Cold-Active Lipases and Esterases: A Review on Recombinant Overexpression and Other Essential Issues
Abstract
:1. Introduction
2. Approaches for Isolation of Genes Encoding Cold-Active Lipase and Esterase
3. Cold-Active Lipase and Esterase Overexpression in Recombinant Heterologous Hosts
| Organisms/Enzymes | Source | Host | Vector | Localization of Expressed Enzyme | Optimum Temp./Residual Activity | References |
|---|---|---|---|---|---|---|
| Alkalibacterium sp. SL3/esterase | Uncultured | E. coli BL21 (DE3) | pET-28a (+) | Soluble | 30 °C and 68% at 0 °C | [84] |
| Chitinophaga pinensis-like/esterase | Uncultured | E. coli RosettaTM (Novagen) | pGEX-6P-2 | Insoluble inclusion body | 20 °C and NA | [66] |
| Lactobacillus plantarum/LpLp_2631/esterase | Microbiological Culture | E. coli BL21 (DE3) | pURI3TEV vector | Soluble | 20 °C and 90% at 5 °C | [85] |
| Burkholderia pyrrocinia/BpFae esterase | Microbiological Culture | E. coli BL21 (DE3) | pET28a pCold-TF and pGEX-4T-1. | Insoluble/soluble non inactive form | NA | [86] |
| Candida parapsilosis/esterase | Cultured | S. cerevisiae | pYES2 | Soluble | NA and at 20 °C | [64] |
| Monascus ruber M7/esterase | Cultured | E. coli BL21(DE3) | pET-30a (+) | Soluble | 40 °C and 50% at 4–10 °C | [87] |
| Alcanivorax dieselolei/lipase | Cultured | E. coli BL21(DE3) | pGEX-6p-1 (GE) | Soluble | 20 °C and 95% at 10 °C | [88,89] |
| Pseudomonas fluorescens KE38/lipase | Uncultured | E. coli BL21(DE3) | pET28a | Insoluble inclusion body | 25 °C and NA | [90] |
| Aphanizomenon flos-aquae/esterase | Uncultured | E. coli BL21(DE3) | pET28a | Insoluble inclusion body | 5–15 °C | [91] |
| Bacillus halodurans/lipase | Uncultured | E. coli BL21 (DE3) | pET-28a (+) | Soluble | 30 °C | [92] |
| Bacillus licheniformis/esterase | Cultured | E. coli BL21 (DE3) | pET-28a (+) | Soluble | 30 °C and 35% at 0 °C | [63] |
| G. antarctica PI12/esterase | Expressed sequence tag | BL21 (DE3) | pET200_GaDlh | Soluble | 10 °C and 50% at 0–30 °C | [93] |
| Paenibacillus sp. R4/esterase | Cultured | BL21 (DE3) | pET-22b (+) | Soluble | 35 °C and 45% at 10 °C | [94] |
| Pseudomonas sp./lipase | Uncultured | BL21(DE3) | pET32b (+) | Insoluble inclusion body | 35 °C and 50% at 15–40 °C | [27] |
| Yarrowia lipolytica(LIPY8)/lipase | Cultured | Insect (Sf9) | pFastBac1 | Soluble | 17 °C and 70% at 8–30 °C | [65] |
4. Purification of Cold-Active Lipolytic Enzymes
| Enzymes | Type of Purification | Purification Steps | Buffer | Column/Resin | Fold/Yield | Molecular Mass | References |
|---|---|---|---|---|---|---|---|
| GaDlh | Complete | Single-step/Ni-affinity chromatography | Tris–HCl | Ni–NTA column | 1.9/7.7% | 28 kDa | [106] |
| AMBL-20 | Partial | Single step/ammonium sulfate precipitation | Tris–HCl | NA | NA | NA | [107] |
| HaSGNH1 | Complete | Single-step/Ni2+-affinity | Tris–HCl | HisTrap HP | 2.5/~5 mg/g | 24 kDa | [108] |
| LSK25 | Complete | Single-step/Ni-Sepharose affinity | Tris–HCl | Ni Sepharose® 6Fast Flow column | 1.3/44% | 65 kDa | [27] |
| AaSGNH1 | Complete | Single-step/Ni-Sepharose affinity | Tris–HCl | Ni-NTA agarose | 0.6–0.7 mg/mL | 43.9 kDa | [109] |
| B8W22 | Complete | Double-step/Ni-Sepharose affinityand ion-exchange | Tris–HCl | DEAE FF column/Octyl Sepharose FF column | 59.03/20% | 35 kDa | [110] |
| ERMR1:04 | Complete | Triple-step/ammonium sulfate precipitation, Size exclusion, and hydrophobic interaction | Tris–HCl | Sephadex G-100 column, Octyl-Sepharose fast flow column | 21.3/NA | 250 kDa (hexameric) 39.8 kD (monomeric) | [111] |
| estHIJ | Complete | Single-step/Ni-affinity | Phosphate buffer | Ni-NTA affinity column. | 3.5/47.5% | 29 kDa | [112] |
| ZY124 | Complete | Double step/ammonium sulfate precipitation and hydrophobic chromatography | Tris–HCl | Phenyl Sepharose FF column andmicrocolumn reversed-phase LC-1MS | 1.34/NA | 37.9 kDa. | [105] |
| AMS8 | Complete | Reverse Micelle Extraction | Sodium phosphate | NA | NA/58.84% | NA | [113] |
| KM12 | Complete | Double-step/ammonium sulfate precipitation and ion-exchange | Tris–HCl | Q-Sepharose FF column | 15.63/36.0% | 33 kDa | [114] |
| KCTC 22881 | Complete | Double-step/affinity chromatography and size-exclusion chromatography | Tris–HCl | HisTrap FF, PD-10 and Sephacryl S200 HR | NA | 31.0 kDa | [104] |
| EstN7 | Complete | Single-step/Ni-affinity | Potassium Phosphate | Ni–NTA affinity column | 5/94.5% | 37.0 kDa | [48] |
| GlaEst12-like | Complete | Single-step/Ni-sepharose affinity | Sodium Phosphate | Nickel-Sepharose HP | 1.7/40% | 63 kDa | [115] |
| RSAP17 | Complete | Double-step/ammonium sulfate precipitation and ion-exchange | Tris–HCl | DEAE-cellulose anion exchanger | NA | 103.8 kDa | [116] |
| PsEst3 | Complete | Double-step/nickel-affinity and size-exclusion chromatography | Tris–HCl | Ni-affinity and HiLoad 16/60 Superdex 200 column | NA | 29 kDa | [103] |
5. Three-Dimensional (3D) Structure and Functional Mechanisms of Cold-Active Lipase and Esterase
6. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- De Maayer, P.; Anderson, D.; Cary, C.; Cowan, D.A. Some like it cold: Understanding the survival strategies of psychrophiles. EMBO Rep. 2014, 15, 508–517. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Liu, G.; Huang, K. Cold adaptation mechanisms of a snow alga Chlamydomonas nivalis during temperature fluctuations. Front. Microbiol. 2020, 11, 611080. [Google Scholar] [CrossRef] [PubMed]
- Petrovskaya, L.; Novototskaya-Vlasova, K.; Komolova, A.; Rivkina, E. 6 Biochemical adaptations to the permafrost environment: Lipolytic enzymes from Psychrobacter cryohalolentis K5T. In Microbial Life in the Cryosphere and Its Feedback on Global Change; De Gruyter: Berlin, Germany, 2021; pp. 141–152. [Google Scholar] [CrossRef]
- Boetius, A.; Anesio, A.M.; Deming, J.W.; Mikucki, J.A.; Rapp, J.Z. Microbial ecology of the cryosphere: Sea ice and glacial habitats. Nat. Rev. Microbiol. 2015, 13, 677–690. [Google Scholar] [CrossRef] [PubMed]
- Deming, J.W. Psychrophiles and polar regions. Curr. Opin. Microbiol. 2002, 5, 301–309. [Google Scholar] [CrossRef]
- Yang, Y.; Levick, D.T.; Just, C.K. Halophilic, thermophilic, and psychrophilic archaea: Cellular and molecular adaptations and potential applications. J. Young Investig. 2007, 17. Available online: https://www.jyi.org/2007-october/2007/10/10/halophilic-thermophilic-and-psychrophilic-archaea-cellular-and-molecular-adaptations-and-potential-applications (accessed on 24 November 2022).
