Abstract
The peptide hormone insulin-like 3 (INSL3) is produced almost exclusively by Leydig cells of the male gonad. INSL3 has several functions such as fetal testis descent and bone metabolism in adults. Insl3 gene expression in Leydig cells is not hormonally regulated but rather is constitutively expressed. The regulatory region of the Insl3 gene has been described in various species; moreover, functional studies have revealed that the Insl3 promoter is regulated by various transcription factors that include the nuclear receptors AR, NUR77, COUP-TFII, LRH1, and SF1, as well as the Krüppel-like factor KLF6. However, these transcription factors are also found in several tissues that do not express Insl3, indicating that other, yet unidentified factors, must be involved to drive Insl3 expression specifically in Leydig cells. Through a fine functional promoter analysis, we have identified a 35-bp region that is responsible for conferring 70% of the activity of the mouse Insl3 promoter in Leydig cells. All tri- and dinucleotide mutations introduced dramatically reduced Insl3 promoter activity, indicating that the entire 35-bp sequence is required. Nuclear proteins from MA-10 Leydig cells bound specifically to the 35-bp region. The 35-bp sequence contains GC- and GA-rich motifs as well as potential binding elements for members of the CREB, C/EBP, AP1, AP2, and NF-κB families. The Insl3 promoter was indeed activated 2-fold by NF-κB p50 but not by other transcription factors tested. These results help to further define the regulation of Insl3 gene transcription in Leydig cells.
1. Introduction
Leydig cells located in the testis produce testosterone and insulin-like 3 (INSL3), two hormones essential for male sex differentiation and reproductive function. More specifically, testosterone is required to masculinize the male embryo during fetal life, and postnatally to complete internal and external male organ development as well as for the initiation and maintenance of spermatogenesis []. INSL3 regulates the inguino-scrotal phase of testicular descent during fetal life [,] and bone metabolism in adults []. INSL3 has also been proposed to reduce germ cell apoptosis [,], modulate skeletal muscle metabolism and function [], regulate motor and sensory brain functions [], and contribute to corneal wound healing []. INSL3 is an exclusive marker for the differentiation and functional status of Leydig cells in the testis [,].
The Insl3 gene is composed of two exons (219 and 404 bp) separated by a 739-bp intron. The Insl3 gene is atypically located entirely within the last intron of the Jak3 gene [,,]. Consequently, the regulatory elements controlling Insl3 gene expression in Leydig cells are believed to be located within a short promoter region of about 1000 bp. The genomic region corresponding to the Insl3 promoter has been isolated from rat [], mouse [,], human [], dog [], and pig []. Functional studies have identified several transcription factors regulating Insl3 promoter activity in Leydig cells. These include the Krüppel-like factor KLF6 [] as well as the nuclear receptors SF1 (Ad4BP, NR5A1) [,,,], LRH1 (NR5A2) [], NUR77 (NGFI-B, NR4A1) [,], testosterone-activated androgen receptor (AR, NR3C4) [,], COUP-TFII (NR2F2) [], and DAX1 (NR0B1) []. Transcriptional cooperation between COUP-TFII and SF1 [,], and between KLF6 and NUR77 and SF1 [] on the Insl3 promoter has also been reported. However, most of these transcription factors are also found in cell types that do not express the Insl3 gene, such as Sertoli and adrenal cells, indicating that additional transcription factors are likely involved in directing Insl3 expression in Leydig cells. In the present study, we have identified a 35-bp regulatory region bound by nuclear proteins that is essential for mouse Insl3 promoter activity in MA-10 Leydig cells.
2. Results
2.1. A 35-bp Region Is Responsible for 70% of Mouse Insl3 Promoter Activity
To identify the regions responsible for the activity of the mouse Insl3 promoter in Leydig cells, several 5′ progressive deletions of the promoter were generated and transfected in MA-10 Leydig cells. The MA-10 cell line expresses the Insl3 gene and has been validated as an appropriate model to study Insl3 gene expression [,,]. As shown in Figure 1, Insl3 promoter deletions from −1082 bp to −186 bp had no significant effect on Insl3 promoter activity. However, further deletion to −111 bp led to a 70% reduction in Insl3 promoter activity (Figure 1). Finally, a further reduction in Insl3 promoter activity to 15% was observed with a deletion to −79 bp, which is considered a minimal promoter (Figure 1). These data, therefore, identify a critical region located between −186 and −111 bp that is responsible for 70% of Insl3 promoter activity.
