Comparative Transcriptome Analysis of CCCH Family in Roles of Flower Opening and Abiotic Stress in Osmanthus fragrans
Abstract
1. Introduction
2. Results
2.1. Genome-Wide Identification of the OfCCCHs
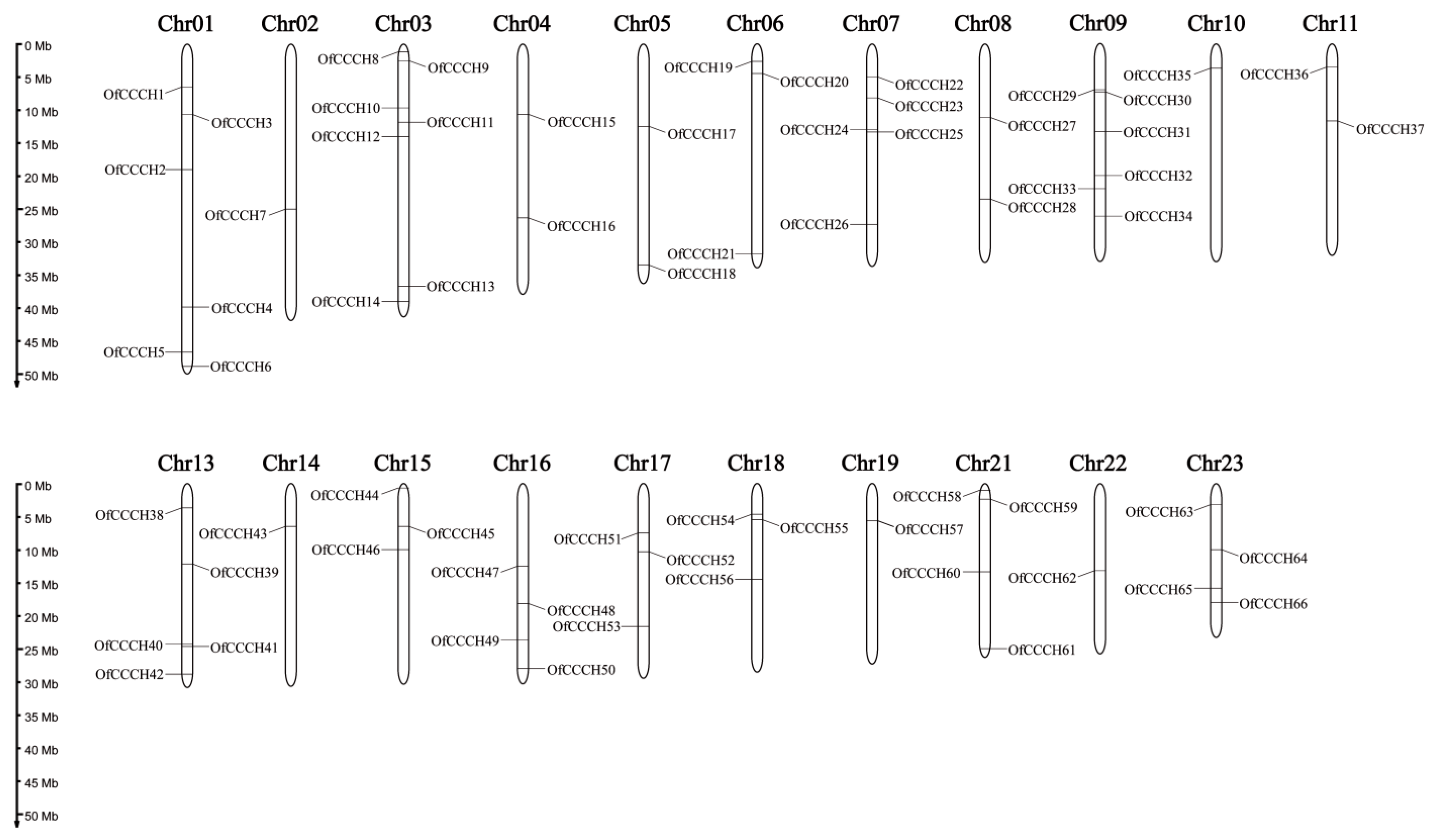
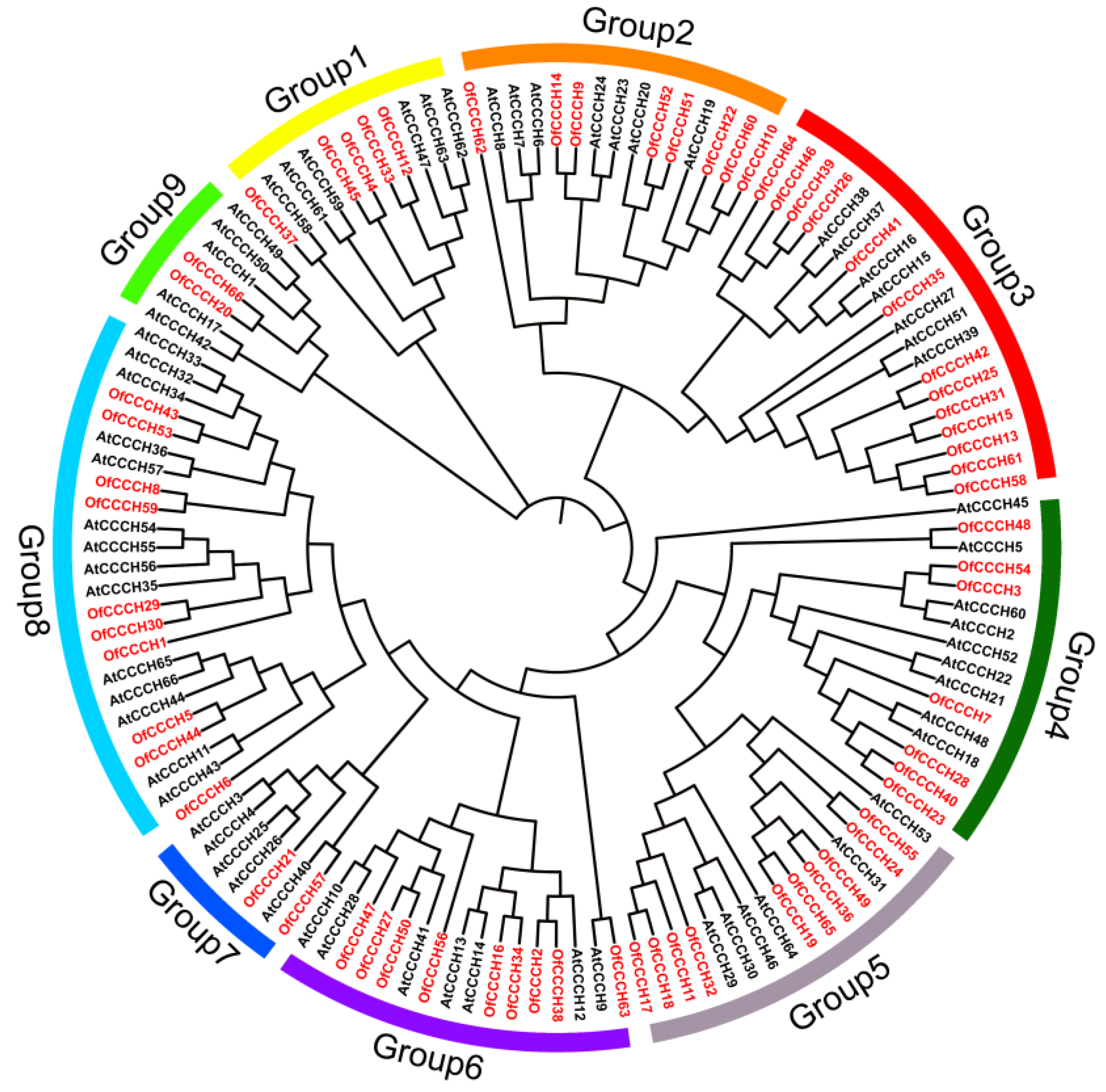
2.2. Phylogenetic Analysis, Gene Duplication, and Synteny Analysis
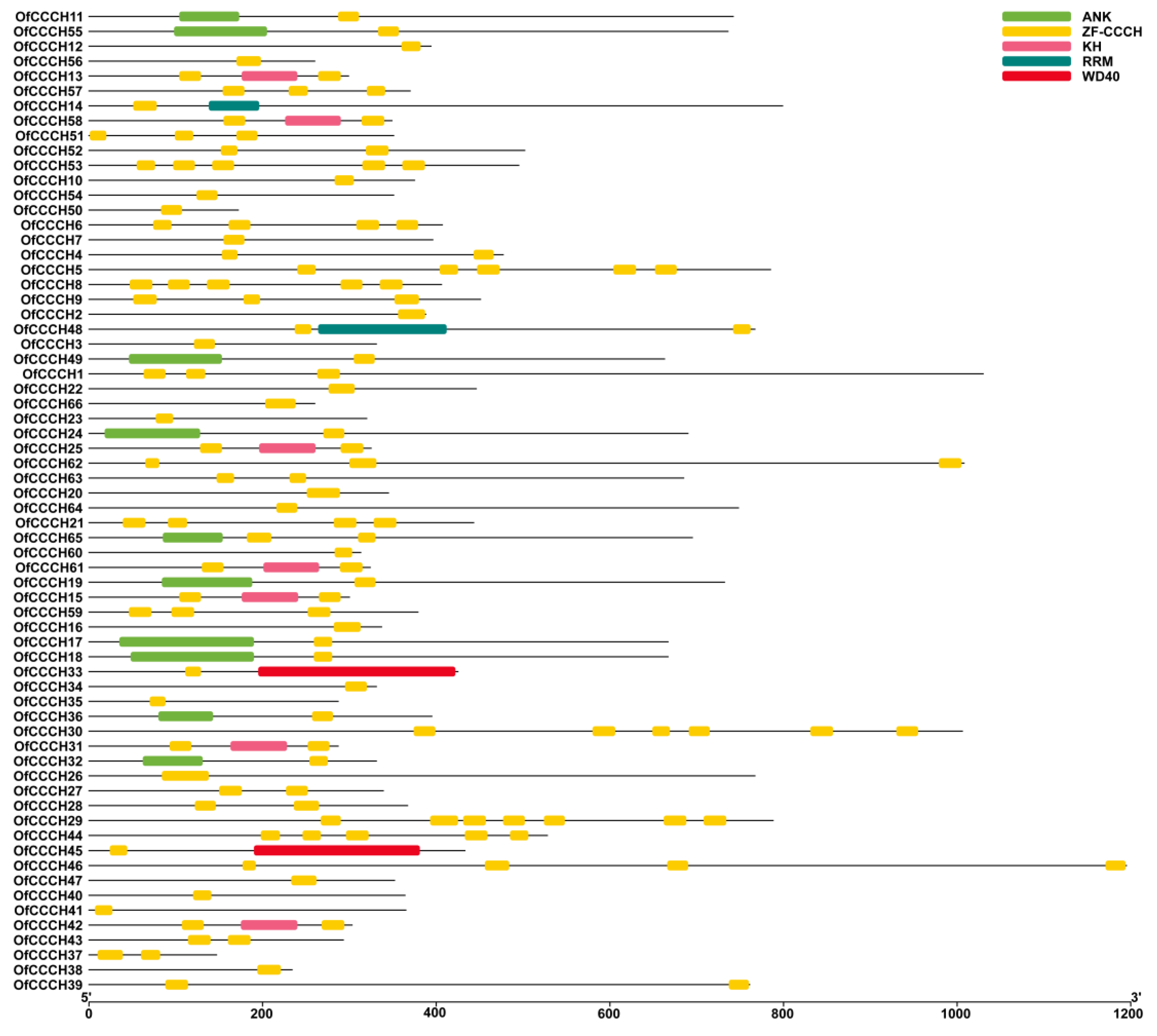
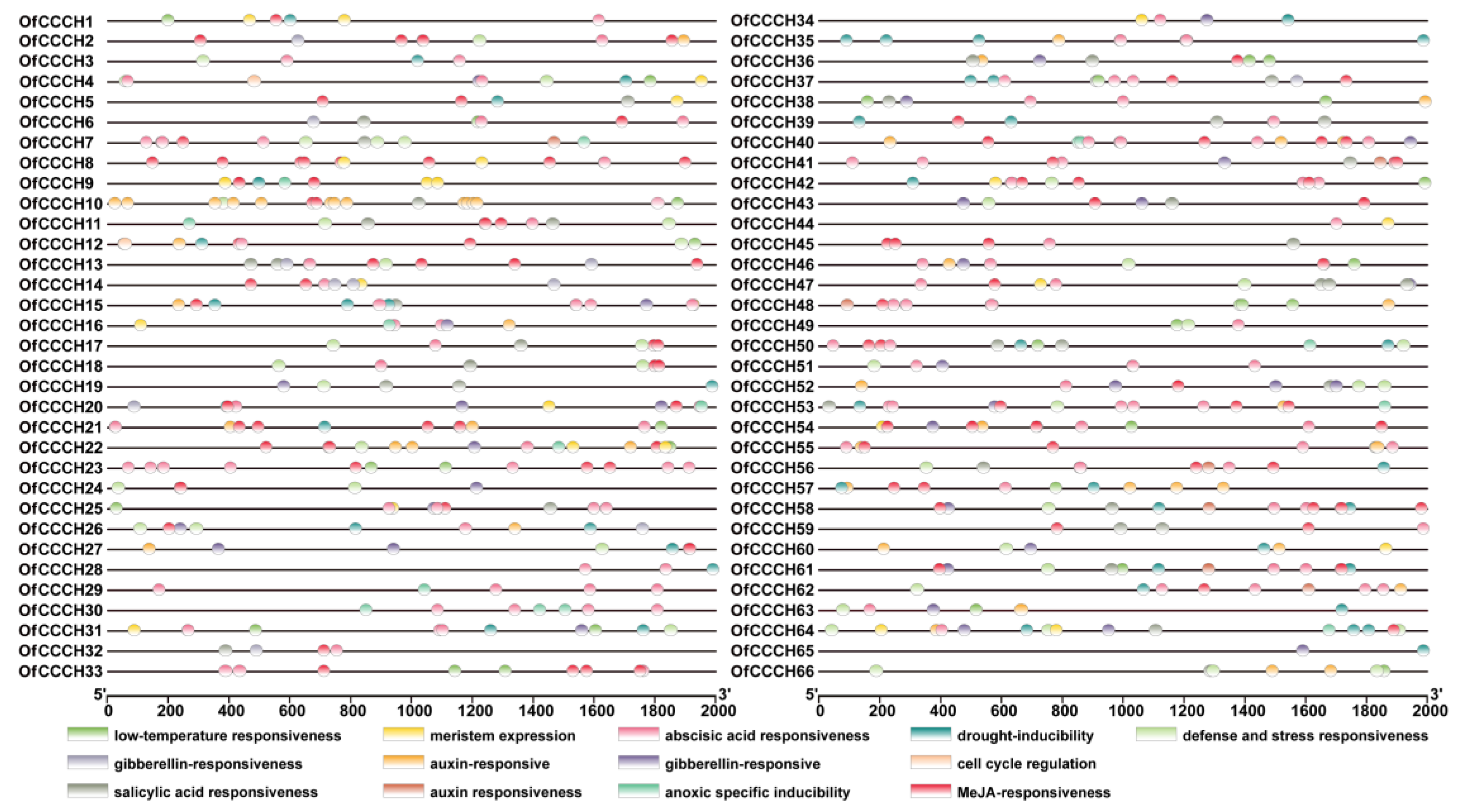
2.3. Gene Structure, Domain, and CREs Analysis
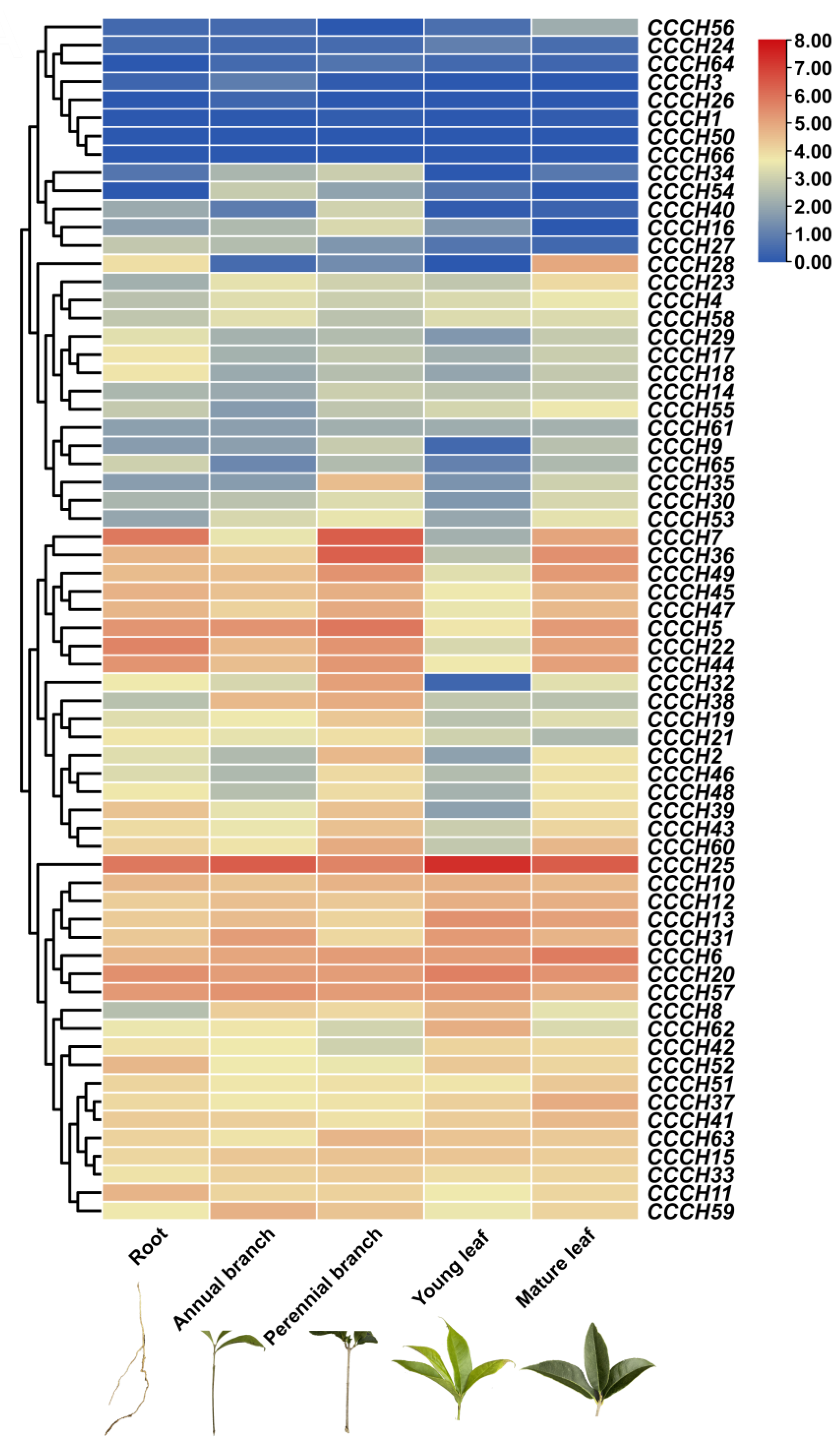
2.4. Transcriptome Analysis of OfCCCH Genes in Different Tissues
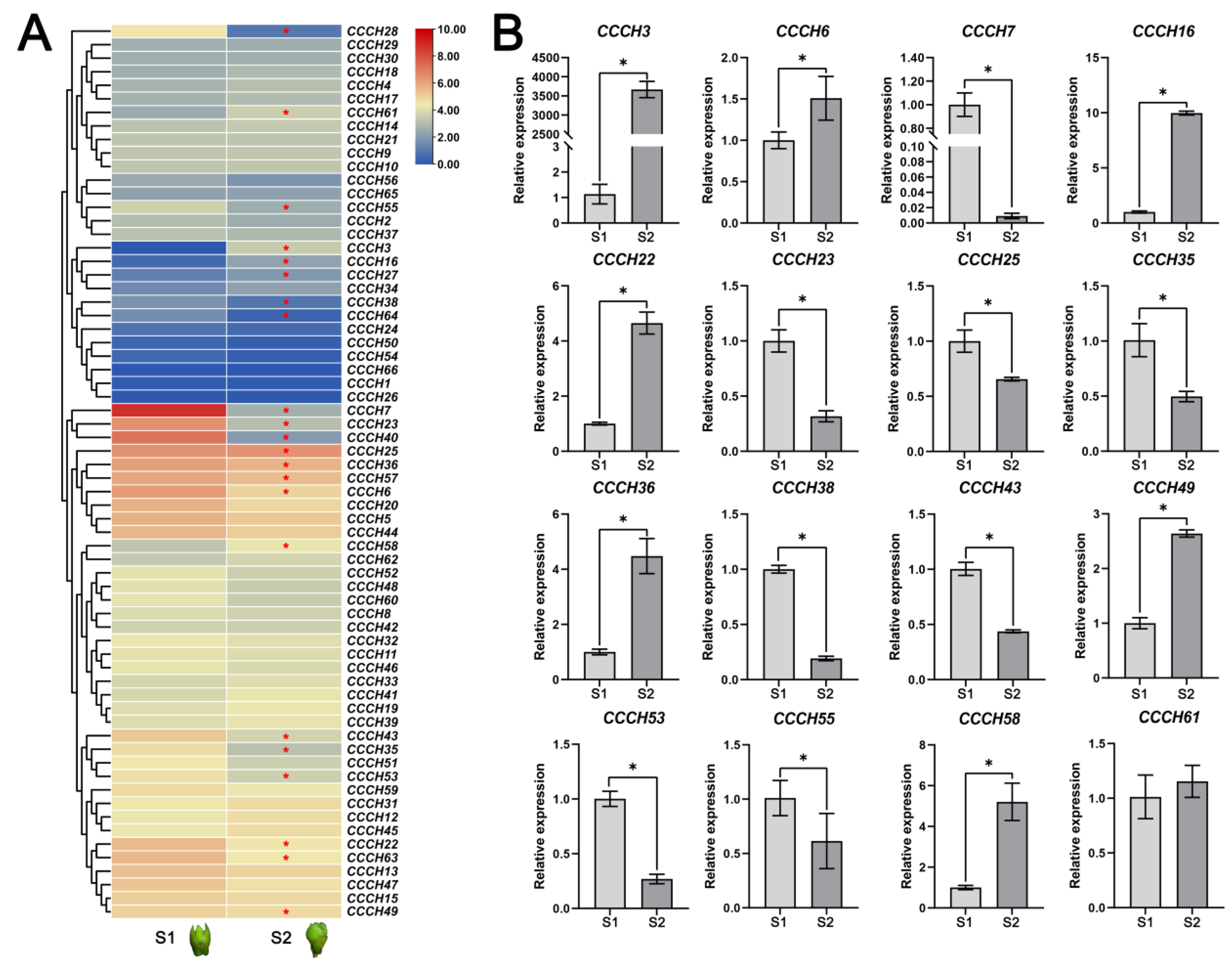
2.5. Transcriptome and qRT-PCR Analysis of OfCCCH Genes during the Flower Opening Processes
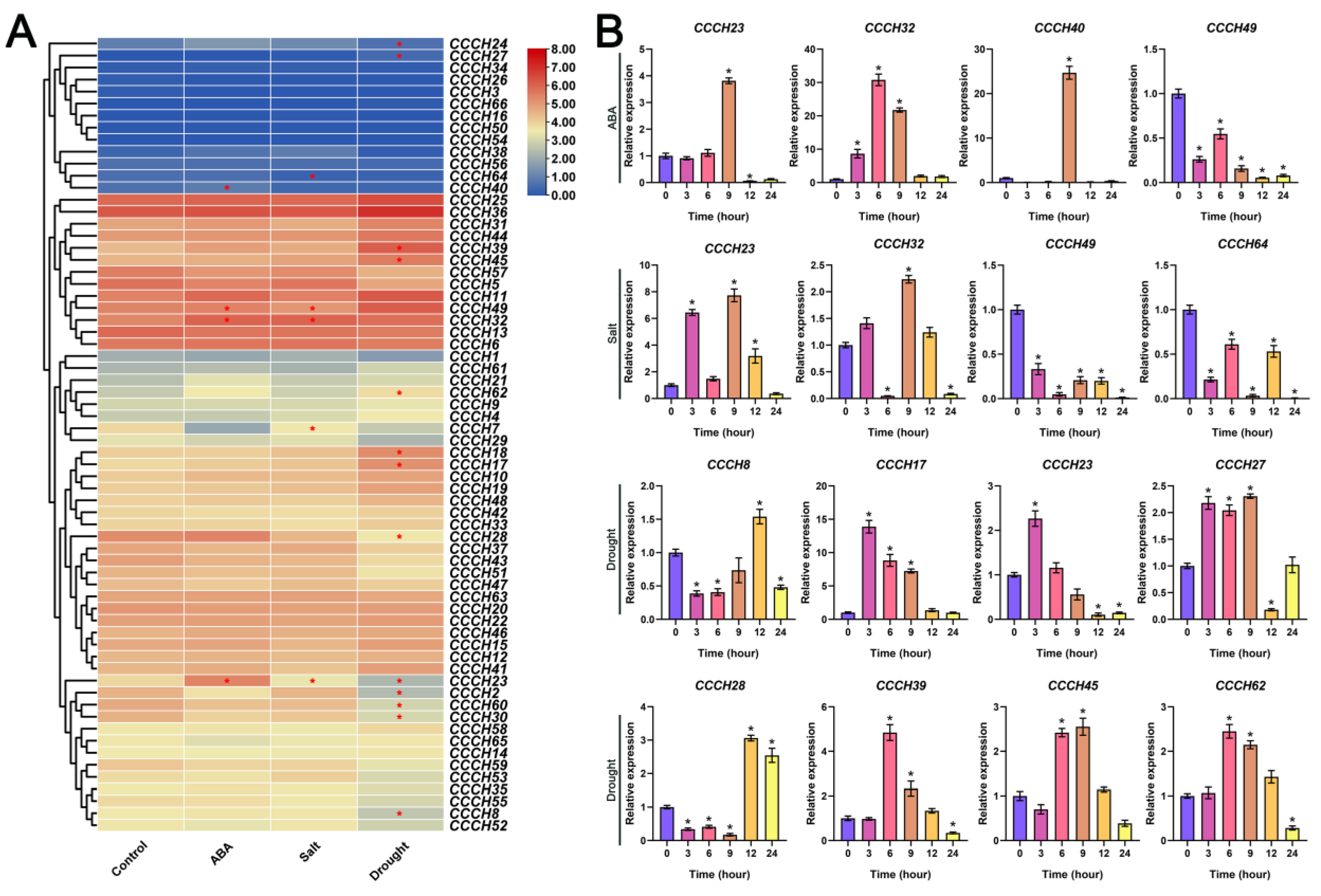
2.6. Transcriptome and qRT-PCR Analysis of OfCCCH Genes in Response to Abiotic Stress
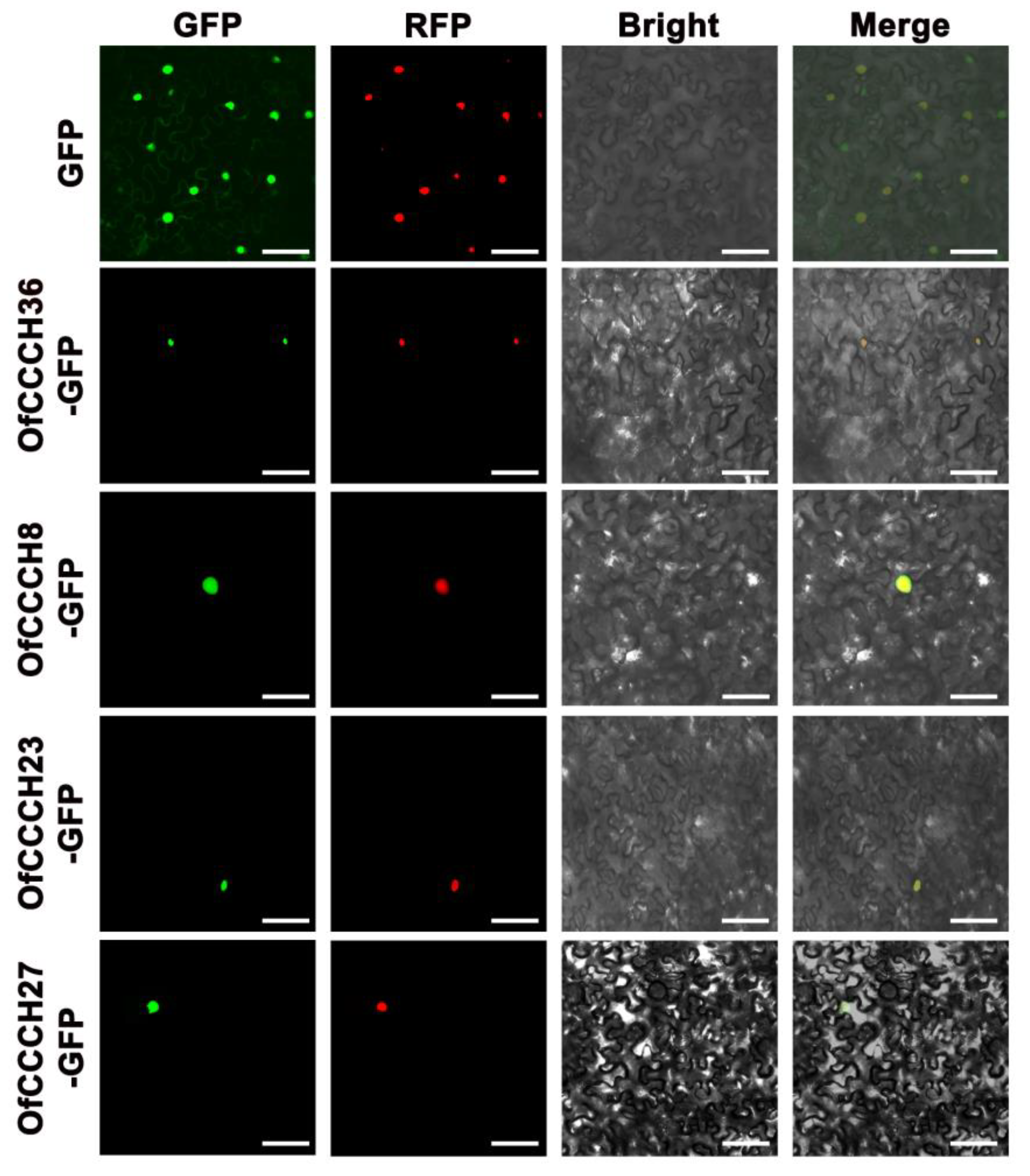
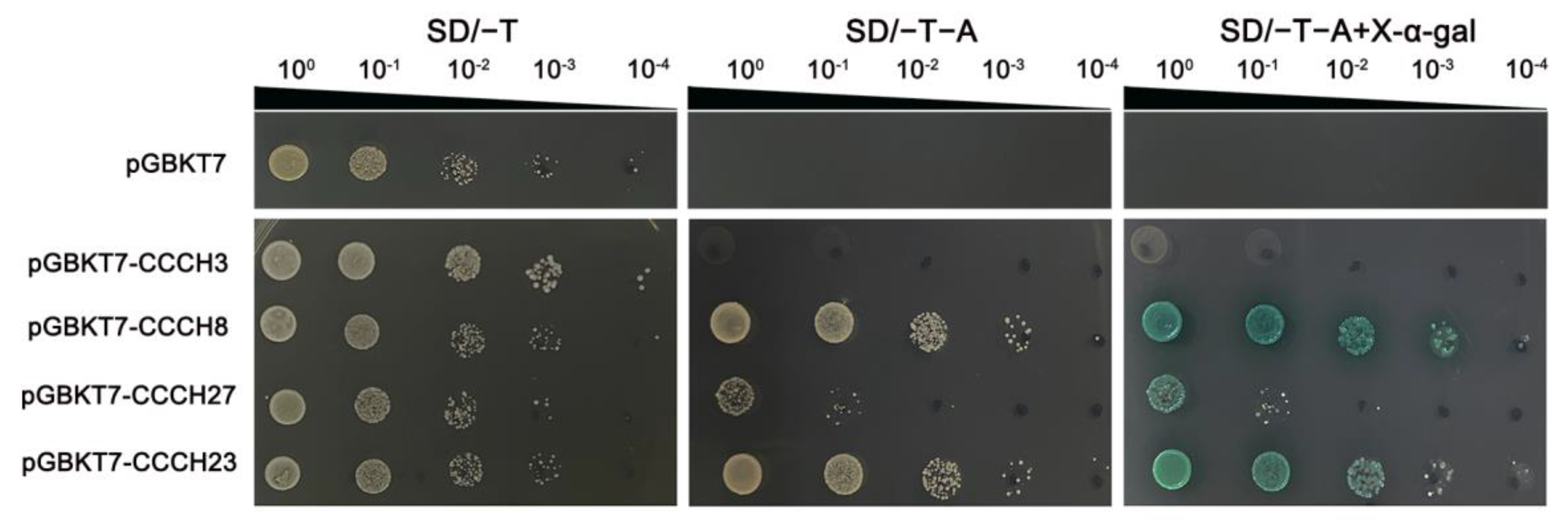
2.7. Characterization of the OfCCCH Proteins
3. Discussion
3.1. Characteristic Analysis of OfCCCH TFs in Osmanthus Fragrans
3.2. OfCCCHs Involved in Flower Opening and Abiotic Stress Tolerance in Osmanthus fragrans
4. Materials and Methods
4.1. Plant Materials and Treatments
4.2. Genome-Wide Identification of the CCCHs
4.3. Chromosomal Localization, Gene Duplication, and Synteny Analysis
4.4. Phylogenetic Tree Construction and Sequence Analysis of CCCHs
4.5. Gene Structure, Domain, and CREs (Cis-Regulatory Elements) Prediction for the OfCCCHs
4.6. Total RNA Extraction and RNA-Sequencing
4.7. Quantitative Real-Time PCR Analysis
4.8. Subcellular Localization of the OfCCCH Proteins