- Struvay, C.; Feller, G. Optimization to low temperature activity in psychrophilic enzymes. Int. J. Mol. Sci. 2012, 13, 11643–11665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- D’Amico, S.; Collins, T.; Marx, J.C.; Feller, G.; Gerday, C. Psychrophilic microorganisms: Challenges for life. EMBO Rep. 2006, 7, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Collins, T.; Margesin, R. Psychrophilic lifestyles: Mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871. [Google Scholar] [CrossRef]
- Collins, T.; Matzapetakis, M.; Santos, H. Backbone and side chain 1H, 15N and 13C assignments for a thiol-disulphide oxidoreductase from the Antarctic bacterium Pseudoalteromonas haloplanktis TAC125. Biomol. NMR Assign. 2010, 4, 151–154. [Google Scholar] [CrossRef] [Green Version]
- Santiago, M.; Ramírez-Sarmiento, C.A.; Zamora, R.A.; Parra, L.P. Discovery, molecular mechanisms, and industrial applications of cold-active enzymes. Front. Microbiol. 2016, 7, 1408. [Google Scholar] [CrossRef]
- Gerday, C. Fundamentals of Cold-Active Enzymes. In Cold-adapted Yeasts: Biodiversity, Adaptation Strategies and Biotechnological Significance; Buzzini, P., Margesin, R., Eds.; Springer: Berlin, Heidelberg, 2014; pp. 325–350. [Google Scholar] [CrossRef]
- Feller, G. Psychrophilic enzymes: From folding to function and biotechnology. Scientifica 2013, 2013, 512840. [Google Scholar] [CrossRef] [PubMed]
- Smalås, A.; Leiros, H.; Os, V.; Willassen, N. Cold adapted enzymes. Biotechnol. Annu. Rev. 2000, 6, 1–57. [Google Scholar] [CrossRef]
- Baeza, M.; Alcaíno, J.; Cifuentes, V.; Turchetti, B.; Buzzini, P. Cold-active enzymes from cold-adapted yeasts. In Biotechnology of Yeasts and Filamentous Fungi; Springer: Berlin/Heidelberg, Germany, 2017; pp. 297–324. [Google Scholar] [CrossRef]
- Joseph, B.; Ramteke, P.W.; Thomas, G.; Shrivastava, N. Cold-active microbial lipases: A versatile tool for industrial applications. Biotechnol. Mol. Biol. Rev. 2007, 2, 39–48. [Google Scholar] [CrossRef]
- Mangiagalli, M.; Brocca, S.; Orlando, M.; Lotti, M. The “cold revolution”. Present and future applications of cold-active enzymes and ice-binding proteins. New Biotechnol. 2020, 55, 5–11. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Mukhia, S.; Kumar, R. Industrial applications of cold-adapted enzymes: Challenges, innovations and future perspective. 3 Biotech 2021, 11, 426. [Google Scholar] [CrossRef] [PubMed]
- Hamid, B.; Bashir, Z.; Yatoo, A.M.; Mohiddin, F.; Majeed, N.; Bansal, M.; Poczai, P.; Almalki, W.H.; Sayyed, R.; Shati, A.A. Cold-Active Enzymes and Their Potential Industrial Applications—A Review. Molecules 2022, 27, 5885. [Google Scholar] [CrossRef]
- Gurung, N.; Ray, S.; Bose, S.; Rai, V. A broader view: Microbial enzymes and their relevance in industries, medicine, and beyond. BioMed Res. Int. 2013, 2013, 329121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramnath, L.; Sithole, B.; Govinden, R. Classification of lipolytic enzymes and their biotechnological applications in the pulping industry. Can. J. Microbiol. 2017, 63, 179–192. [Google Scholar] [CrossRef]
- Lee, C.W.; Kwon, S.; Park, S.-H.; Kim, B.-Y.; Yoo, W.; Ryu, B.H.; Kim, H.-W.; Shin, S.C.; Kim, S.; Park, H. Crystal structure and functional characterization of an esterase (Ea EST) from Exiguobacterium antarcticum. PLoS ONE 2017, 12, e0169540. [Google Scholar] [CrossRef] [Green Version]
- Kumar, K.; Mhetre, A.; Ratnaparkhi, G.S.; Kamat, S.S. A Superfamily-wide Activity Atlas of Serine Hydrolases in Drosophila melanogaster. Biochemistry 2021, 60, 1312–1324. [Google Scholar] [CrossRef] [PubMed]
- Ali, Y.B.; Verger, R.; Abousalham, A. Lipases or esterases: Does it really matter? Toward a new bio-physico-chemical classification. In Lipases and Phospholipases; Springer: Berlin/Heidelberg, Germany, 2012; pp. 31–51. [Google Scholar] [CrossRef]
- Shin, W.-R.; Um, H.-J.; Kim, Y.-C.; Kim, S.C.; Cho, B.-K.; Ahn, J.-Y.; Min, J.; Kim, Y.-H. Biochemical characterization and molecular docking analysis of novel esterases from Sphingobium chungbukense DJ77. Int. J. Biol. Macromol. 2021, 168, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Chahinian, H.; Sarda, L. Distinction between esterases and lipases: Comparative biochemical properties of sequence-related carboxylesterases. Protein Pept. Lett. 2009, 16, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Salwoom, L.; Salleh, A.B.; Convey, P.; Mohamad Ali, M.S. New recombinant cold-adapted and organic solvent tolerant lipase from psychrophilic Pseudomonas sp. LSK25, isolated from Signy Island Antarctica. Int. J. Mol. Sci. 2019, 20, 1264. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, S.; Kanwar, S.S. Organic Solvent Tolerant Lipases and Applications. Sci. World J. 2014, 2014, 625258. [Google Scholar] [CrossRef] [Green Version]
- Mu, R.; Wang, Z.; Wamsley, M.C.; Duke, C.N.; Lii, P.H.; Epley, S.E.; Todd, L.C.; Roberts, P.J. Application of enzymes in regioselective and stereoselective organic reactions. Catalysts 2020, 10, 832. [Google Scholar] [CrossRef]
- Kuddus, M. Cold-active enzymes in food biotechnology: An updated mini review. J. Appl. Biol. Biotechnol. 2018, 6, 3–5. [Google Scholar] [CrossRef] [Green Version]
