenic Potential of Dientamoeba fragilis in Experimentally Infected Mice. Parasite Epidemiol. Control 2016, 1, 136–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stensvold, C.R.; Clark, C.G.; Röser, D. Limited Intra-Genetic Diversity in Dientamoeba fragilis Housekeeping Genes. Infect. Genet. Evol. 2013, 18, 284–286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cacciò, S.M.; Sannella, A.R.; Bruno, A.; Stensvold, C.R.; David, E.B.; Guimarães, S.; Manuali, E.; Magistrali, C.; Mahdad, K.; Beaman, M.; et al. Multilocus Sequence Typing of Dientamoeba fragilis Identified a Major Clone with Widespread Geographical Distribution. Int. J. Parasitol. 2016, 46, 793–798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stark, D.J.; Beebe, N.; Marriott, D.; Ellis, J.T.; Harkness, J. Dientamoebiasis: Clinical Importance and Recent Advances. Trends Parasitol. 2006, 22, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Vandenberg, O.; Peek, R.; Souayah, H.; Dediste, A.; Buset, M.; Scheen, R.; Retore, P.; Zissis, G.; van Gool, T. Clinical and Microbiological Features of Dientamoebiasis in Patients Suspected of Suffering from a Parasitic Gastrointestinal Illness: A Comparison of Dientamoeba fragilis and Giardia lamblia Infections. Int. J. Infect. Dis. 2006, 10, 255–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lhotská, Z.; Jirků, M.; Hložková, O.; Brožová, K.; Jirsová, D.; Stensvold, C.R.; Kolísko, M.; Jirků-Pomajbíková, K. A Study on the Prevalence and Subtype Diversity of the Intestinal Protist Blastocystis sp. in a Gut-Healthy Human Population in the Czech Republic. Front. Cell. Infect. Microbiol. 2020, 10, 544335. [Google Scholar] [CrossRef] [PubMed]
- Šloufová, M.; Lhotská, Z.; Jirků, M.; Petrželková, K.J.; Stensvold, C.R.; Cinek, O.; Jirků-Pomajbíková, K. Comparison of Molecular Diagnostic Approaches for the Detection and Differentiation of the Intestinal Protist Blastocystis sp. in Humans. Parasite 2022, 29, 30. [Google Scholar] [CrossRef]
- Stensvold, C.R.; Arendrup, M.C.; Mølbak, K.; Nielsen, H.V. The Prevalence of Dientamoeba fragilis in Patients with Suspected Enteroparasitic Disease in a Metropolitan Area in Denmark. Clin. Microbiol. Infect. 2007, 13, 839–842. [Google Scholar] [CrossRef] [Green Version]
- Sarzhanov, F.; Dogruman-Al, F.; Santin, M.; Maloney, J.G.; Gureser, A.S.; Karasartova, D.; Taylan-Ozkan, A. Investigation of Neglected Protists Blastocystis sp. And Dientamoeba fragilis in Immunocompetent and Immunodeficient Diarrheal Patients Using Both Conventional and Molecular Methods. PLoS Negl. Trop. Dis. 2021, 15, e0009779. [Google Scholar] [CrossRef]
- Bruijnesteijn van Coppenraet, L.E.S.; Dullaert-de Boer, M.; Ruijs, G.J.H.M.; van der Reijden, W.A.; van der Zanden, A.G.M.; Weel, J.F.L.; Schuurs, T.A. Case-Control Comparison of Bacterial and Protozoan Microorganisms Associated with Gastroenteritis: Application of Molecular Detection. Clin. Microbiol. Infect. 2015, 21, 592.e9–592.e19. [Google Scholar] [CrossRef]
- Heusinkveld, M.; Mughini-Gras, L.; Pijnacker, R.; Vennema, H.; Scholts, R.; van Huisstede-Vlaanderen, K.W.; Kortbeek, T.; Kooistra-Smid, M.; van Pelt, W. Potential Causative Agents of Acute Gastroenteritis in Households with Preschool Children: Prevalence, Risk Factors, Clinical Relevance and Household Transmission. Eur. J. Clin. Microbiol. Infect. Dis. 2016, 35, 1691–1700. [Google Scholar] [CrossRef] [PubMed]