4.9. Transcriptional Activation Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Hu, X.; Zuo, J. The CCCH zinc finger family of soybean (Glycine max L.): Genome-wide identification, expression, domestication, GWAS and haplotype analysis. BMC Genom. 2021, 22, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 1–20. [Google Scholar] [CrossRef]
- Han, G.; Lu, C.; Guo, J.; Qiao, Z.; Sui, N.; Qiu, N.; Wang, B. C2H2 zinc finger proteins: Master regulators of abiotic stress responses in plants. Front. Plant Sci. 2020, 11, 115. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Qiao, Z.; Li, Y.; Wang, C.; Wang, B. The roles of CCCH zinc-finger proteins in plant abiotic stress tolerance. Int. J. Mol. Sci. 2021, 22, 8327. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Hu, R.; Zhang, D.; Qi, G.; Zuo, R.; Cao, Y.; Chen, P.; Kong, Y.; Zhou, G. Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genom. 2012, 13, 1–22. [Google Scholar] [CrossRef]
- Su, L.-Y.; Xiao, X.-C.; Jiang, M.-Q.; Huang, S.-Q.; Xue, X.-D.; Xue, L.; Lai, Z.-X.; Lin, Y.-L. Genome-wide analysis of the CCCH zinc finger family in longan: Characteristic identification and expression profiles in Dimocarpus longan Lour. J. Integr. Agric. 2022, 21, 113–130. [Google Scholar] [CrossRef]
- Cheng, X.; Cao, J.; Gao, C.; Gao, W.; Yan, S.; Yao, H.; Xu, K.; Liu, X.; Xu, D.; Pan, X. Identification of the wheat C3H gene family and expression analysis of candidates associated with seed dormancy and germination. Plant Physiol. Biochem. 2020, 156, 524–537. [Google Scholar] [CrossRef] [PubMed]