- Mhetras, N.; Mapare, V.; Gokhale, D. Cold Active Lipases: Biocatalytic Tools for Greener Technology. Appl. Biochem. Biotechnol. 2021, 193, 1–22. [Google Scholar] [CrossRef]
- Bhatia, R.K.; Ullah, S.; Hoque, M.Z.; Ahmad, I.; Yang, Y.-H.; Bhatt, A.K.; Bhatia, S.K. Psychrophiles: A source of cold-adapted enzymes for energy efficient biotechnological industrial processes. J. Environ. Chem. Eng. 2021, 9, 104607. [Google Scholar] [CrossRef]
- Esakkiraj, P.; Bharathi, C.; Ayyanna, R.; Jha, N.; Panigrahi, A.; Karthe, P.; Arul, V. Functional and molecular characterization of a cold-active lipase from Psychrobacter celer PU3 with potential antibiofilm property. Int. J. Biol. Macromol. 2022, 211, 741–753. [Google Scholar] [CrossRef]
- Xiang, M.; Wang, L.; Yan, Q.; Jiang, Z.; Yang, S. Heterologous expression and biochemical characterization of a cold-active lipase from Rhizopus microsporus suitable for oleate synthesis and bread making. Biotechnol. Lett. 2021, 43, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Jeon, S.; Hwang, J.; Yoo, W.; Chang, J.W.; Do, H.; Kim, H.-W.; Kim, K.K.; Lee, J.H.; Kim, T.D. Purification and Crystallographic Analysis of a Novel Cold-Active Esterase (Ha Est1) from Halocynthiibacter arcticus. Crystals 2021, 11, 170. [Google Scholar] [CrossRef]
- Boyko, K.M.; Kryukova, M.V.; Petrovskaya, L.E.; Kryukova, E.A.; Nikolaeva, A.Y.; Korzhenevsky, D.A.; Lomakina, G.Y.; Novototskaya-Vlasova, K.A.; Rivkina, E.M.; Dolgikh, D.A.; et al. Structural and Biochemical Characterization of a Cold-Active PMGL3 Esterase with Unusual Oligomeric Structure. Biomolecules 2021, 11, 57. [Google Scholar] [CrossRef] [PubMed]
- Al-Ghanayem, A.A.; Joseph, B. Current prospective in using cold-active enzymes as eco-friendly detergent additive. Appl. Microbiol. Biotechnol. 2020, 104, 2871–2882. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, N.; Revathi, D.; Sheelu, G.; Yamuna Rani, K.; Sridhar, S.; Mehtab, V.; Sumana, C. Recent advances on sources and industrial applications of lipases. Biotechnol. Prog. 2018, 34, 5–28. [Google Scholar] [CrossRef]
- Kavitha, M. Cold active lipases—An update. Front. Life Sci. 2016, 9, 226–238. [Google Scholar] [CrossRef] [Green Version]
- Al-Maqtari, Q.A.; Waleed, A.; Mahdi, A.A. Cold-active enzymes and their applications in industrial fields—A review. Int. J. Res. Stud. Agric. Sci. 2019, 6, 107–123. [Google Scholar]
- da Silva, T.H.; Câmara, P.E.; Pinto, O.H.B.; Carvalho-Silva, M.; Oliveira, F.S.; Convey, P.; Rosa, C.A.; Rosa, L.H. Diversity of Fungi Present in Permafrost in the South Shetland Islands, Maritime Antarctic. Microb. Ecol. 2021, 83, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Houde, A.; Kademi, A.; Leblanc, D. Lipases and their industrial applications. Appl. Biochem. Biotechnol. 2004, 118, 155–170. [Google Scholar] [CrossRef] [PubMed]
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and hot extremozymes: Industrial relevance and current trends. Front. Bioeng. Biotechnol. 2015, 3, 148. [Google Scholar] [CrossRef] [Green Version]
- Chandra, P.; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microb. Cell Factories 2020, 19, 1–42. [Google Scholar] [CrossRef]
- Lashani, E.; Shahnavaz, B.; Makhdoumi, A. Characterization of Psychrophilic and Psychrotolerant Cultivable Bacteria in Alpine Soil in Iran. Biol. J. Microorg. 2020, 9, 47–57. [Google Scholar] [CrossRef]
- Jain, R.; Pandey, A.; Pasupuleti, M.; Pande, V. Prolonged production and aggregation complexity of cold-active lipase from Pseudomonas proteolytica (GBPI_Hb61) isolated from cold Desert Himalaya. Mol. Biotechnol. 2017, 59, 34–45. [Google Scholar] [CrossRef] [PubMed]
- De Santi, C.; Tedesco, P.; Ambrosino, L.; Altermark, B.; Willassen, N.-P.; de Pascale, D. A new alkaliphilic cold-active esterase from the psychrophilic marine bacterium Rhodococcus sp.: Functional and structural studies and biotechnological potential. Appl. Biochem. Biotechnol. 2014, 172, 3054–3068. [Google Scholar] [CrossRef] [PubMed]
- Noby, N.; Saeed, H.; Embaby, A.M.; Pavlidis, I.V.; Hussein, A. Cloning, expression and characterization of cold active esterase (EstN7) from Bacillus cohnii strain N1: A novel member of family IV. Int. J. Biol. Macromol. 2018, 120, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Bakermans, C.; Ayala-del-Río, H.L.; Ponder, M.A.; Vishnivetskaya, T.; Gilichinsky, D.; Thomashow, M.F.; Tiedje, J.M. Psychrobacter cryohalolentis sp. nov. and Psychrobacter arcticus sp. nov., isolated from Siberian permafrost. Int. J. Syst. Evol. Microbiol. 2006, 56, 1285–1291. [Google Scholar] [CrossRef]
- De Santi, C.; Altermark, B.; Pierechod, M.M.; Ambrosino, L.; de Pascale, D.; Willassen, N.-P. Characterization of a cold-active and salt tolerant esterase identified by functional screening of Arctic metagenomic libraries. BMC Biochem. 2016, 17, 1. [Google Scholar] [CrossRef] [Green Version]
- Jain, R.; Pandey, N.; Pandey, A. Aggregation properties of cold-active lipase produced by a psychrotolerant strain of Pseudomonas palleroniana (GBPI_508). Biocatal. Biotransform. 2020, 38, 263–273. [Google Scholar] [CrossRef]
- Roumpeka, D.D.; Wallace, R.J.; Escalettes, F.; Fotheringham, I.; Watson, M. A review of bioinformatics tools for bio-prospecting from metagenomic sequence data. Front. Genet. 2017, 8, 23. [Google Scholar] [CrossRef]