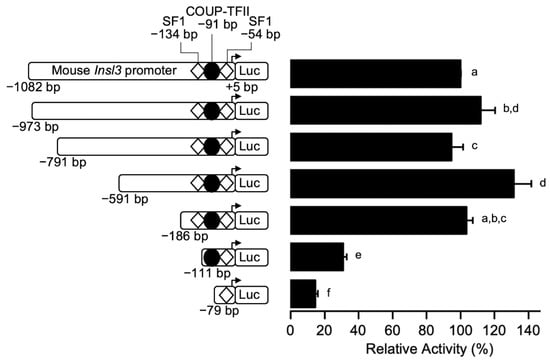
Figure 1.
Identification of a critical regulatory region within the proximal mouse Insl3 promoter. MA-10 Leydig cells were transfected with various 5’ deletion constructs of the mouse Insl3 promoter; the 5′ end point of each construct is indicated on the left of the graph. The position of previously identified binding sites for SF1 (white diamonds) and COUP-TFII (black circle) is indicated. Results are shown as % Relative Activity (±SEM) relative to the activity of the −1132 bp reporter, which was set to 100%. Different letters indicate a statistically significant difference between groups (p < 0.05).
To more precisely locate the region conferring 70% of Insl3 promoter activity, additional 5’ deletion constructs to −151, −141, −131, and −120 bp were generated and transfected in MA-10 Leydig cells. As shown in Figure 2, deletion of a 35-bp region between −186 and −151 bp led to a 70% reduction in Insl3 promoter activity. Promoter activity remained similar with all other deletion constructs, except for the minimal −79 bp which was reduced to 15% (Figure 2). Together, these results indicate that a 35-bp region located between −186 and −151 bp is responsible for conferring 70% of activity to the mouse Insl3 promoter in MA-10 Leydig cells.
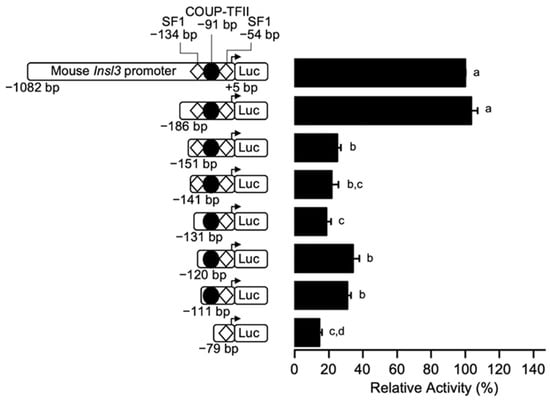
Figure 2.
The region between −186 and −151 bp confers 70% of mouse Insl3 promoter activity in Leydig cells. MA-10 Leydig cells were transfected with a series of fine 5’ deletion constructs of the mouse Insl3 promoter; the 5′ end point of each construct is indicated on the left of the graph. The position of previously identified binding sites for SF1 (white diamonds) and COUP-TFII (black circle) is indicated. Results are shown as % Relative Activity (± SEM) relative to the activity of the −1132 bp reporter, which was set to 100%. Different letters indicate a statistically significant difference between groups (p < 0.05).
2.2. Fine Mapping of the 35-bp Region Conferring 70% of Insl3 Promoter Activity
To identify the sequences responsible for conferring 70% of mouse Insl3 promoter activity within the −186 and −151 bp region, a linker-scanning approach was used. Eight sequential mutations were introduced in the 35-bp region in the context of the −1182 bp Insl3 promoter (Figure 3) and the reporter constructs were transfected in MA-10 Leydig cells. As shown in Figure 3, all the mutations (M1 to M8) led to a loss of 60–65% in Insl3 promoter activity compared to the wild type −1182 bp promoter. This indicates that the entire 35-bp region is necessary for full mouse Insl3 promoter activity.