- Pi, B.; Pan, J.; Xiao, M.; Hu, X.; Zhang, L.; Chen, M.; Liu, B.; Ruan, Y.; Huang, Y. Systematic analysis of CCCH zinc finger family in Brassica napus showed that BnRR-TZFs are involved in stress resistance. BMC Plant Biol. 2021, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- D’Orso, F.; De Leonardis, A.M.; Salvi, S.; Gadaleta, A.; Ruberti, I.; Cattivelli, L.; Morelli, G.; Mastrangelo, A.M. Conservation of AtTZF1, AtTZF2, and AtTZF3 homolog gene regulation by salt stress in evolutionarily distant plant species. Front. Plant Sci. 2015, 6, 394. [Google Scholar] [PubMed]
- Li, J.; Jia, D.; Chen, X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 2001, 13, 2269–2281. [Google Scholar] [CrossRef]
- Wang, B.; Fang, R.; Chen, F.; Han, J.; Liu, Y.G.; Chen, L.; Zhu, Q. A novel CCCH‐type zinc finger protein SAW1 activates OsGA20ox3 to regulate gibberellin homeostasis and anther development in rice. J. Integr. Plant Biol. 2020, 62, 1594–1606. [Google Scholar] [CrossRef]
- Zhang, D.; Xu, Z.; Cao, S.; Chen, K.; Li, S.; Liu, X.; Gao, C.; Zhang, B.; Zhou, Y. An uncanonical CCCH-tandem zinc-finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mol. Plant 2018, 11, 163–174. [Google Scholar] [CrossRef]
- Grabowska, A.; Wisniewska, A.; Tagashira, N.; Malepszy, S.; Filipecki, M. Characterization of CsSEF1 gene encoding putative CCCH-type zinc finger protein expressed during cucumber somatic embryogenesis. J. Plant Physiol. 2009, 166, 310–323. [Google Scholar] [CrossRef]