- Lorenz, P.; Schleper, C. Metagenome—A challenging source of enzyme discovery. J. Mol. Catal. B Enzym. 2002, 19, 13–19. [Google Scholar] [CrossRef]
- Dias, R.; Silva, L.C.; Eller, M.R.; Oliveira, V.M.; De Paula, S.; Silva, C.C. Metagenomics: Library construction and screening methods. V Metagenom. Methods Appl. Perspect 2014, 5, 28–34. [Google Scholar]
- Thomas, T.; Gilbert, J.; Meyer, F. Metagenomics—A guide from sampling to data analysis. Microb. Inform. Exp. 2012, 2, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chauhan, N.S. Metagenome analysis and interpretation. In Data Processing Handbook for Complex Biological Data Sources; Elsevier: Amsterdam, The Netherlands, 2019; pp. 139–160. [Google Scholar] [CrossRef]
- Dhanjal, D.S.; Chopra, R.S.; Chopra, C. Metagenomics and Enzymes: The Novelty Perspective. In Metagenomics: Techniques, Applications, Challenges and Opportunities; Chopra, R.S., Chopra, C., Sharma, N.R., Eds.; Springer: Singapore, 2020; pp. 109–131. [Google Scholar] [CrossRef]
- Park, J.-E.; Jeong, G.-S.; Lee, H.-W.; Kim, H. Molecular Characterization of Novel Family IV and VIII Esterases from a Compost Metagenomic Library. Microorganisms 2021, 9, 1614. [Google Scholar] [CrossRef] [PubMed]
- Rai, A.; Bhattacharjee, A. Molecular profiling of microbial community structure and their CAZymes via metagenomics, from Tsomgo lake in the Eastern Himalayas. Arch. Microbiol. 2021, 203, 3135–3146. [Google Scholar] [CrossRef] [PubMed]
- Parages, M.L.; Gutiérrez-Barranquero, J.A.; Reen, F.J.; Dobson, A.D.W.; O’Gara, F. Integrated (Meta) Genomic and Synthetic Biology Approaches to Develop New Biocatalysts. Mar. Drugs 2016, 14, 62. [Google Scholar] [CrossRef] [PubMed]
- Kleiner-Grote, G.R.M.; Risse, J.M.; Friehs, K. Secretion of recombinant proteins from E. coli. Eng. Life Sci. 2018, 18, 532–550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kastenhofer, J.; Rettenbacher, L.; Feuchtenhofer, L.; Mairhofer, J.; Spadiut, O. Inhibition of E. coli host RNA polymerase allows efficient extracellular recombinant protein production by enhancing outer membrane leakiness. Biotechnol. J. 2021, 16, 2000274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Xu, H.; Wu, Y.; Zeng, J.; Guo, Z.; Wang, L.; Shen, C.; Qiao, D.; Cao, Y. A new cold-adapted, alkali-stable and highly salt-tolerant esterase from Bacillus licheniformis. Int. J. Biol. Macromol. 2018, 111, 1183–1193. [Google Scholar] [CrossRef]
- Xue, D.; Yao, D.; You, X.; Gong, C. Green Synthesis of the Flavor Esters with a Marine Candida parapsilosis Esterase Expressed in Saccharomyces cerevisiae. Indian J. Microbiol. 2020, 60, 175–181. [Google Scholar] [CrossRef]
- Li, T.; Zhang, W.; Hao, J.; Sun, M.; Lin, S.X. Cold-active extracellular lipase: Expression in Sf9 insect cells, purification, and catalysis. Biotechnol. Rep. 2019, 21, e00295. [Google Scholar] [CrossRef]
- Hu, X.P.; Heath, C.; Taylor, M.P.; Tuffin, M.; Cowan, D. A novel, extremely alkaliphilic and cold-active esterase from Antarctic desert soil. Extremophiles 2012, 16, 79–86. [Google Scholar] [CrossRef]
- Yoshida, K.; Konishi, K.; Magana-Mora, A.; Rougny, A.; Yasutake, Y.; Muramatsu, S.; Murata, S.; Kumagai, T.; Aburatani, S.; Sakasegawa, S.-i. Production of recombinant extracellular cholesterol esterase using consistently active promoters in Burkholderia stabilis. Biosci. Biotechnol. Biochem. 2019, 83, 1974–1984. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.; Gupta, N.; Rathi, P. Bacterial lipases: An overview of production, purification and biochemical properties. Appl. Microbiol. Biotechnol. 2004, 64, 763–781. [Google Scholar] [CrossRef] [PubMed]
- Ingram, Z.; Patkar, A.; Oh, D.; Zhang, K.K.; Chung, C.; Lin-Cereghino, J.; LinCereghino, G.P. Overcoming Obstacles in Protein Expression in the Yeast Pichia pastoris: Interviews of Leaders in the Pichia Field. Pac. J. Health 2021, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell Physiol. 2020, 235, 5867–5881. [Google Scholar] [CrossRef]
- Jarvis, D.L. Developing baculovirus-insect cell expression systems for humanized recombinant glycoprotein production. Virology 2003, 310, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Wu, C.P.; Chang, C.J.; Li, C.H.; Wu, Y.L. The influence of serial passage on the stability of an exogenous gene expression in recombinant baculovirus. Entomol. Res. 2021, 51, 168–175. [Google Scholar] [CrossRef]
- Gomes, A.; Byregowda, S.; Veeregowda, B.; Balamurugan, V. An overview of heterologous expression host systems for the production of recombinant proteins. Adv. Anim. Vet. Sci 2016, 4, 346–356. [Google Scholar] [CrossRef]
- Miyauchi, Y.; Kimura, A.; Sawai, M.; Fujimoto, K.; Hirota, Y.; Tanaka, Y.; Takechi, S.; Mackenzie, P.I.; Ishii, Y. Use of a Baculovirus-Mammalian Cell Expression-System for Expression of Drug-Metabolizing Enzymes: Optimization of Infection With a Focus on Cytochrome P450 3A4. Front. Pharmacol. 2022, 13, 832931. [Google Scholar] [CrossRef]
- Lozano Terol, G.; Gallego-Jara, J.; Sola Martínez, R.A.; Martínez Vivancos, A.; Cánovas Díaz, M.; de Diego Puente, T. Impact of the Expression System on Recombinant Protein Production in Escherichia coli BL21. Front. Microbiol. 2021, 12, 682001. [Google Scholar] [CrossRef]
