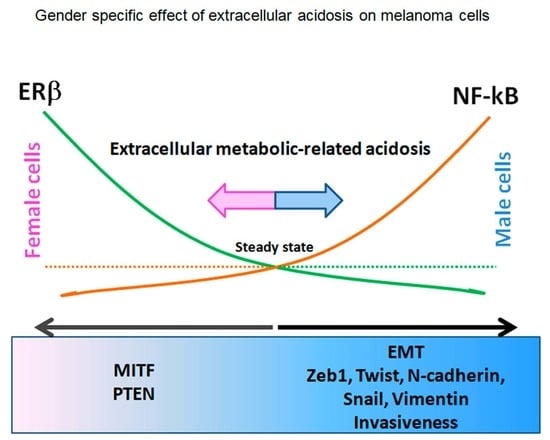
Extracellular Acidosis Differentially Regulates Estrogen Receptor β-Dependent EMT Reprogramming in Female and Male Melanoma Cells
Abstract
1. Introduction
2. Results
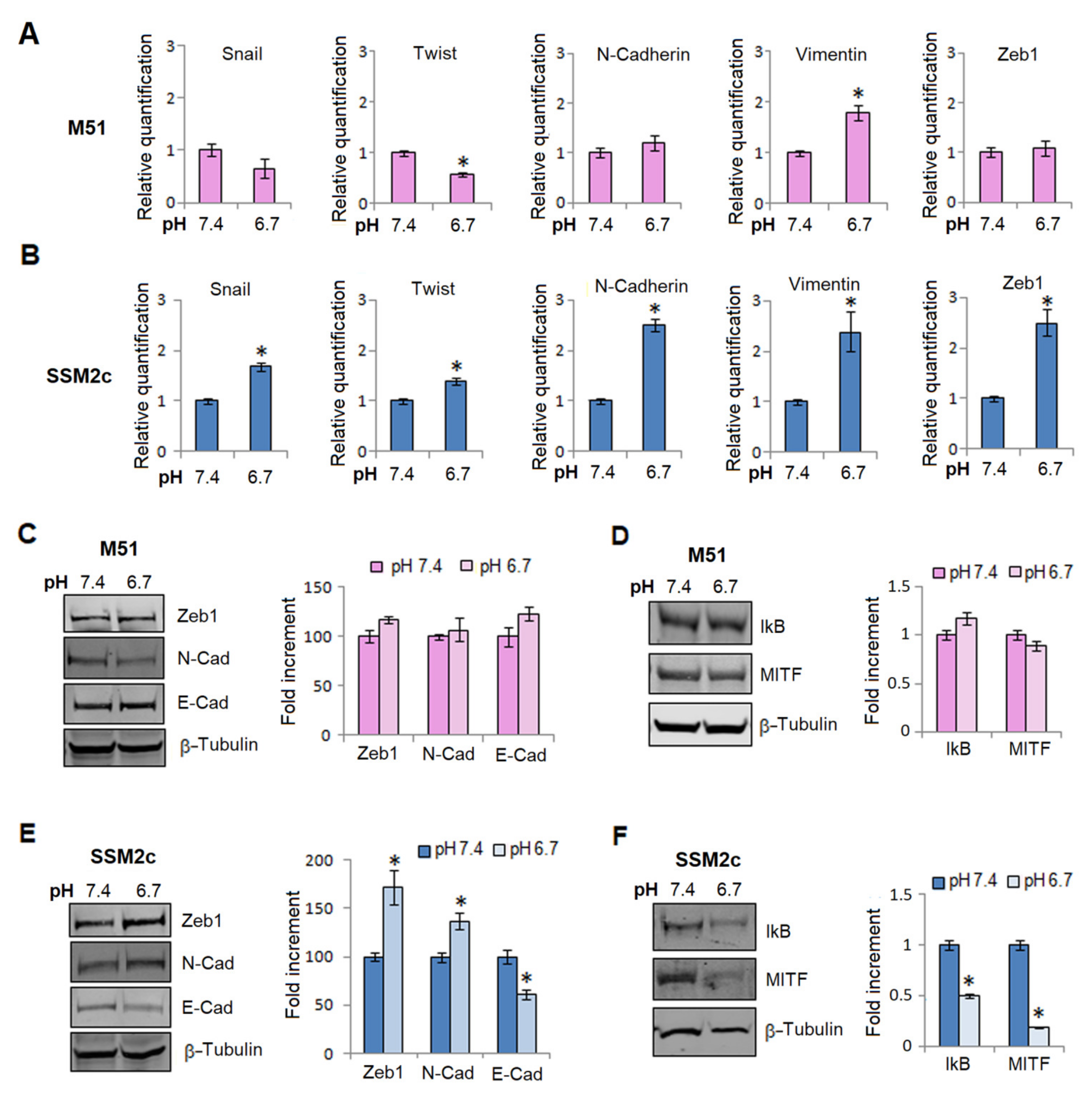
2.1. ERβ Expression and EMT Reprogramming in Melanoma Cells Exposed to Extracellular Acidosis
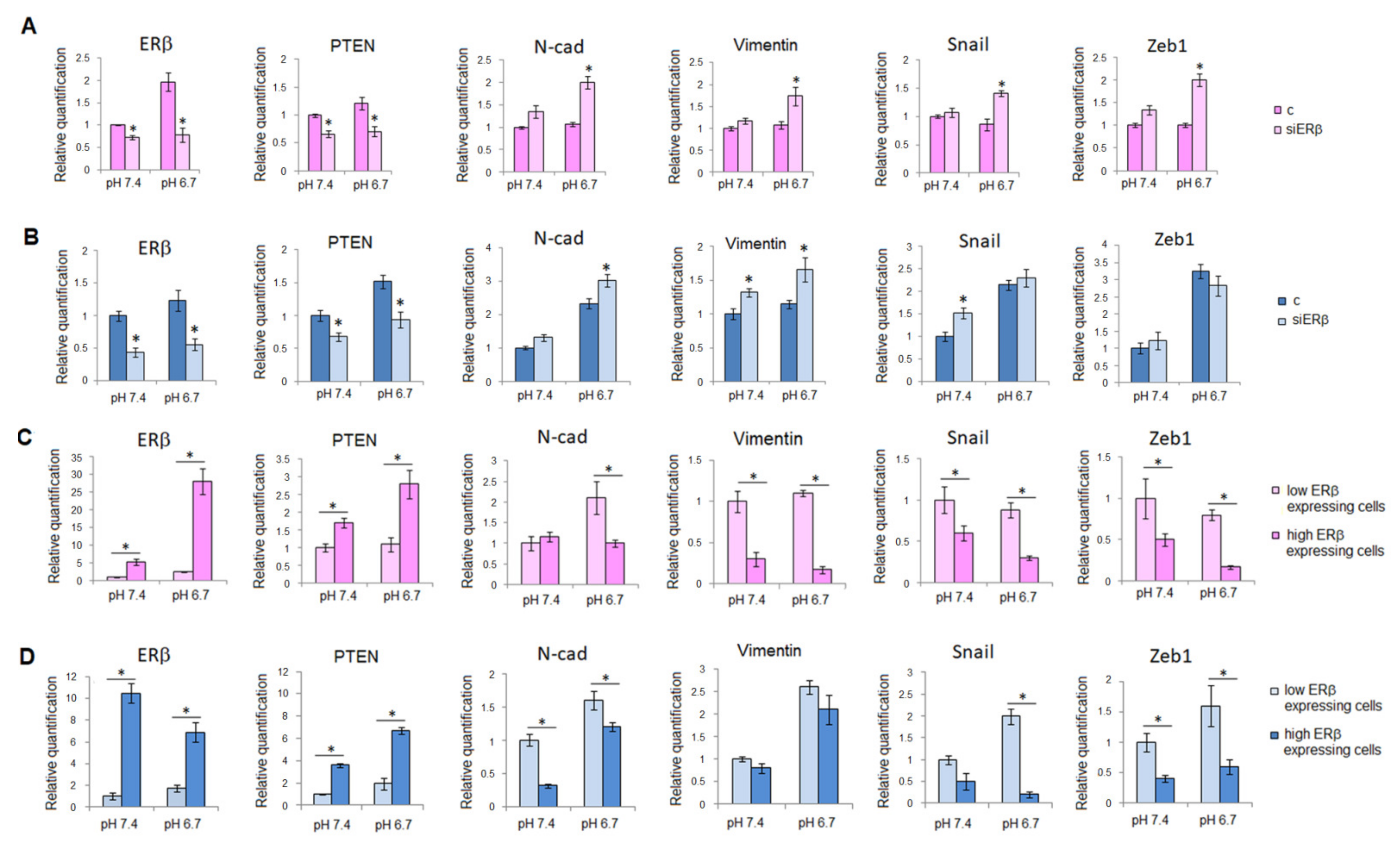
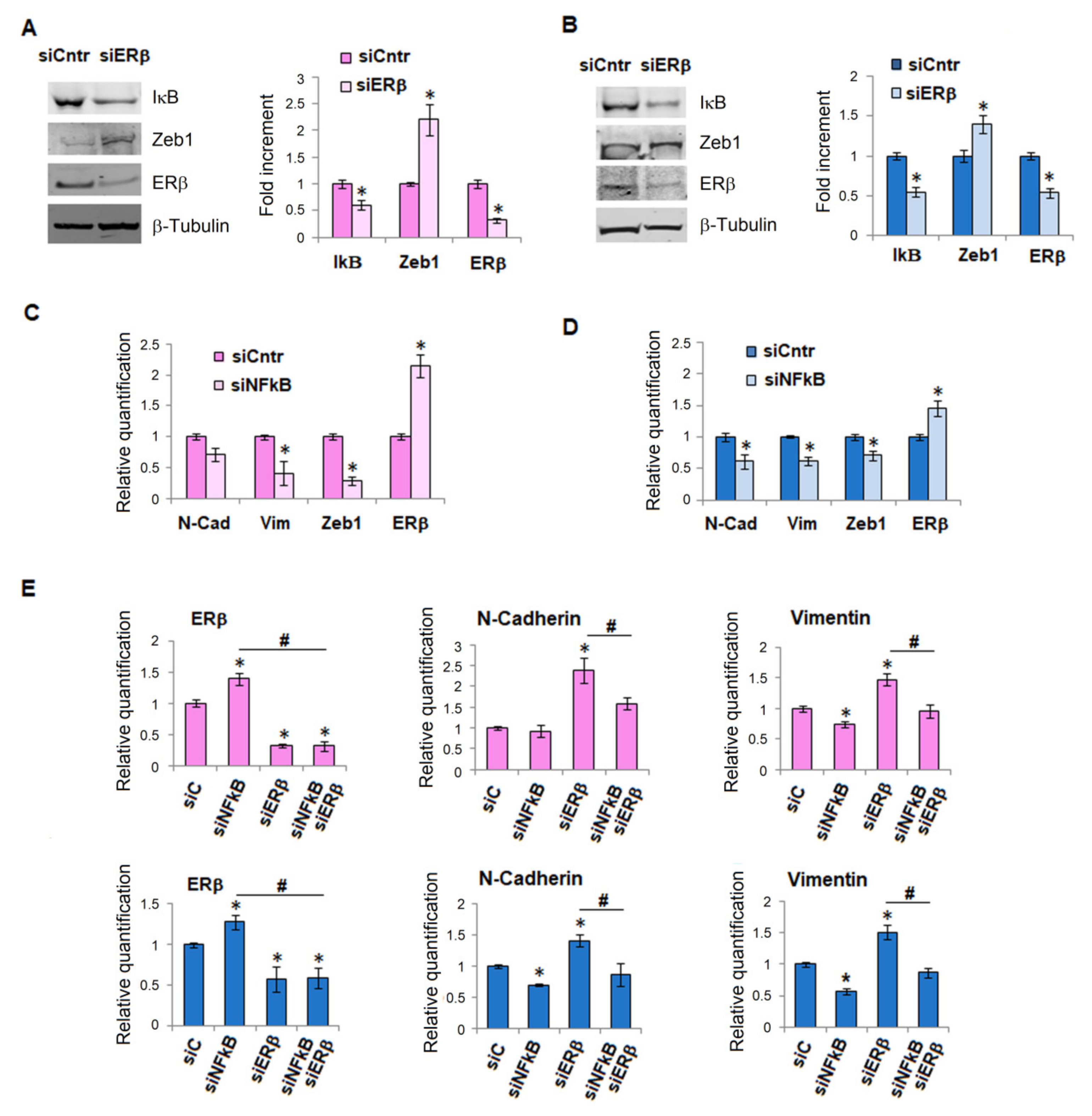
2.2. ERβ as a Potent Inhibitor of EMT in Human Melanoma Cells
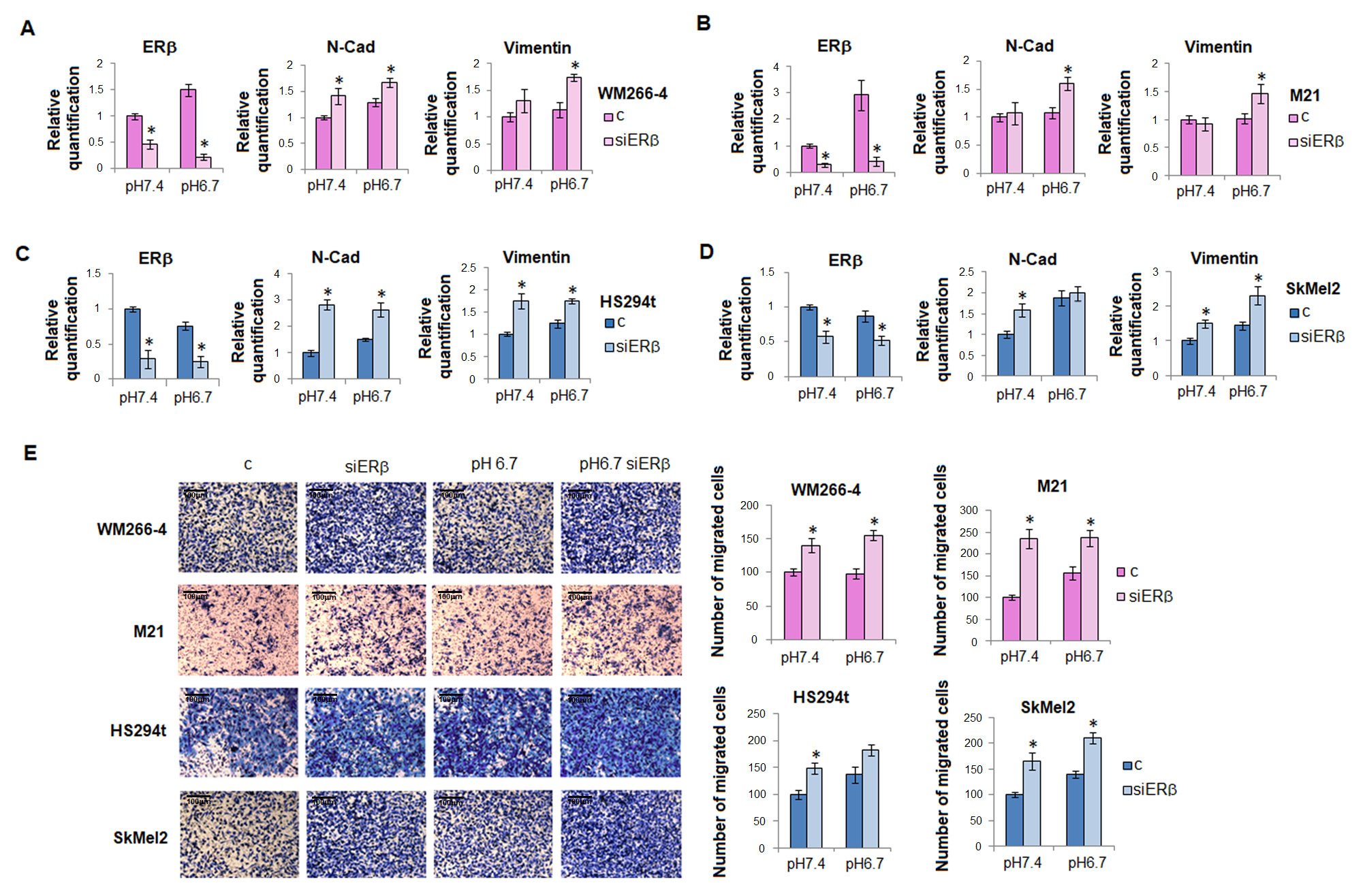
2.3. ERβ-NF-κB Crosstalk in EMT of Melanoma Cells
3. Discussion
4. Materials and Methods
4.1. Cell Cultures
4.2. Quantitative Real-Time PCR (qPCR)
4.3. Western Blotting Analysis
4.4. Flow Cytometry and Cell Sorting Experiments
4.5. siRNA Transfection
4.6. Invasion Assay
4.7. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Maldonado, J.L.; Fridlyand, J.; Patel, H.; Jain, A.N.; Busam, K.; Kageshita, T.; Ono, T.; Albertson, D.G.; Pinkel, D.; Bastian, B.C. Determinants of BRAF Mutations in Primary Melanomas. J. Natl. Cancer Inst. 2003, 95, 1878–1890. [Google Scholar] [CrossRef] [PubMed]
- Bellenghi, M.; Puglisi, R.; Pontecorvi, G.; De Feo, A.; Carè, A.; Mattia, G. Sex and Gender Disparities in Melanoma. Cancers 2020, 12, 1819. [Google Scholar] [CrossRef] [PubMed]
- Joosse, A.; de Vries, E.; Eckel, R.; Nijsten, T.; Eggermont, A.M.M.; Hölzel, D.; Coebergh, J.W.W.; Engel, J. Gender Differences in Melanoma Survival: Female Patients Have a Decreased Risk of Metastasis. J. Investig. Dermatol. 2011, 131, 719–726. [Google Scholar] [CrossRef] [PubMed]