- Bogamuwa, S.; Jang, J.C. The A rabidopsis tandem CCCH zinc finger proteins AtTZF4, 5 and 6 are involved in light-, abscisic acid-and gibberellic acid‐mediated regulation of seed germination. Plant Cell Environ. 2013, 36, 1507–1519. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Yu, G.; Lei, S.; Zhang, C.; Bin Xu, B.X.; Huang, B. CCCH protein-PvCCCH69 acted as a repressor for leaf senescence through suppressing ABA-signaling pathway. Hortic. Res. 2021, 8, 165. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Gao, X.; Zhi, Y.; Li, X.; Zhang, Q.; Niu, J.; Wang, J.; Zhai, H.; Zhao, N.; Li, J. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 2019, 223, 1918–1936. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, M.G.; Jan, A.; Ishizaki, T.; Valencia, M.; Dedicova, B.; Maruyama, K.; Ogata, T.; Todaka, D.; Yamaguchi-Shinozaki, K.; Nakashima, K. Expression of the CCCH-tandem zinc finger protein gene OsTZF5 under a stress-inducible promoter mitigates the effect of drought stress on rice grain yield under field conditions. Plant Biotechnol. J. 2020, 18, 1711–1721. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Liu, H.-L.; Wang, K.; Gao, Y.-M.; Wu, M.; Xiang, Y. Identification of CCCH zinc finger proteins family in Moso bamboo (Phyllostachys edulis), and PeC3H74 confers drought tolerance to transgenic plants. Front. Plant Sci. 2020, 11, 1697. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Lin, W.; Yu, G.; Cheng, Q.; Xu, B.; Huang, B. Improved cold tolerance in switchgrass by a novel CCCH-type zinc finger transcription factor gene, PvC3H72, associated with ICE1–CBF–COR regulon and ABA-responsive genes. Biotechnol. Biofuels 2019, 12, 1–11. [Google Scholar] [CrossRef]