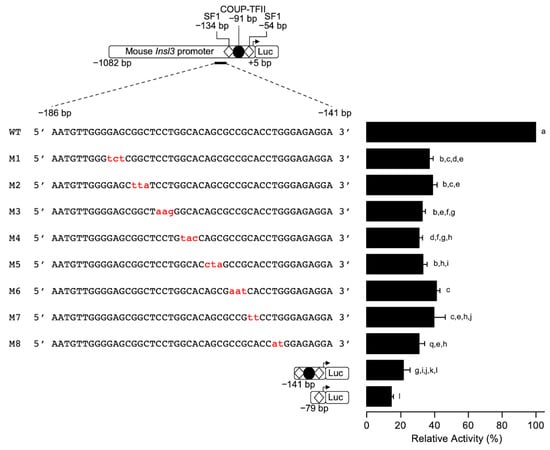
Figure 3.
Fine mapping of the 35-bp region in the proximal mouse Insl3 promoter. MA-10 Leydig cells were transfected with various −1132 bp mouse Insl3 promoter constructs: a wild-type promoter and a series of trinucleotide or dinucleotide mutated constructs (M1 to M8; the mutations are in red lowercase). The position of previously identified binding sites for SF1 (white diamonds) and COUP-TFII (black circle) is indicated. Results are shown as % Relative Activity (± SEM) relative to the activity of the −1132 bp wild-type reporter, which was set to 100%. Different letters indicate a statistically significant difference between groups (p < 0.05).
2.3. Binding of Nuclear Proteins to the 35-bp Regulatory Region
The linker-scanning site-directed mutagenesis data (Figure 3) established that the full 35-bp region is important and that the activity of the Insl3 promoter does not depend on any specific motif within this region. This suggests that binding of more than one transcription factor may be involved. A DNA–protein interaction approach (electromobility shift assay (EMSA)) was used to determine if protein(s) from Leydig cell nuclear extracts can bind to the 35-bp region. As shown in Figure 4, binding was detected (Figure 4, lane 2) that could be competed with increasing molar excess (5- and 25-fold) of unlabelled wild-type (WT) oligonucleotides (Figure 4, lanes 3 and 4). Due to the size of the band, it is possible that more than one protein binds to this region. To assess the specificity of the protein–DNA interaction, competition experiments were performed using unlabelled oligonucleotides (5× and 25× molar excess) containing the same mutations as those described in Figure 3 for promoter activity. As shown in Figure 4, oligonucleotides corresponding to mutants M1 (lanes 5 and 6), M3 (lanes 9 and 10), and M4 (lanes 11 and 12) were unable to compete the binding complex. Oligonucleotides corresponding to mutants M2 (Figure 4, lanes 7 and 8) and M5 (Figure 4, lanes 13 and 14) were as efficient as the WT oligonucleotide (Figure 4, lanes 3 and 4) at displacing the binding complex. On the other hand, oligonucleotides for mutants M6 only partially competed the binding complex (Figure 4, lanes 15 and 16). Together, these data indicate that protein(s) do bind to the 35-bp Insl3 promoter region and that the nucleotides mutated in mutants M1, M3, M4, and to a lesser extent M6, are important for this binding.
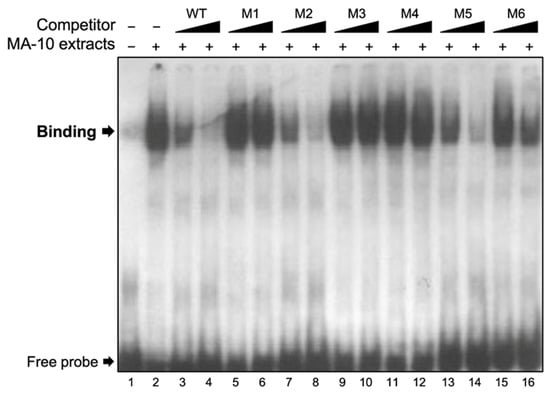
Figure 4.
Nuclear proteins from MA-10 cells bind specifically to the 35-bp region. EMSA was used to determine the binding of nuclear extracts from MA-10 Leydig cells (MA-10 extracts) to a double-stranded 32P-labelled oligonucleotide corresponding to the 35-bp region (−186 to −151 bp) of the Insl3 promoter. Protein binding was challenged by increasing concentrations (black triangles; molar excesses of 5× and 25×) of unlabelled oligonucleotides corresponding to the wild-type 35-bp region (WT) or oligonucleotides containing linker-scanning mutations (M1, M2, M3, M4, M5, M6; defined in Figure 3) in the 35-bp region.