- Roh, M.R.; Eliades, P.; Gupta, S.; Grant-Kels, J.M.; Tsao, H. Cutaneous melanoma in women. Int. J. Women’s Dermatol. 2017, 3, S11–S15. [Google Scholar] [CrossRef] [PubMed]
- Marzagalli, M.; Montagnani Marelli, M.; Casati, L.; Fontana, F.; Moretti, R.M.; Limonta, P. Estrogen Receptor β in Melanoma: From Molecular Insights to Potential Clinical Utility. Front. Endocrinol. 2016, 7, 140. [Google Scholar] [CrossRef]
- McQuade, J.L.; Daniel, C.R.; Hess, K.R.; Davies, M.A. Sex as a predictor of response to cancer immunotherapy. Lancet Oncol. 2018, 19, e376. [Google Scholar] [CrossRef]
- Schneider, G.; Kirschner, M.A.; Berkowitz, R.; Ertel, N.H. Increased Estrogen Production in Obese Men. J. Clin. Endocrinol. Metab. 1979, 48, 633–638. [Google Scholar] [CrossRef]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10, 52. [Google Scholar] [CrossRef]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial–mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Najem, A.; Soumoy, L.; Sabbah, M.; Krayem, M.; Awada, A.; Journe, F.; Ghanem, G.E. Understanding Molecular Mechanisms of Phenotype Switching and Crosstalk with TME to Reveal New Vulnerabilities of Melanoma. Cells 2022, 11, 1157. [Google Scholar] [CrossRef]
- Vaupel, P.; Kallinowski, F.; Okunieff, P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: A review. Cancer Res. 1989, 49, 6449–6465. [Google Scholar]
- Wike-Hooley, J.L.; Haveman, J.; Reinhold, H.S. The relevance of tumour pH to the treatment of malignant disease. Radiother. Oncol. 1984, 2, 343–366. [Google Scholar] [CrossRef]