- Rameneni, J.J.; Dhandapani, V.; Paul, P.; Devaraj, S.P.; Choi, S.R.; Yi, S.Y.; Kim, M.-S.; Hong, S.; Oh, S.H.; Oh, M.-H. Comprehensive analysis of CCCH zinc-finger-type transcription factors in the Brassica rapa genome. Hortic. Environ. Biotechnol. 2018, 59, 729–747. [Google Scholar] [CrossRef]
- Lopez-Ortiz, C.; Peña-Garcia, Y.; Natarajan, P.; Bhandari, M.; Abburi, V.; Dutta, S.K.; Yadav, L.; Stommel, J.; Nimmakayala, P.; Reddy, U.K. The ankyrin repeat gene family in Capsicum spp: Genome-wide survey, characterization and gene expression profile. Sci. Rep. 2020, 10, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Palzer, K.-A.; Bolduan, V.; Käfer, R.; Kleinert, H.; Bros, M.; Pautz, A. The Role of KH-Type Splicing Regulatory Protein (KSRP) for Immune Functions and Tumorigenesis. Cells 2022, 11, 1482. [Google Scholar] [CrossRef] [PubMed]
- Brien, C.; Jewell, N.; Watts-Williams, S.J.; Garnett, T.; Berger, B. Smoothing and extraction of traits in the growth analysis of noninvasive phenotypic data. Plant Methods 2020, 16, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wang, Y.; Shen, J.; Yin, J.; Li, D.; Gao, Y.; Xu, W.; Liang, J. OsRACK1A, encodes a circadian clock-regulated WD40 protein, negatively affect salt tolerance in rice. Rice 2018, 11, 1–15. [Google Scholar] [CrossRef]
- Kim, D.H.; Yamaguchi, S.; Lim, S.; Oh, E.; Park, J.; Hanada, A.; Kamiya, Y.; Choi, G. SOMNUS, a CCCH-Type Zinc Finger Protein in Arabidopsis, Negatively Regulates Light-Dependent Seed Germination Downstream of PIL5. Plant Cell 2008, 20, 1260–1277. [Google Scholar] [CrossRef] [PubMed]