2.4. Binding Assessment of Various Transcription Factors to the 35-bp Region
The 35-bp nucleotide sequence (5′- AAT GTT GGG GAG CGG CTC CTG GCA CAG CGC CGC AC) contains potential binding sites for zinc finger-containing transcription factors known to bind GC- and GA-rich motifs. These include RBP2 (retinoblastoma-binding protein 2), GLI (GLI-Krüppel family zinc finger), IKZF1 (IKAROS family zinc finger 1), SP1 (specificity protein 1), KLF6 (Krüppel-like factor 6), HLTF (helicase-like transcription factor), ZEB1 (zinc finger E-box binding homeobox 1), and HAND1 (heart and neural crest derivatives-expressed protein 1). We, therefore, used oligonucleotides containing a consensus binding sequence for each of these transcription factors in EMSAs. As shown in Figure 5, binding of proteins from MA-10 Leydig cell nuclear extract was detected on the 35-bp region used as a probe (Figure 5A,B, lane 2). As expected, this binding was competed with unlabelled oligonucleotides corresponding the WT sequence and M1 mutant, but not M2 mutant (Figure 5A,B, lanes 3–8). Surprisingly, oligonucleotides containing a consensus binding site for RBP2, GLI, IKZP, SP1 (Figure 5A, lanes 9–16) and KLF6, HLTF, ZEB1, and HAND1 (Figure 5B, lanes 9–16) were unable to compete the binding complex. This indicates that these transcription factors do not bind to the 35-bp region of the mouse Insl3 promoter.
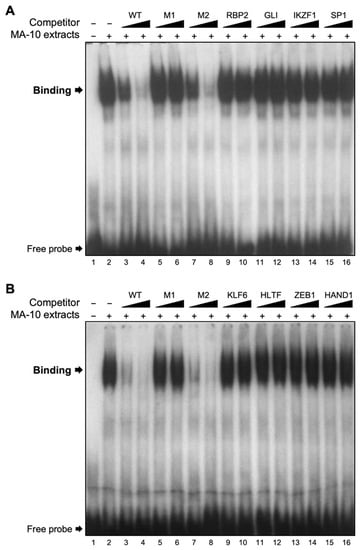
Figure 5.
Assessment of potential transcription factors binding to the 35-bp region. EMSA was used to challenge the binding of nuclear protein(s) from MA-10 Leydig cells (MA-10 extracts) to a double-stranded 32P-labelled oligonucleotide corresponding to the 35-bp region (−186 to −151 bp) of the Insl3 promoter. Protein binding was challenged by increasing concentrations (black triangles; molar excesses of 5× and 25×) of unlabelled oligonucleotides corresponding to the wild-type 35-bp region (WT), mutants M1 and M2, or oligonucleotides containing a consensus binding sequence for transcription factors (A) RBP2, GLI, IKZF1, SP1, and (B) KLF6, HLTF, ZEB1, HAND1.
2.5. NF-κB p50 Activates the Mouse Insl3 Promoter via the 35-bp Region
A closer analysis of the M1 sequence suggested that this might represent a potential binding site for NF-κB p50, or for members of the CREB or C/EBP families of transcription factors. In silico analysis of the sequences in mutants M3 and M4 predicts that they could be recognized by members of the AP1 and AP2 families. Several AP1 members are known to be present in Leydig cells, including cJUN, a well-characterized activator of several genes (reviewed in []). We, therefore, tested whether members of these families could activate the Insl3 promoter in MA-10 Leydig cells. As shown in Figure 6A,B, the −1082 bp Insl3 promoter was activated by SF1 and COUP-TFII as previously reported [,,,,,]. On the other hand, cJUN, CREB, and C/EBPβ failed to activate the Insl3 promoter construct (Figure 6A,B). In the presence of NF-κB p50, a 2-fold activation of the −1082 bp Insl3 promoter was observed (Figure 6B). Combining two transcription factors did not result in any functional cooperation (Figure 6A,B). However, in the presence of CREB or C/EBPβ, activation by the nuclear receptors SF1 and COUP-TFII (Figure 6A), and by NF-κB p50 (Figure 6B), was abolished, a phenomenon that occurs when transcription factors compete for a limited amount of common co-factors.