- Burgaña, A.; Abellana, R.; Yordanov, S.Z.; Kazan, R.; Pérez Ortiz, A.M.; Ramos, C.C.; Hernández, C.G.; Rivero, M.M.; Gonçalves, A.Q.; Padilla, E.; et al. Paromomycin Is Superior to Metronidazole in Dientamoeba fragilis Treatment. Int. J. Parasitol. Drugs Drug Resist. 2019, 11, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Venturini, E.; Scarso, S.; Prelazzi, G.A.; Niccolai, C.; Bianchi, L.; Montagnani, C.; Lapini, M.; Chiappini, E.; Antonelli, A.; Rossolini, G.M.; et al. Epidemiology and Clinical Features of Intestinal Protozoan Infections Detected by Real-Time PCR in Non-Native Children within an Italian Tertiary Care Children’s Hospital: A Cross-Sectional Study. Travel Med. Infect. Dis. 2021, 43, 102107. [Google Scholar] [CrossRef] [PubMed]
- Noble, G.A.; Noble, E.R. Entamoebae in Farm Mammals. J. Parasitol. 1952, 38, 571–595. [Google Scholar] [CrossRef]
- Fletcher, S.; Caprarelli, G.; Merif, J.; Andresen, D.; Van Hal, S.; Stark, D.; Ellis, J. Epidemiology and Geographical Distribution of Enteric Protozoan Infections in Sydney, Australia. J. Public Health Res. 2014, 3, 83–91. [Google Scholar] [CrossRef]
- Lagacé-Wiens, P.R.; Van Caeseele, P.G.; Koschik, C. Dientamoeba fragilis: An Emerging Role in Intestinal Disease. Can. Med. Assoc. J. 2006, 175, 468–469. [Google Scholar] [CrossRef] [Green Version]
- Röser, D.; Simonsen, J.; Nielsen, H.V.; Stensvold, C.R.; Mølbak, K. Dientamoeba fragilis in Denmark: Epidemiological Experience Derived from Four Years of Routine Real-Time PCR. Eur. J. Clin. Microbiol. Infect. Dis. 2013, 32, 1303–1310. [Google Scholar] [CrossRef]
- Johnson, J.A.; Clark, C.G. Cryptic Genetic Diversity in Dientamoeba fragilis. J. Clin. Microbiol. 2000, 38, 4653–4654. [Google Scholar] [CrossRef] [Green Version]
- Peek, R.; Reedeker, F.R.; van Gool, T. Direct Amplification and Genotyping of Dientamoeba fragilis From Human Stool Specimens. J. Clin. Microbiol. 2004, 42, 631–635. [Google Scholar] [CrossRef] [Green Version]
- Verweij, J.J.; Mulder, B.; Poell, B.; van Middelkoop, D.; Brienen, E.A.T.T.; van Lieshout, L. Real-Time PCR for the Detection of Dientamoeba fragilis in Fecal Samples. Mol. Cell. Probes 2007, 21, 400–404. [Google Scholar] [CrossRef]
- Gouy, M.; Guindon, S.; Gascuel, O. SeaView Version 4: A Multiplatform Graphical User Interface for Sequence Alignment and Phylogenetic Tree Building. Mol. Biol. Evol. 2010, 27, 221–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hall, T.A. BioEdit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp. Ser. 1999, 41, 95–98. [Google Scholar]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.F.; von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast Model Selection for Accurate Phylogenetic Estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minh, B.Q.; Schmidt, H.A.; Chernomor, O.; Schrempf, D.; Woodhams, M.D.; von Haeseler, A.; Lanfear, R. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Mol. Biol. Evol. 2020, 37, 1530–1534. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hoang, D.T.; Chernomor, O.; von Haeseler, A.; Minh, B.Q.; Vinh, L.S. UFBoot2: Improving the Ultrafast Bootstrap Approximation. Mol. Biol. Evol. 2018, 35, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Kurt, Ö.; Dogruman, F.; Tanyüsel, M. Eradication of Blastocystis in Humans: Really Necessary for All? Parasitol. Int. 2016, 65, 797–801. [Google Scholar] [CrossRef]
- Billy, V.; Lhotská, Z.; Jirků, M.; Kadlecová, O.; Frgelecová, L.; Wegener Parfrey, L.; Jirků, P. Blastocystis Colonization Alters the Gut Microbiome and, In Some Cases, Promotes Faster Recovery from Induced Colitis. Front. Microbiol. 2021, 12, 641483. [Google Scholar]