- Yan, Z.; Jia, J.; Yan, X.; Shi, H.; Han, Y. Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol. Biol. 2017, 95, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Zhang, H.; Dai, S.; Feng, S.; Gong, S.; Wang, J.; Zhou, A. AaZFP3, a Novel CCCH-Type Zinc Finger Protein from Adonis amurensis, Promotes Early Flowering in by Regulating the Expression of Flowering-Related Genes. Int. J. Mol. Sci. 2022, 23, 8166. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, Y.; Tanaka, R.; Ono, H.; Ogiwara, I.; Kanekatsu, M.; van Doorn, W.; Yamada, T. Length of the dark period affects flower opening and the expression of circadian-clock associated genes as well as xyloglucan endotransglucosylase/hydrolase genes in petals of morning glory (Ipomoea nil). Plant Cell Rep. 2014, 33, 1121–1131. [Google Scholar] [CrossRef] [PubMed]
- Han, G.; Wang, M.; Yuan, F.; Sui, N.; Song, J.; Wang, B. The CCCH zinc finger protein gene AtZFP1 improves salt resistance in Arabidopsis thaliana. Plant Mol. Biol. 2014, 86, 237–253. [Google Scholar] [CrossRef] [PubMed]
- Seong, S.Y.; Shim, J.S.; Bang, S.W.; Kim, J.-K. Overexpression of OsC3H10, a CCCH-zinc finger, improves drought tolerance in rice by regulating stress-related genes. Plants 2020, 9, 1298. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Yue, Y.; Li, H.; Ding, W.; Chen, G.; Shi, T.; Chen, J.; Park, M.S.; Chen, F.; Wang, L. The chromosome-level quality genome provides insights into the evolution of the biosynthesis genes for aroma compounds of Osmanthus fragrans. Hortic. Res. 2018, 5, 72. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Librado, P.; Rozas, J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics 2009, 25, 1451–1452. [Google Scholar] [CrossRef] [PubMed]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Miao, Y.; Zhong, S.; Fang, Q.; Wang, Y.; Dong, B.; Zhao, H. Genome-Wide Identification and Expression Analysis of XTH Gene Family during Flower-Opening Stages in Osmanthus fragrans. Plants 2022, 11, 1015. [Google Scholar] [CrossRef]