- Ruzzolini, J.; Peppicelli, S.; Andreucci, E.; Bianchini, F.; Margheri, F.; Laurenzana, A.; Fibbi, G.; Pimpinelli, N.; Calorini, L. Everolimus selectively targets vemurafenib resistant BRAFV600E melanoma cells adapted to low pH. Cancer Lett. 2017, 408, 43–54. [Google Scholar] [CrossRef]
- Peppicelli, S.; Bianchini, F.; Torre, E.; Calorini, L. Contribution of acidic melanoma cells undergoing epithelial-to-mesenchymal transition to aggressiveness of non-acidic melanoma cells. Clin. Exp. Metastasis 2014, 31, 423–433. [Google Scholar] [CrossRef]
- Andreucci, E.; Peppicelli, S.; Ruzzolini, J.; Bianchini, F.; Biagioni, A.; Papucci, L.; Magnelli, L.; Mazzanti, B.; Stecca, B.; Calorini, L. The acidic tumor microenvironment drives a stem-like phenotype in melanoma cells. J. Mol. Med. 2020, 98, 1431–1446. [Google Scholar] [CrossRef]
- Lindberg, K.; Helguero, L.A.; Omoto, Y.; Gustafsson, J.-Å.; Haldosén, L.-A. Estrogen receptor β represses Akt signaling in breast cancer cells via downregulation of HER2/HER3 and upregulation of PTEN: Implications for tamoxifen sensitivity. Breast Cancer Res. 2011, 13, R43. [Google Scholar] [CrossRef]
- Qi, Y.; Liu, J.; Chao, J.; Scheuerman, M.P.; Rahimi, S.A.; Lee, L.Y.; Li, S. PTEN suppresses epithelial–mesenchymal transition and cancer stem cell activity by downregulating Abi1. Sci. Rep. 2020, 10, 12685. [Google Scholar] [CrossRef]
- Xia, Y.; Shen, S.; Verma, I.M. NF-κB, an active player in human cancers. Cancer Immunol. Res. 2014, 2, 823–830. [Google Scholar] [CrossRef]
- Wu, Y.; Deng, J.; Rychahou, P.G.; Qiu, S.; Evers, B.M.; Zhou, B.P. Stabilization of Snail by NF-κB Is Required for Inflammation-Induced Cell Migration and Invasion. Cancer Cell 2009, 15, 416–428. [Google Scholar] [CrossRef]