- Xu, W.; Klumbys, E.; Ang, E.L.; Zhao, H. Emerging molecular biology tools and strategies for engineering natural product biosynthesis. Metab. Eng. Commun. 2020, 10, e00108. [Google Scholar] [CrossRef]
- Rosano, G.L.; Ceccarelli, E.A. Recombinant protein expression in Escherichia coli: Advances and challenges. Front. Microbiol. 2014, 5, 172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Factories 2015, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kane, J.F.; Hartley, D.L. Formation of recombinant protein inclusion bodies in Escherichia coli. Trends Biotechnol. 1988, 6, 95–101. [Google Scholar] [CrossRef]
- Slouka, C.; Kopp, J.; Hutwimmer, S.; Strahammer, M.; Strohmer, D.; Eitenberger, E.; Schwaighofer, A.; Herwig, C. Custom made inclusion bodies: Impact of classical process parameters and physiological parameters on inclusion body quality attributes. Microb. Cell Factories 2018, 17, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Singhvi, P.; Saneja, A.; Srichandan, S.; Panda, A.K. Bacterial inclusion bodies: A treasure trove of bioactive proteins. Trends Biotechnol. 2020, 38, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Ramón, A.; Señorale-Pose, M.; Marín, M. Inclusion bodies: Not that bad…. Front. Microbiol. 2014, 5, 56. [Google Scholar] [CrossRef] [Green Version]
- Baltà-Foix, R.; Roca-Pinilla, R.; López-Cano, A.; Gifre-Renom, L.; Arís, A.; Garcia-Fruitós, E. Functional Inclusion Bodies. In Microbial Production of High-Value Products; Rehm, B.H.A., Wibowo, D., Eds.; Springer International Publishing: Cham, Switzerland, 2022; pp. 289–308. [Google Scholar] [CrossRef]
- Javed, S.; Azeem, F.; Hussain, S.; Rasul, I.; Siddique, M.H.; Riaz, M.; Afzal, M.; Kouser, A.; Nadeem, H. Bacterial lipases: A review on purification and characterization. Prog. Biophys. Mol. Biol. 2018, 132, 23–34. [Google Scholar] [CrossRef]
- Shepard, C.C.; Tiselius, A. The chromatography of proteins. The effect of salt concentration and pH on the adsorption of proteins to silica gel. Discuss. Faraday Soc. 1949, 7, 275–285. [Google Scholar] [CrossRef]
- Coffman, J.L.; Kramarczyk, J.F.; Kelley, B.D. High-throughput screening of chromatographic separations: I. Method development and column modeling. Biotechnol. Bioeng. 2008, 100, 605–618. [Google Scholar] [CrossRef] [PubMed]
- Bensch, M.; Schulze Wierling, P.; von Lieres, E.; Hubbuch, J. High Throughput Screening of Chromatographic Phases for Rapid Process Development. Chem. Eng. Technol. 2005, 28, 1274–1284. [Google Scholar] [CrossRef]
- Silva, T.C.; Eppink, M.; Ottens, M. Automation and miniaturization: Enabling tools for fast, high-throughput process development in integrated continuous biomanufacturing. J. Chem. Technol. Biotechnol. 2021, 97, 2365–2375. [Google Scholar] [CrossRef]
- Hollander, C.; Walrond, S.C.; Connelly, C.; Megson, L.; Bove, E.; McDonagh, T. A custom ÄKTA avant configuration enabling automated parallel protein purification over a range of process scales. Protein Expr. Purif. 2021, 182, 105842. [Google Scholar] [CrossRef] [PubMed]
- Vorderwuelbecke, S.; Cleverley, S.; Weinberger, S.R.; Wiesner, A. Protein quantification by the SELDI-TOF-MS–based ProteinChip® System. Nat. Methods 2005, 2, 393–395. [Google Scholar] [CrossRef]
- Weiss, A.K.H.; Holzknecht, M.; Cappuccio, E.; Dorigatti, I.; Kreidl, K.; Naschberger, A.; Rupp, B.; Gstach, H.; Jansen-Dürr, P. Expression, Purification, Crystallization, and Enzyme Assays of Fumarylacetoacetate Hydrolase Domain-Containing Proteins. J. Vis. Exp. 2019, 148, e59729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nadeem, H.; Rashid, M.H.; Riaz, M.; Asma, B.; Javed, M.R.; Perveen, R. Invertase from hyper producer strain of Aspergillus niger: Physiochemical properties, thermodynamics and active site residues heat of ionization. Protein Pept. Lett. 2009, 16, 1098–1105. [Google Scholar] [CrossRef] [PubMed]
- Kornberg, A. [1] Why purify enzymes? Methods Enzymol. 1990, 182, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Ventura, S.P.; Coutinho, J.A. Lipase production and purification from fermentation broth using ionic liquids. In Ionic Liquids in Lipid Processing and Analysis; Elsevier: Amsterdam, The Netherlands, 2016; pp. 59–97. [Google Scholar] [CrossRef]
- Zhou, X.; Xia, Y. Expression and characterization of recombinant Locusta migratoria manilensis acetylcholinesterase 1 in Pichia pastoris. Protein Expr. Purif. 2011, 77, 62–67. [Google Scholar] [CrossRef]
- Koteshwara, A.; Philip, N.V.; Aranjani, J.M.; Hariharapura, R.C.; Volety Mallikarjuna, S. A set of simple methods for detection and extraction of laminarinase. Sci. Rep. 2021, 11, 2489. [Google Scholar] [CrossRef]
- Reetz, M.T.; Jaeger, K.-E. Overexpression, immobilization and biotechnological application of Pseudomonas lipases. Chem. Phys. Lipids 1998, 93, 3–14. [Google Scholar] [CrossRef]
- Kimple, M.E.; Brill, A.L.; Pasker, R.L. Overview of affinity tags for protein purification. Curr. Protoc. Protein Sci. 2013, 73, 9.9.1–9.9.23. [Google Scholar] [CrossRef] [Green Version]
- Fakruddin, M.; Mohammad Mazumdar, R.; Bin Mannan, K.S.; Chowdhury, A.; Hossain, M.N. Critical Factors Affecting the Success of Cloning, Expression, and Mass Production of Enzymes by Recombinant E. coli. ISRN Biotechnol. 2013, 2013, 590587. [Google Scholar] [CrossRef] [Green Version]