- Zhang, C.; Fu, J.; Wang, Y.; Bao, Z.; Zhao, H. Identification of suitable reference genes for gene expression normalization in the quantitative real-time PCR analysis of sweet osmanthus (Osmanthus fragrans Lour.). PLoS ONE 2015, 10, e0136355. [Google Scholar] [CrossRef]
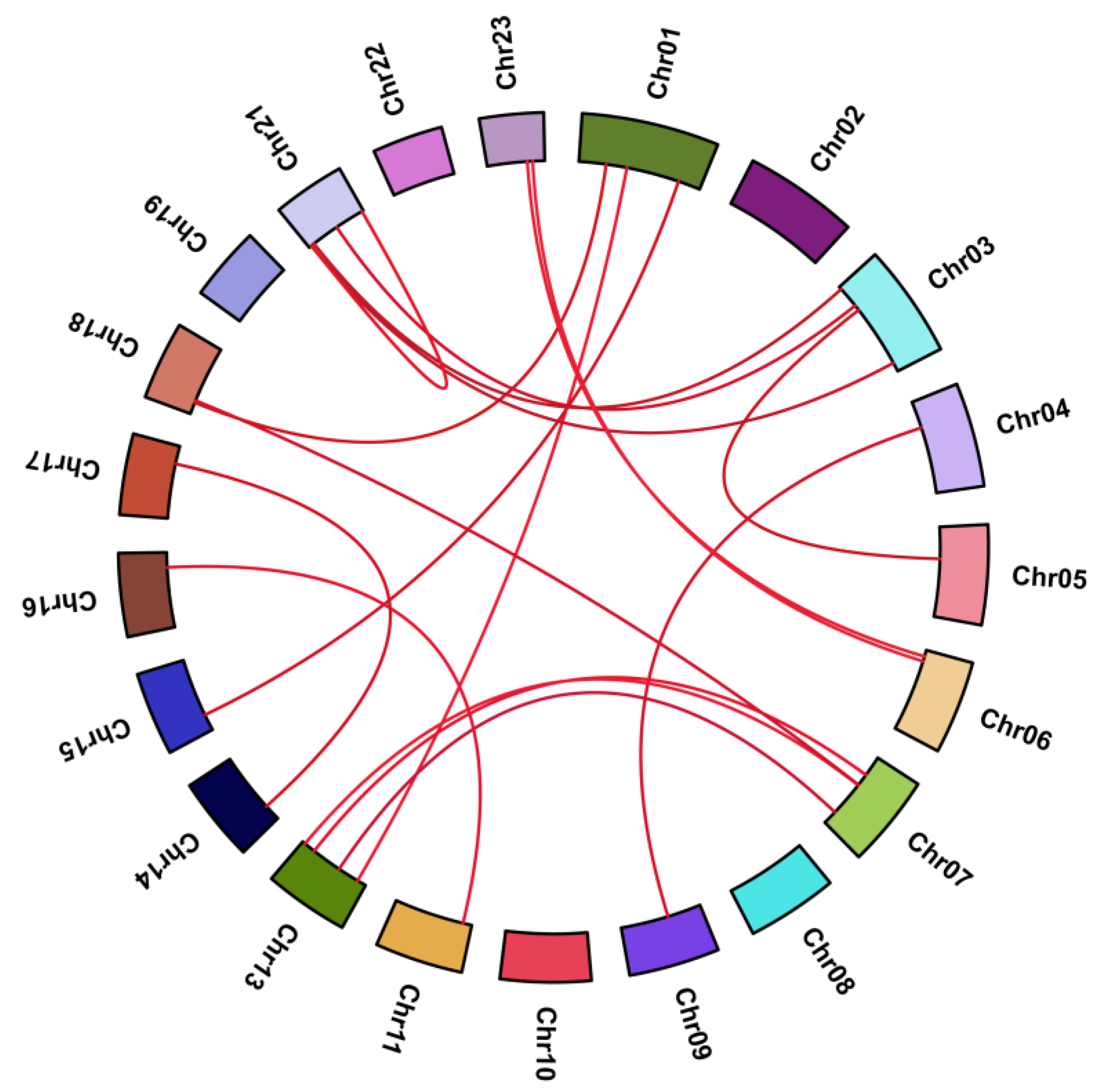

| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Type of Duplication | Type of Selection | Divergene Time (Mya) |
|---|---|---|---|---|---|---|
| CCCH2-CCCH38 | 0.371 | 1.127 | 0.329 | Segmental | Purifying | 37.57 |
| CCCH4-CCCH45 | 0.123 | 0.266 | 0.461 | Segmental | Purifying | 8.87 |
| CCCH3-CCCH54 | 0.053 | 0.311 | 0.171 | Segmental | Purifying | 10.37 |
| CCCH11-CCCH17 | 0.050 | 0.295 | 0.168 | Segmental | Purifying | 9.83 |
| CCCH10-CCCH60 | 0.052 | 0.286 | 0.181 | Segmental | Purifying | 9.53 |
| CCCH8-CCCH59 | 0.141 | 0.243 | 0.581 | Segmental | Purifying | 8.11 |
| CCCH13-CCCH58 | 0.066 | 0.273 | 0.243 | Segmental | Purifying | 9.10 |
| CCCH15-CCCH31 | 0.072 | 0.298 | 0.241 | Segmental | Purifying | 9.93 |
| CCCH19-CCCH65 | 0.086 | 0.237 | 0.364 | Segmental | Purifying | 7.91 |
| CCCH20-CCCH66 | 0.166 | 0.562 | 0.295 | Segmental | Purifying | 18.73 |
| CCCH23-CCCH40 | 0.133 | 0.522 | 0.256 | Segmental | Purifying | 17.41 |
| CCCH26-CCCH39 | 0.126 | 0.258 | 0.491 | Segmental | Purifying | 8.62 |
| CCCH25-CCCH42 | 0.103 | 0.366 | 0.281 | Segmental | Purifying | 12.23 |