- Webb, B.A.; Chimenti, M.; Jacobson, M.P.; Barber, D.L. Dysregulated pH: A perfect storm for cancer progression. Nat. Rev. Cancer 2011, 11, 671–677. [Google Scholar] [CrossRef]
- Corbet, C.; Feron, O. Tumour acidosis: From the passenger to the driver’s seat. Nat. Rev. Cancer 2017, 17, 577–593. [Google Scholar] [CrossRef] [PubMed]
- Fischer, K.; Hoffmann, P.; Voelkl, S.; Meidenbauer, N.; Ammer, J.; Edinger, M.; Gottfried, E.; Schwarz, S.; Rothe, G.; Hoves, S.; et al. Inhibitory effect of tumor cell–derived lactic acid on human T cells. Blood 2007, 109, 3812–3819. [Google Scholar] [CrossRef] [PubMed]
- Thews, O.; Gassner, B.; Kelleher, D.K.; Schwerd, G.; Gekle, M. Impact of Extracellular Acidity on the Activity of P-glycoprotein and the Cytotoxicity of Chemotherapeutic Drugs. Neoplasia 2006, 8, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Walenta, S.; Wetterling, M.; Lehrke, M.; Schwickert, G.; Sundfør, K.; Rofstad, E.K.; Mueller-Klieser, W. High lactate levels predict likelihood of metastases, tumor recurrence, and restricted patient survival in human cervical cancers. Cancer Res. 2000, 60, 916–921. [Google Scholar] [PubMed]
- Gillies, R.J.; Verduzco, D.; Gatenby, R.A. Evolutionary dynamics of carcinogenesis and why targeted therapy does not work. Nat. Rev. Cancer 2012, 12, 487–493. [Google Scholar] [CrossRef]
- De Bem Prunes, B.; Nunes, J.S.; da Silva, V.P.; Laureano, N.K.; Gonçalves, D.R.; Machado, I.S.; Barbosa, S.; Lamers, M.L.; Rados, P.V.; Kurth, I.; et al. The role of tumor acidification in aggressiveness, cell dissemination and treatment resistance of oral squamous cell carcinoma. Life Sci. 2022, 288, 120163. [Google Scholar] [CrossRef]