- de Almeida, J.M.; Moure, V.R.; Müller-Santos, M.; de Souza, E.M.; Pedrosa, F.O.; Mitchell, D.A.; Krieger, N. Tailoring recombinant lipases: Keeping the His-tag favors esterification reactions, removing it favors hydrolysis reactions. Sci. Rep. 2018, 8, 10000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, L.; Liu, Y.; Yin, X.; Zhou, H.; Wu, J.; Wu, M.; Yang, L. Effects of His-tag on Catalytic Activity and Enantioselectivity of Recombinant Transaminases. Appl. Biochem. Biotechnol. 2020, 190, 880–895. [Google Scholar] [CrossRef]
- Booth, W.T.; Schlachter, C.R.; Pote, S.; Ussin, N.; Mank, N.J.; Klapper, V.; Offermann, L.R.; Tang, C.; Hurlburt, B.K.; Chruszcz, M. Impact of an N-terminal Polyhistidine Tag on Protein Thermal Stability. ACS Omega 2018, 3, 760–768. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.; Park, A.K.; Lee, J.H.; Shin, S.C.; Park, H.; Kim, H.W. PsEst3, a new psychrophilic esterase from the Arctic bacterium Paenibacillus sp. R4: Crystallization and X-ray crystallographic analysis. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2018, 74, 367–372. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.W.; Yoo, W.; Park, S.-H.; Le, L.T.H.L.; Jeong, C.-S.; Ryu, B.H.; Shin, S.C.; Kim, H.-W.; Park, H.; Kim, K.K. Structural and functional characterization of a novel cold-active S-formylglutathione hydrolase (Sf SFGH) homolog from Shewanella frigidimarina, a psychrophilic bacterium. Microb. Cell Factories 2019, 18, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Ji, F.; Wang, J.; Pu, Z.; Jiang, B.; Bao, Y. Purification and characterization of a novel organic solvent-tolerant and cold-adapted lipase from Psychrobacter sp. ZY124. Extremophiles 2018, 22, 287–300. [Google Scholar] [CrossRef] [PubMed]
- Hashim, N.H.F.; Mahadi, N.M.; Illias, R.M.; Feroz, S.R.; Abu Bakar, F.D.; Murad, A.M.A. Biochemical and structural characterization of a novel cold-active esterase-like protein from the psychrophilic yeast Glaciozyma antarctica. Extremophiles 2018, 22, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Yasin, M.T.; Ali, Y.; Ahmad, K.; Ghani, A.; Amanat, K.; Basheir, M.M.; Faheem, M.; Hussain, S.; Ahmad, B.; Hussain, A.; et al. Alkaline lipase production by novel meso-tolerant psychrophilic Exiguobacterium sp. strain (AMBL-20) isolated from glacier of northeastern Pakistan. Arch. Microbiol. 2021, 203, 1309–1320. [Google Scholar] [CrossRef]
- Le, L.T.H.L.; Yoo, W.; Jeon, S.; Lee, C.; Kim, K.K.; Lee, J.H.; Kim, T.D. Biodiesel and flavor compound production using a novel promiscuous cold-adapted SGNH-type lipase (HaSGNH1) from the psychrophilic bacterium Halocynthiibacter arcticus. Biotechnol. Biofuels 2020, 13, 55. [Google Scholar] [CrossRef] [Green Version]
- Knepp, Z.J.; Ghaner, A.; Root, K.T. Purification and refolding protocol for cold-active recombinant esterase AaSGNH1 from Aphanizomenon flos-aquae expressed as insoluble inclusion bodies. Prep. Biochem. Biotechnol. 2021, 52, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.-J.; Chen, C.-S.; Liu, H.-T.; Chen, J.-L.; Xia, Y.; Wu, S.-J. Purification, identification and characterization of an esterase with high enantioselectivity to (S)-ethyl indoline-2-carboxylate. Biotechnol. Lett. 2019, 41, 1223–1232. [Google Scholar] [CrossRef]
- Kumar, A.; Mukhia, S.; Kumar, N.; Acharya, V.; Kumar, S.; Kumar, R. A Broad Temperature Active Lipase Purified From a Psychrotrophic Bacterium of Sikkim Himalaya With Potential Application in Detergent Formulation. Front. Bioeng. Biotechnol. 2020, 8, 642. [Google Scholar] [CrossRef] [PubMed]
- Noby, N.; Hussein, A.; Saeed, H.; Embaby, A.M. Recombinant cold-adapted halotolerant, organic solvent-stable esterase (estHIJ) from Bacillus halodurans. Anal. Biochem. 2020, 591, 113554. [Google Scholar] [CrossRef] [PubMed]
- Salleh, A.B.; Mohamad Ali, M.S. Optimization and in silico analysis of a cold-adapted lipase from an antarctic Pseudomonas sp. strain ams8 reaction in triton x-100 reverse micelles. Catalysts 2018, 8, 289. [Google Scholar] [CrossRef]
- Malekabadi, S.; Badoei-dalfard, A.; Karami, Z. Biochemical characterization of a novel cold-active, halophilic and organic solvent-tolerant lipase from B. licheniformis KM12 with potential application for biodiesel production. Int. J. Biol. Macromol. 2018, 109, 389–398. [Google Scholar] [CrossRef]
- Mohamad Tahir, H.; Raja Abd Rahman, R.N.Z.; Chor Leow, A.T.; Mohamad Ali, M.S. Expression, Characterisation and Homology Modelling of a Novel Hormone-Sensitive Lipase (HSL)-Like Esterase from Glaciozyma antarctica. Catalysts 2020, 10, 58. [Google Scholar] [CrossRef] [Green Version]
- Uddin, M.R.; Roy, P.; Mandal, S. Production of extracellular lipase from psychrotrophic bacterium Oceanisphaera sp. RSAP17 isolated from arctic soil. Antonie Leeuwenhoek 2021, 114, 2175–2188. [Google Scholar] [CrossRef]
- Zhong, X.-L.; Tian, Y.-Z.; Jia, M.-L.; Liu, Y.-D.; Cheng, D.; Li, G. Characterization and purification via nucleic acid aptamers of a novel esterase from the metagenome of paper mill wastewater sediments. Int. J. Biol. Macromol. 2020, 153, 441–450. [Google Scholar] [CrossRef]