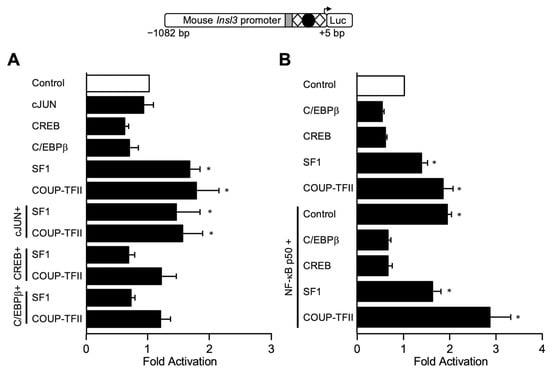
Figure 6.
The mouse Insl3 promoter is activated by NF-κB p50 but not CREB, cJUN, or C/EBPβ. MA-10 Leydig cells were transfected with a −1132 bp mouse Insl3 promoter along with an empty expression vector (open bar) or expression vectors for cJUN, CREB, C/EBPβ, NF-κB p50, SF1, and COUP-TFII either alone or in combination as indicated in (A,B) (black bars). The position of previously identified binding sites for SF1 (white diamonds) and COUP-TFII (black circle) is indicated. The grey box represents the 35-bp sequence responsible for 70% of Insl3 promoter activity. Results are shown as Fold Activation over control ± SEM. An asterisk (*) represents a statistically significant activation compared to control (empty expression vector, value set at 1, p < 0.05).
Next, to locate the NF-κB p50 responsive region, MA-10 Leydig cells were transfected with Insl3 promoter deletion constructs. As shown in Figure 7, a deletion to −186 bp that retains the 35-bp region was still activated ~2-fold by NF-κB p50. However, a deletion construct to −151 bp that removes the 35-bp region was no longer activated by NF-κB p50 (Figure 7).
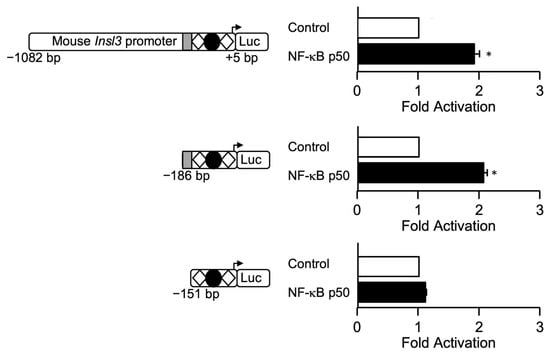
Figure 7.
NF-κB p50-dependent activation of the mouse Insl3 promoter requires the 35-bp region. MA-10 Leydig cells were transfected with three Insl3 promoter constructs (−1132 bp, −186 bp, −151 bp) along with an empty expression vector (open bar) or an expression vector for NF-κB p50 (black bars). The position of previously identified binding sites for SF1 (white diamonds) and COUP-TFII (black circle) is indicated. The grey box represents the 35-bp sequence responsible for 70% of Insl3 promoter activity. Results are shown as Fold Activation over control ± SEM. An asterisk (*) represents a statistically significant activation compared to control (empty expression vector, value set at 1, p < 0.05).
3. Discussion
The peptide hormone INSL3 produced by Leydig cells plays important roles in male reproductive development and function (reviewed in []). Although some transcription factors have been implicated in the regulation of Insl3 gene expression in Leydig cells, most are also present in cells that do not express Insl3 indicating that additional, yet unidentified transcription factors are also required to direct Insl3 expression in Leydig cells. In the present study, we have located and characterized new regulatory elements important for Insl3 promoter activity in Leydig cells.