- Schmidt, A.N.; Nanney, L.B.; Boyd, A.S.; King, L.E.; Ellis, D.L. Oestrogen receptor-beta expression in melanocytic lesions. Exp. Dermatol. 2006, 15, 971–980. [Google Scholar] [CrossRef]
- De Giorgi, V.; Gori, A.; Grazzini, M.; Rossari, S.; Scarfì, F.; Corciova, S.; Verdelli, A.; Lotti, T.; Massi, D. Estrogens, estrogen receptors and melanoma. Expert Rev. Anticancer Ther. 2011, 11, 739–747. [Google Scholar] [CrossRef]
- De Giorgi, V.; Mavilia, C.; Massi, D.; Gozzini, A.; Aragona, P.; Tanini, A.; Sestini, S.; Paglierani, M.; Boddi, V.; Brandi, M.L.; et al. Estrogen Receptor Expression in Cutaneous Melanoma: A Real-Time Reverse Transcriptase–Polymerase Chain Reaction and Immunohistochemical Study. Arch. Dermatol. 2009, 145, 30–36. [Google Scholar] [CrossRef]
- Pietrobono, S.; Anichini, G.; Sala, C.; Manetti, F.; Almada, L.L.; Pepe, S.; Carr, R.M.; Paradise, B.D.; Sarkaria, J.N.; Davila, J.I.; et al. ST3GAL1 is a target of the SOX2-GLI1 transcriptional complex and promotes melanoma metastasis through AXL. Nat. Commun. 2020, 11, 5865. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Polyak, K.; Weinberg, R.A. Transitions between epithelial and mesenchymal states: Acquisition of malignant and stem cell traits. Nat. Rev. Cancer 2009, 9, 265–273. [Google Scholar] [CrossRef]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial–mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef]
- Hazan, R.B.; Phillips, G.R.; Qiao, R.F.; Norton, L.; Aaronson, S.A. Exogenous Expression of N-Cadherin in Breast Cancer Cells Induces Cell Migration, Invasion, and Metastasis. J. Cell Biol. 2000, 148, 779–790. [Google Scholar] [CrossRef]