- Rege, K.; Heng, M. Miniaturized parallel screens to identify chromatographic steps required for recombinant protein purification. Nat. Protoc. 2010, 5, 408–417. [Google Scholar] [CrossRef]
- Fountoulakis, M.; Takács, B. Design of Protein Purification Pathways: Application to the Proteome ofHaemophilus influenzaeUsing Heparin Chromatography. Protein Expr. Purif. 1998, 14, 113–119. [Google Scholar] [CrossRef] [PubMed]
- Fu, J.; Leiros, H.-K.S.; de Pascale, D.; Johnson, K.A.; Blencke, H.-M.; Landfald, B. Functional and structural studies of a novel cold-adapted esterase from an Arctic intertidal metagenomic library. Appl. Microbiol. Biotechnol. 2013, 97, 3965–3978. [Google Scholar] [CrossRef] [PubMed]
- Huo, Y.-Y.; Li, S.; Huang, J.; Rong, Z.; Wang, Z.; Li, Z.; Ji, R.; Kuang, S.; Cui, H.-L.; Li, J.; et al. Crystal structure of Pelagibacterium halotolerans PE8: New insight into its substrate-binding pattern. Sci. Rep. 2017, 7, 4422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lemak, S.; Tchigvintsev, A.; Petit, P.; Flick, R.; Singer, A.U.; Brown, G.; Evdokimova, E.; Egorova, O.; Gonzalez, C.F.; Chernikova, T.N. Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica. Biochem. J. 2012, 445, 193–203. [Google Scholar] [CrossRef] [Green Version]
- Cen, Y.; Singh, W.; Arkin, M.; Moody, T.S.; Huang, M.; Zhou, J.; Wu, Q.; Reetz, M.T. Artificial cysteine-lipases with high activity and altered catalytic mechanism created by laboratory evolution. Nat. Commun. 2019, 10, 3198. [Google Scholar] [CrossRef]
- Park, A.; Kim, S.; Park, J.; Joe, S.; Min, B.; Oh, J.; Song, J.; Park, S.; Park, S.; Lee, H. Structural and experimental evidence for the enantiomeric recognition toward a bulky sec-alcohol by Candida antarctica lipase B. ACS Catal. 2016, 6, 7458–7465. [Google Scholar] [CrossRef]
- Stauch, B.; Fisher, S.J.; Cianci, M. Open and closed states of Candida antarctica lipase B: Protonation and the mechanism of interfacial activation1. J. Lipid Res. 2015, 56, 2348–2358. [Google Scholar] [CrossRef] [Green Version]
- Tang, Q.; Popowicz, G.M.; Wang, X.; Liu, J.; Pavlidis, I.V.; Wang, Y. Lipase-driven epoxidation is a two-stage synergistic process. ChemistrySelect 2016, 1, 836–839. [Google Scholar] [CrossRef]
- Zhang, Y.; Ding, H.-T.; Jiang, W.-X.; Zhang, X.; Cao, H.-Y.; Wang, J.-P.; Li, C.-Y.; Huang, F.; Zhang, X.-Y.; Chen, X.-L. Active site architecture of an acetyl xylan esterase indicates a novel cold adaptation strategy. J. Biol. Chem. 2021, 297, 100841. [Google Scholar] [CrossRef]
- van der Ent, F.; Lund, B.A.; Svalberg, L.; Purg, M.; Chukwu, G.; Widersten, M.; Isaksen, G.V.; Brandsdal, B.O.; Åqvist, J. Structure and Mechanism of a Cold-Adapted Bacterial Lipase. Biochemistry 2022, 61, 933–942. [Google Scholar] [CrossRef]
- Hedstrom, L. Serine Protease Mechanism and Specificity. Chem. Rev. 2002, 102, 4501–4524. [Google Scholar] [CrossRef]
- Jegannathan, K.R.; Abang, S.; Poncelet, D.; Chan, E.S.; Ravindra, P. Production of Biodiesel Using Immobilized Lipase—A Critical Review. Crit. Rev. Biotechnol. 2008, 28, 253–264. [Google Scholar] [CrossRef] [PubMed]
- Ben Hlima, H.; Dammak, M.; Karray, A.; Drira, M.; Michaud, P.; Fendri, I.; Abdelkafi, S. Molecular and Structural Characterizations of Lipases from Chlorella by Functional Genomics. Mar. Drugs 2021, 19, 70. [Google Scholar] [CrossRef] [PubMed]
- Khersonsky, O.; Fleishman, S.J. Why reinvent the wheel? Building new proteins based on ready-made parts. Protein Sci. 2016, 25, 1179–1187. [Google Scholar] [CrossRef] [Green Version]
- Feller, G.; Gerday, C. Psychrophilic enzymes: Molecular basis of cold adaptation. Cell. Mol. Life Sci. CMLS 1997, 53, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Brininger, C.; Spradlin, S.; Cobani, L.; Evilia, C. The more adaptive to change, the more likely you are to survive: Protein adaptation in extremophiles. In Cell & Developmental Biology; Elsevier: Amsterdam, The Netherlands, 2018; pp. 158–169. [Google Scholar]
- Noby, N.; Auhim, S.; Johnson, R.; Worthy, H.; Saeed, H.; Hussein, A.; Rizkallah, P.; Wells, S.; Jones, D. Structure and dynamics of a cold-active esterase reveals water entropy and active site accessibility as the likely drivers for cold-adaptation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Bagchi, B. The entropy of water. In Water in Biological and Chemical Processes: From Structure and Dynamics to Function; Cambridge University Press: Cambridge, UK, 2013; pp. 287–304. [Google Scholar] [CrossRef]
- Noby, N.; Auhim, H.S.; Winter, S.; Worthy, H.L.; Embaby, A.M.; Saeed, H.; Hussein, A.; Pudney, C.R.; Rizkallah, P.J.; Wells, S.A. Structure and in silico simulations of a cold-active esterase reveals its prime cold-adaptation mechanism. Open Biol. 2021, 11, 210182. [Google Scholar] [CrossRef]
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Nussinov, R. Different roles of electrostatics in heat and in cold: Adaptation by citrate synthase. ChemBioChem 2004, 5, 280–290. [Google Scholar] [CrossRef]
- Parvizpour, S.; Hussin, N.; Shamsir, M.S.; Razmara, J. Psychrophilic enzymes: Structural adaptation, pharmaceutical and industrial applications. Appl. Microbiol. Biotechnol. 2021, 105, 899–907. [Google Scholar] [CrossRef]