| CCCH24-CCCH55 | 0.201 | 0.596 | 0.338 | Segmental | Purifying | 19.87 |
| CCCH36-CCCH49 | 0.061 | 0.333 | 0.184 | Segmental | Purifying | 11.12 |
| CCCH43-CCCH53 | 0.043 | 0.211 | 0.201 | Segmental | Purifying | 7.03 |
| CCCH58- CCCH61 | 0.005 | 0.011 | 0.438 | Segmental | Purifying | 0.37 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Y.; Cao, S.; Shen, L.; Wang, Y.; Zhong, S.; Yang, L.; Xiao, Z.; Fang, Q.; Zhao, H.; Dong, B. Comparative Transcriptome Analysis of CCCH Family in Roles of Flower Opening and Abiotic Stress in Osmanthus fragrans. Int. J. Mol. Sci. 2022, 23, 15363. https://doi.org/10.3390/ijms232315363
Ye Y, Cao S, Shen L, Wang Y, Zhong S, Yang L, Xiao Z, Fang Q, Zhao H, Dong B. Comparative Transcriptome Analysis of CCCH Family in Roles of Flower Opening and Abiotic Stress in Osmanthus fragrans. International Journal of Molecular Sciences. 2022; 23(23):15363. https://doi.org/10.3390/ijms232315363
Chicago/Turabian StyleYe, Yong, Shanshan Cao, Lixiao Shen, Yiguang Wang, Shiwei Zhong, Liyuan Yang, Zheng Xiao, Qiu Fang, Hongbo Zhao, and Bin Dong. 2022. "Comparative Transcriptome Analysis of CCCH Family in Roles of Flower Opening and Abiotic Stress in Osmanthus fragrans" International Journal of Molecular Sciences 23, no. 23: 15363. https://doi.org/10.3390/ijms232315363
APA StyleYe, Y., Cao, S., Shen, L., Wang, Y., Zhong, S., Yang, L., Xiao, Z., Fang, Q., Zhao, H., & Dong, B. (2022). Comparative Transcriptome Analysis of CCCH Family in Roles of Flower Opening and Abiotic Stress in Osmanthus fragrans. International Journal of Molecular Sciences, 23(23), 15363. https://doi.org/10.3390/ijms232315363