- Alonso, S.R.; Tracey, L.; Ortiz, P.; Pérez-Gómez, B.; Palacios, J.; Pollán, M.; Linares, J.; Serrano, S.; Sáez-Castillo, A.I.; Sánchez, L.; et al. A High-Throughput Study in Melanoma Identifies Epithelial-Mesenchymal Transition as a Major Determinant of Metastasis. Cancer Res. 2007, 67, 3450–3460. [Google Scholar] [CrossRef]
- Krengel, S.; Grotelüschen, F.; Bartsch, S.; Tronnier, M. Cadherin expression pattern in melanocytic tumors more likely depends on the melanocyte environment than on tumor cell progression: Cadherin expression pattern in melanocytic tumors. J. Cutan. Pathol. 2004, 31, 1–7. [Google Scholar] [CrossRef]
- Lin, K.; Baritaki, S.; Militello, L.; Malaponte, G.; Bevelacqua, Y.; Bonavida, B. The Role of B-RAF Mutations in Melanoma and the Induction of EMT via Dysregulation of the NF- B/Snail/RKIP/PTEN Circuit. Genes Cancer 2010, 1, 409–420. [Google Scholar] [CrossRef]
- Pires, B.R.B.; Mencalha, A.L.; Ferreira, G.M.; de Souza, W.F.; Morgado-Díaz, J.A.; Maia, A.M.; Corrêa, S.; Abdelhay, E.S.F.W. NF-kappaB Is Involved in the Regulation of EMT Genes in Breast Cancer Cells. PLoS ONE 2017, 12, e0169622. [Google Scholar] [CrossRef]
- Zhang, Y.; Yin, C.; Zhou, X.; Wu, Y.; Wang, L. Silencing of estrogen receptor β promotes the invasion and migration of osteosarcoma cells through activating Wnt signaling pathway. OncoTargets Ther. 2019, 12, 6779–6788. [Google Scholar] [CrossRef]
- Frasor, J.; Weaver, A.; Pradhan, M.; Dai, Y.; Miller, L.D.; Lin, C.-Y.; Stanculescu, A. Positive Cross-Talk between Estrogen Receptor and NF-κB in Breast Cancer. Cancer Res. 2009, 69, 8918–8925. [Google Scholar] [CrossRef]