- Paredes, D.I.; Watters, K.; Pitman, D.J.; Bystroff, C.; Dordick, J.S. Comparative void-volume analysis of psychrophilic and mesophilic enzymes: Structural bioinformatics of psychrophilic enzymes reveals sources of core flexibility. BMC Struct. Biol. 2011, 11, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreini, C.; Bertini, I.; Cavallaro, G.; Holliday, G.L.; Thornton, J.M. Metal ions in biological catalysis: From enzyme databases to general principles. JBIC J. Biol. Inorg. Chem. 2008, 13, 1205–1218. [Google Scholar] [CrossRef]
- Horitani, M.; Kusubayashi, K.; Oshima, K.; Yato, A.; Sugimoto, H.; Watanabe, K. X-ray crystallography and electron paramagnetic resonance spectroscopy reveal active site rearrangement of cold-adapted inorganic pyrophosphatase. Sci. Rep. 2020, 10, 4368. [Google Scholar] [CrossRef] [Green Version]
- Marchetti, A.; Orlando, M.; Mangiagalli, M.; Lotti, M. A cold-active esterase enhances mesophilic properties through Mn2+ binding. FEBS J. 2022. early version. [Google Scholar] [CrossRef]
- Furnham, N.; Dawson, N.L.; Rahman, S.A.; Thornton, J.M.; Orengo, C.A. Large-Scale Analysis Exploring Evolution of Catalytic Machineries and Mechanisms in Enzyme Superfamilies. J. Mol. Biol. 2016, 428, 253–267. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rizzello, A.; Romano, A.; Kottra, G.; Acierno, R.; Storelli, C.; Verri, T.; Daniel, H.; Maffia, M. Protein cold adaptation strategy via a unique seven-amino acid domain in the icefish (Chionodraco hamatus) PEPT1 transporter. Proc. Natl. Acad. Sci. USA 2013, 110, 7068–7073. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angelin, J.; Kavitha, M. Chapter 5—Molecular mechanisms behind the cold and hot adaptation in extremozymes. In Extremozymes and Their Industrial Applications; Arora, N.K., Agnihotri, S., Mishra, J., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 141–176. [Google Scholar] [CrossRef]
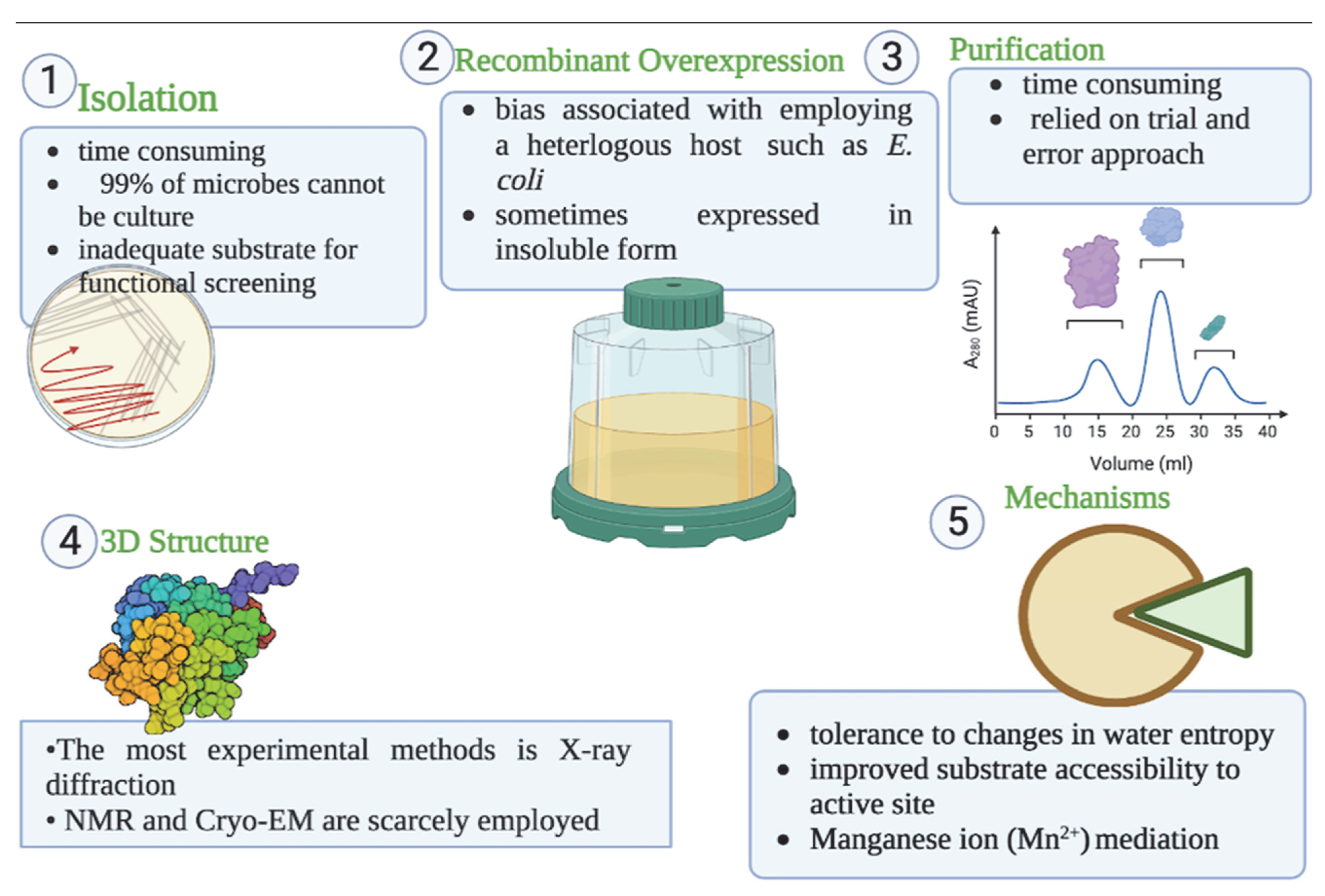
| Enzymes | PDB Code | Organism | Expression System | Experimental Method | Resolution (Å) | Ligand | References |
|---|---|---|---|---|---|---|---|
| Esterase | 4V2I | Thalassospira sp. | Escherichia coli BL21(DE3) | X-ray Diffraction | 1.69 | Magnesium ion | [50] |
| Esterase | 4AO8 | Arctic Intertidal Metagenomic Library. | Escherichia coli K-12 | X-ray Diffraction | 1.61 | Dihydroxyethyl Ester | [120] |
| Esterase | 5DWD | Pelagibacterium halotolerans PE8 | Escherichia coli | X-ray Diffraction | 1.66 | 2-(2-{2-[2-(2-Methoxy-Ethoxy)-Eth0xy]-Ethoxy}-Ethoxy)-Ethanol | [121] |
| Esterase | 3I6Y | Oleispira antarctica | Escherichia coli BL21(DE3) | X-ray Diffraction | 1.75 | Dihydroxyethyl Ester | [122] |
| Lipase | 6ISP | Laboratory Evolution of Moesziomyces antarcticus | Escherichia coli BL21(DE3) | X-ray Diffraction | 1.88 | N, N-Bis(3-D-Gluconamidopropyl) Deoxycholamide and Calcium Ion | [123] |
| Lipase | 6ISR | Laboratory Evolution of Moesziomyces antarcticus | Escherichia coli BL21(DE3) | X-ray Diffraction | 2.60 | Tetraethylene Glycol | [123] |
| Lipase | 6ISQ | Laboratory Evolution of Moesziomyces antarcticus | Escherichia coli BL21(DE3) | X-ray Diffraction | 1.86 | 1,2-Ethanediol | [123] |
| Lipase | 5GV5 | Moesziomyces antarcticus | Aspergillus niger | X-ray Diffraction | 2.89 | [(1s)-2-(Methoxycarbonylamino)-1-Phenyl-Ethoxy]-Propyl-Phosphinic Acid | [124] |
| Lipase | 5A6V | Moesziomyces antarcticus | Aspergillus oryzae | X-ray Diffraction | 2.28 | Xenon | [125] |
| Lipase | 5A71 | Moesziomyces antarcticus | Aspergillus oryzae | X-ray Diffraction | 0.91 | Isopropyl alcohol | [125] |
| Lipase | 5CH8 | Penicillium cyclopium | Komagataella pastoris | X-ray Diffraction | 1.62 | Glycerol | [126] |
| Esterase | 7B1X | uncultured bacterium | Escherichia coli | X-ray Diffraction | 2.30 | None | [36] |
| Esterase | 7DDY | Arcticibacterium luteifluviistationis | Escherichia coli BL21(DE3) | X-ray Diffraction | 2.50 | None | [127] |
| EsteraseD | 6JZL | Shewanella frigidimarina | Escherichia coli | X-ray Diffraction | 2.32 | None | [40] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Matinja, A.I.; Kamarudin, N.H.A.; Leow, A.T.C.; Oslan, S.N.; Ali, M.S.M. Cold-Active Lipases and Esterases: A Review on Recombinant Overexpression and Other Essential Issues. Int. J. Mol. Sci. 2022, 23, 15394. https://doi.org/10.3390/ijms232315394
Matinja AI, Kamarudin NHA, Leow ATC, Oslan SN, Ali MSM. Cold-Active Lipases and Esterases: A Review on Recombinant Overexpression and Other Essential Issues. International Journal of Molecular Sciences. 2022; 23(23):15394. https://doi.org/10.3390/ijms232315394
Chicago/Turabian StyleMatinja, Adamu Idris, Nor Hafizah Ahmad Kamarudin, Adam Thean Chor Leow, Siti Nurbaya Oslan, and Mohd Shukuri Mohamad Ali. 2022. "Cold-Active Lipases and Esterases: A Review on Recombinant Overexpression and Other Essential Issues" International Journal of Molecular Sciences 23, no. 23: 15394. https://doi.org/10.3390/ijms232315394
APA StyleMatinja, A. I., Kamarudin, N. H. A., Leow, A. T. C., Oslan, S. N., & Ali, M. S. M. (2022). Cold-Active Lipases and Esterases: A Review on Recombinant Overexpression and Other Essential Issues. International Journal of Molecular Sciences, 23(23), 15394. https://doi.org/10.3390/ijms232315394