In our study, we made use of the mouse MA-10 Leydig cell line [], which are immortalized cells corresponding to immature Leydig cells from the adult population [,]. Contrary to primary Leydig cells, MA-10 cells proliferate and contain an aberrant chromosome number []. Despite these issues, MA-10 cells nonetheless respond to hormonal stimulation, like primary Leydig cells, with an increase in steroid hormone production [,,,,], indicating that the signalling cascades, kinases, and transcription factors required for this response are present in MA-10 Leydig cells. More relevant to our current work, MA-10 Leydig cells constitutively express Insl3 [,,,,,,,] and have been validated as a suitable model to study Insl3 gene transcription [].
3.1. Identification of an Essential 35-bp Region within the Proximal Insl3 Promoter
For the promoter functional assays, we used a genomic fragment of −1082 bp upstream of the mouse Insl3 transcription start site. Because the entire Insl3 gene is located within the last intron of the Jak3 gene [,,], the −1082 bp fragment corresponds to the longest sequence that can be used before entering the coding region of the Jak3 gene. Our 5′ progressive deletion analysis of the mouse Insl3 −1082 bp regulatory region revealed that deletion of up to −186 bp did not significantly affect Insl3 promoter activity in MA-10 Leydig cells. This is similar to a previous study performed in another Leydig cell line, the MLTC-1 cells, where a construct of −188 bp retained all mouse Insl3 promoter activity []. Another study also reported that a short proximal region of the mouse Insl3 promoter contained within −157 bp was required for Leydig-specific transcription []. Deletion analysis of the human INSL3 promoter also showed that truncation to −132 bp still maintained full promoter activity in both MA-10 and MLTC-1 Leydig cells but not non-steroidogenic cells []. Although the minimal required region appears shorter in the human promoter, the Insl3 promoter from various rodents contains an additional ~50 bp compared to the INSL3 promoter from primates, as revealed by the alignment of the INSL3 promoter sequence from various species (Figure 8). Therefore, −132 bp in the human INSL3 promoter is equivalent to −178 bp in the mouse Insl3 promoter (Figure 8). Although the Insl3 promoter has been isolated from rat, pig, and dog [,,], a 5′ progressive promoter deletion approach to locate species-specific regulatory elements required for its activity in Leydig cells was not performed.
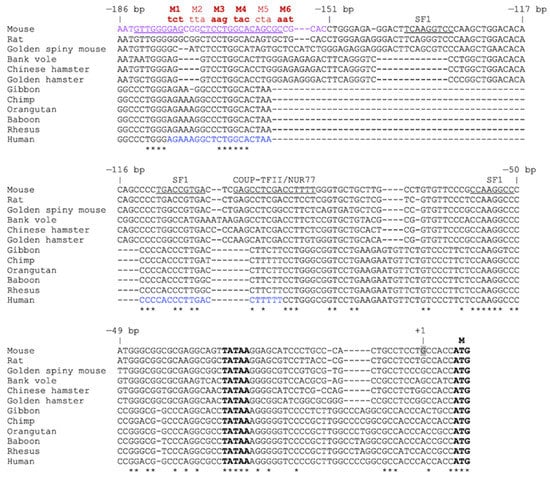
Figure 8.
Alignment of the proximal Insl3 gene promoter from different species. The sequences of the proximal Insl3 promoter (−186 bp to the ATG of the mouse promoter) from various species indicated on the left were aligned using Clustal Omega multiple sequence alignment tool. The numbering corresponds to the position in the mouse Insl3 promoter. The 35-bp region important for mouse Insl3 promoter activity identified in this study is shown in purple. The mutations generated in the 35-bp region are shown in red (M1–M6); the bolded ones (M1, M3, M4, M6) were unable to compete the binding in EMSA. The underlined sequences correspond to potential binding sites for CREB, NF-κB, AP1, AP2, and C/EBP family members. The sequence shown in blue corresponds to an approximately 40-bp region (−132 and −93 bp) previously identified as important for the activity of the human INSL3 promoter []. The positions of previously identified binding sites for transcription factors SF1, COUP-TFII, and NUR77 are shown. The TATA-box, transcription start site (+1), and ATG (M) are also indicated. An asterisk indicates a nucleotide conserved across all 12 species.