- Mak, P.; Li, J.; Samanta, S.; Mercurio, A.M. ERβ regulation of NF-κB activation in prostate cancer is mediated by HIF-1. Oncotarget 2015, 6, 40247–40254. [Google Scholar] [CrossRef] [PubMed]
- Xiao, L.; Luo, Y.; Tai, R.; Zhang, N. Estrogen receptor β suppresses inflammation and the progression of prostate cancer. Mol. Med. Rep. 2019, 19, 3555–3563. [Google Scholar] [CrossRef]
- Saleiro, D.; Murillo, G.; Lubahn, D.B.; Kopelovich, L.; Korach, K.S.; Mehta, R.G. Enhanced Induction of Mucin-Depleted Foci in Estrogen Receptor β Knockout Mice. Cancer Prev. Res. 2010, 3, 1198–1204. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Eghbali-Webb, M. Gender-Related Differences in Basal and Hypoxia-Induced Activation of Signal Transduction Pathways Controlling Cell Cycle Progression and Apoptosis, in Cardiac Fibroblasts. Endocrine 2002, 18, 137–146. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.R. In situ detection of mycoplasma contamination in cell cultures by fluorescent Hoechst 33258 stain. Exp. Cell Res. 1977, 104, 255–262. [Google Scholar] [CrossRef]

| Gene | Primer Forward | Primer Reverse | Amplicon Length |
|---|---|---|---|
| β-actin | CCAACCGCGAGAAGATGA | CCAGAGGCGTACAGGGATAG | 97 bp |
| ERβ | TGATGTCCTTGACCAAGCTG | CACTTGGTCGTACAGGCTCGA | 101 bp |
| N-Cadherin | CACTGCTCAGGACCCAGAT | TAAGCCGAGTGATGGTCC | 419 bp |
| PTEN | TGAGTTCCCTCAGCCGTTACCT | GAGGTTTCCTCTGGTCCTGGTA | 138 bp |
| Snail | CCCAGTGCCTCGACCACTAT | CCAGATGAGCATTGGCAG | 201 bp |
| Twist | CGGGAGTCCGCAGTCTTA | TGAATCTTGCTCAGCTTGTC | 150 bp |
| Vimentin | TGTCCAAATCGATGTGGATGTTTC | TTGTACCATTCTTCTGCCTCCTG | 117 bp |