- Sobotková, K.; Parker, W.; Levá, J.; Růžková., J.; Lukeš, J.; Jirků, P. Helminth Therapy—From the Parasite Perspective. Trends Parasitol. 2019, 35, 501–515. [Google Scholar] [CrossRef] [PubMed]
- Deng, L.; Wojciech, L.; Gascoigne, N.R.J.; Peng, G.; Tan, K.S.W. New Insights into the Interactions between Blastocystis, the Gut Microbiota, and Host Immunity. PLoS Pathog. 2021, 17, e1009253. [Google Scholar] [CrossRef] [PubMed]
| Host | Category | N | N Positive in cPCR | N Positive in qPCR |
|---|---|---|---|---|
| Homo sapiens | human | 296 | 22 | 72 |
| Canis lupus familiaris | dog | 54 | 0 | 4 |
| Felis silvestris catus | cat | 19 | 0 | 1 |
| Equus caballus | horse | 15 | 0 | 2 |
| Oryctolagus cuniculus domesticus | rabbit | 13 | 1 | 5 |
| Ovis aries | sheep | 6 | 0 | 1 |
| Capra aegagrus hircus | goat | 4 | 0 | 1 |
| Cavia aperea porcellus Bos primigenius taurus | guinea pig cow | 2 2 | 0 0 | 1 0 |
| Sus scrofa domestica | pig | 3 | 0 | 0 |
| Gallus gallus domesticus | poultry | 8 | 0 | 0 |
| Testudo horsfieldii | turtle | 2 | 0 | 0 |
| Psittacus erithacus | parrot | 1 | 0 | 0 |
| Columba livia domestica | pigeon | 1 | 0 | 0 |
| Phodopus sungorus | hamster | 1 | 0 | 0 |
| Atelerix albiventris | hedgehog | 1 | 0 | 0 |
| Anas platyrhynchos domesticus | duck | 1 | 0 | 0 |
| Anser anser domesticus | goose | 1 | 0 | 0 |
| Phelsuma madagascariensis | felsuma | 1 | 0 | 0 |
| Age | N | N Positive | Incidence (%) | SD |
|---|---|---|---|---|
| 0–3 | 18 | 2 | 11 | 0.323 |
| 4–6 | 14 | 7 | 50 | 0.519 |
| 7–12 | 16 | 7 | 44 | 0.512 |
| 13–17 | 4 | 1 | 25 | 0.500 |
| 18–30 | 73 | 15 | 21 | 0.407 |
| 31–49 | 88 | 21 | 24 | 0.429 |
| 50–60 | 36 | 9 | 25 | 0.439 |
| 60+ | 47 | 10 | 21 | 0.414 |
| Sample | Methods | Sample | Methods | ||||||
|---|---|---|---|---|---|---|---|---|---|
| cPCR | qPCR | Ct Value | Host | cPCR | qPCR | Ct Value | Host | ||
| B45 | − | + | 15 | human | B437 | − | + | 34 | human |
| B9 | + | + | 16 | human | B286 | − | + | 35 | human |
| B13 | + | + | 17 | human | B289 | − | + | 35 | human |
| B126 | + | + | 17 | human | B372 | − | + | 35 | human |
| B226 | − | + | 17 | human | B375 | − | + | 35 | human |
| B288 | + | + | 17 | human | B49 | − | + | 36 | human |
| B312 | − | + | 17 | human | B34 | − | + | 37 | human |
| B328 | + | + | 18 | human | B378 | − | + | 37 | human |
| B24 | + | + | 19 | human | B246 | − | + | 37 | human |
| B373 | + | + | 19 | human | B31 | − | + | 38 | human |
| B224 | + | + | 20 | human | B150 | − | + | 38 | human |
| B339 | + | + | 20 | human | B287 | − | + | 38 | human |
| B371 | + | + | 20 | human | B41 | − | + | 39 | human |
| B32 | − | + | 22 | human | B121 | − | + | 39 | human |
| B345 | + | + | 22 | human | B227 | − | + | 39 | human |
| B418 | + | + | 23 | human | B234 | − | + | 39 | human |
| B82 | + | + | 24 | human | B245 | − | + | 39 | human |
| B113 | + | + | 24 | human | B291 | − | + | 39 | human |
| B238 | + | + | 24 | human | B325 | − | + | 39 | human |
| B347 | − | + | 24 | human | B327 | − | + | 39 | human |
| B414 | − | + | 24 | human | B331 | − | + | 39 | human |
| B329 | + | + | 25 | human | B416 | − | + | 39 | human |
| B343 | − | + | 25 | human | B3 | − | + | 40 | human |
| B413 | + | + | 25 | human | B23 | − | + | 40 | human |
| B326 | + | + | 26 | human | B35 | − | + | 40 | human |
| B341 | + | + | 26 | human | B42 | − | + | 40 | human |
| B393 | + | + | 26 | human | B106 | − | + | 40 | human |
| B225 | − | + | 27 | human | B232 | − | + | 40 | human |
| B361 | − | + | 27 | human | B174 | − | + | 23 | rabbit |
| B338 | + | + | 28 | human | B263 | − | + | 28 | rabbit |
| B377 | + | + | 28 | human | B262 | − | + | 29 | guinea pig |
| B424 | − | + | 28 | human | B151 | − | + | 31 | rabbit |
| B346 | − | + | 29 | human | B163 | − | + | 32 | rabbit |
| B367 | − | + | 29 | human | B422 | − | + | 33 | goat |
| B124 | − | + | 30 | human | B285 | − | + | 34 | rabbit |
| B231 | − | + | 30 | human | B229 | − | + | 38 | cat |
| B338 | − | + | 31 | human | B351 | − | + | 38 | sheep |
| B374 | − | + | 31 | human | B137 | − | + | 39 | dog |
| B425 | − | + | 31 | human | B152 | − | + | 39 | rabbit |
| B430 | − | + | 31 | human | B171 | − | + | 39 | horse |
| B431 | − | + | 31 | human | B337 | − | + | 39 | dog |
| B406 | − | + | 32 | human | B369 | − | + | 39 | dog |
| B280 | − | + | 33 | human | B205 | − | + | 40 | horse |
| B419 | − | + | 33 | human | B273 | − | + | 40 | dog |
| Category | Samples N * | Prevalence | Category | Samples N * | Prevalence | Category | Samples N * | Prevalence |
|---|---|---|---|---|---|---|---|---|
| living locality | non-travelers | |||||||
 village | 83/296 | 29% |  city | 213/296 | 23% |  only in the Czechia | 52/296 | 15% # |
| (24/83) | (48/213) | (8/52) | ||||||
| contact with animals | travelers | |||||||
 in contact with animals | 252/296 | 25% |  no contact with animals | 44/296 | 23% |  inside Europe | 140/296 | 24% # |
| (62/252) | (10/44) | (34/140) | ||||||
 in contact with pets | 180/252 * | 20% # |  in contact with farm animals | 72/252 * | 36% # |  outside Europe | 104/296 | 29% # |
| (36/180) | (26/72) | (30/104) | ||||||
| N Family Members | N Positive Members (Sample) | Contact Animal (Sample) |
|---|---|---|
| 3 | 3 (B371, B372, B373) | dog (B369), horse (B337) |
| 4 | 3 (B338, B339, B341) | sheep (B351) |
| 3 | 2 (B418, B419) | goat (B422) |
| 4 | 1 (B150) | rabbit (B151) |
| 4 | 1 (B49) | rabbit (B285) |
| 1 | 1 (B227) | cat (B229) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jirků, M.; Kašparová, A.; Lhotská, Z.; Oborník, M.; Brožová, K.; Petrželková, K.J.; Samaš, P.; Kadlecová, O.; Stensvold, C.R.; Jirků, K. A Cross-Sectional Study on the Occurrence of the Intestinal Protist, Dientamoeba fragilis, in the Gut-Healthy Volunteers and Their Animals. Int. J. Mol. Sci. 2022, 23, 15407. https://doi.org/10.3390/ijms232315407
Jirků M, Kašparová A, Lhotská Z, Oborník M, Brožová K, Petrželková KJ, Samaš P, Kadlecová O, Stensvold CR, Jirků K. A Cross-Sectional Study on the Occurrence of the Intestinal Protist, Dientamoeba fragilis, in the Gut-Healthy Volunteers and Their Animals. International Journal of Molecular Sciences. 2022; 23(23):15407. https://doi.org/10.3390/ijms232315407
Chicago/Turabian StyleJirků, Milan, Andrea Kašparová, Zuzana Lhotská, Miroslav Oborník, Kristýna Brožová, Klára J. Petrželková, Peter Samaš, Oldřiška Kadlecová, Christen Rune Stensvold, and Kateřina Jirků. 2022. "A Cross-Sectional Study on the Occurrence of the Intestinal Protist, Dientamoeba fragilis, in the Gut-Healthy Volunteers and Their Animals" International Journal of Molecular Sciences 23, no. 23: 15407. https://doi.org/10.3390/ijms232315407
APA StyleJirků, M., Kašparová, A., Lhotská, Z., Oborník, M., Brožová, K., Petrželková, K. J., Samaš, P., Kadlecová, O., Stensvold, C. R., & Jirků, K. (2022). A Cross-Sectional Study on the Occurrence of the Intestinal Protist, Dientamoeba fragilis, in the Gut-Healthy Volunteers and Their Animals. International Journal of Molecular Sciences, 23(23), 15407. https://doi.org/10.3390/ijms232315407