| Zeb1 | GGCATACACCTACTCAACTACGG | TGGGCGGTGTAGAATCAGAGTC | 155 bp |
| 18s | CGCCGCTAGAGGTGAAATTCT | CGAACCTCCGA CTTTCGTTCT | 101 bp |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Peppicelli, S.; Ruzzolini, J.; Lulli, M.; Biagioni, A.; Bianchini, F.; Caldarella, A.; Nediani, C.; Andreucci, E.; Calorini, L. Extracellular Acidosis Differentially Regulates Estrogen Receptor β-Dependent EMT Reprogramming in Female and Male Melanoma Cells. Int. J. Mol. Sci. 2022, 23, 15374. https://doi.org/10.3390/ijms232315374
Peppicelli S, Ruzzolini J, Lulli M, Biagioni A, Bianchini F, Caldarella A, Nediani C, Andreucci E, Calorini L. Extracellular Acidosis Differentially Regulates Estrogen Receptor β-Dependent EMT Reprogramming in Female and Male Melanoma Cells. International Journal of Molecular Sciences. 2022; 23(23):15374. https://doi.org/10.3390/ijms232315374
Chicago/Turabian StylePeppicelli, Silvia, Jessica Ruzzolini, Matteo Lulli, Alessio Biagioni, Francesca Bianchini, Adele Caldarella, Chiara Nediani, Elena Andreucci, and Lido Calorini. 2022. "Extracellular Acidosis Differentially Regulates Estrogen Receptor β-Dependent EMT Reprogramming in Female and Male Melanoma Cells" International Journal of Molecular Sciences 23, no. 23: 15374. https://doi.org/10.3390/ijms232315374
APA StylePeppicelli, S., Ruzzolini, J., Lulli, M., Biagioni, A., Bianchini, F., Caldarella, A., Nediani, C., Andreucci, E., & Calorini, L. (2022). Extracellular Acidosis Differentially Regulates Estrogen Receptor β-Dependent EMT Reprogramming in Female and Male Melanoma Cells. International Journal of Molecular Sciences, 23(23), 15374. https://doi.org/10.3390/ijms232315374