Through fine promoter deletions, we identified a 35-bp region located between −186 and −151 bp that is essential for maximal Insl3 promoter activity in Leydig cells. Site-directed mutagenesis in the context of the −1082 bp Insl3 promoter further confirmed the importance of the entire 35-bp region since any trinucleotide or dinucleotide mutation within this region reduced promoter activity. This suggests that more than one transcription factor may bind to this region. The 35-bp region contains several GC- and GA-rich boxes, some of which have been conserved across species (Figure 8). In addition, a potential E-box motif (CAnnTG) for the binding of bHLH transcription factors is also present. The significant reduction in Insl3 promoter activity that occurred when mutations were introduced in either half-site of this palindromic E-box motif (mutations M7 and M8) strongly supports the involvement of bHLH family members. In addition, mutations that target GC/GA-rich motifs reduce the activity of both the mouse (present work) and human [] INSL3 promoter in Leydig cells. This prompted us to assess whether proteins known to bind to GC/GA-rich motifs could bind to the 35-bp region of the mouse Insl3 promoter. Although a protein complex from MA-10 Leydig cells specifically bound to the 35-bp region as identified by EMSA, competition experiments using several oligonucleotides containing binding sites for various transcription factors known to bind GC/GA-rich sequences such as RBP2, GLI, IKZF1, SP1, KLF6, HLTF, ZEB1, and HAND1, failed to displace the binding. This indicates that the protein complex binding to the 35-bp region does not contain members of these transcription factor families.
3.2. Potential Transcription Factors Acting via the 35-bp Element
Our protein–DNA interaction assays indicated that the protein(s) responsible for the activity of the 35-bp region might not be zinc finger-containing transcription factors recognizing GC/GA-rich motifs. Of all the mutations introduced in the 35-bp region and tested by EMSA, mutants M1, M3, M4, and M6 failed displace the binding complex. This indicates that the integrity of these sequences is essential for protein binding, and thus, provides clues regarding the nature of the transcription factor that could bind to these sequences.
In silico analysis of the sequence surrounding mutant M1 identified potential binding sites for members of the CREB, C/EBP, and NF-κB families of transcription factors, which are expressed in Leydig cells where they regulate gene expression [,,,,,]. However, when assayed in functional promoter assays, CREB and C/EBPβ failed to activate the Insl3 promoter, either by themselves or in combination with other transcription factors. On the other hand, activity of the −1082 bp Insl3 promoter was increased by 2-fold in the presence of NF-κB p50, supporting a role for this transcription factor in Insl3 gene transcription in Leydig cells. Furthermore, the NF-κB p50-dependent activation of the Insl3 promoter required the 35-bp region as a deletion construct lacking this region was no longer activated by NF-κB p50. Additional work such as performing EMSAs and competition assays using a consensus NF-κB p50 motif, using mutated Insl3 promoter constructs, as well as testing other potential partners, would be needed to fully understand the mechanism of NF-κB p50 action in the regulation of Insl3 promoter activity in Leydig cells.
Another sequence we identified within the 35-bp element as important for the binding of nuclear proteins and for the activity of the mouse Insl3 promoter in MA-10 Leydig cells was the sequence 5′-TCCTGGCACA-3′ located between −170 and −161 bp. Mutation of this sequence (mutants M3 and M4) prevented protein binding and reduced Insl3 promoter activity by 70%. In silico analysis of this sequence predicted that it could be recognized by members of the AP1 and AP2 families. Several AP1 members are known to be expressed in Leydig cells, including cJUN, a well-characterized activator of several genes (reviewed in []). AP2 factors are present in Leydig cells and regulate the activity of the luteinizing hormone receptor promoter []. Despite the fact that cJUN failed to activate the Insl3 promoter in our assays, we cannot exclude the possibility that other AP1 or AP2 family members might contribute to Insl3 promoter activity. At this time, the nature of the factor binding to this sequence remains to be established. This could be determined using a DNA–protein precipitation assay where a double-stranded oligonucleotide containing the sequence of interest is biotinylated and incubated with nuclear extracts. DNA-bound proteins are then isolated using avidin beads, eluted, and analyzed by LC-MS/MS.
In conclusion, although our present work has identified a key 35-bp regulatory region, additional work is needed to fully decipher the transcription factors acting via this 35-bp element responsible for 70% of Insl3 promoter activity in Leydig cells.
4. Materials and Methods
4.1. Plasmids
The mouse Insl3 promoter constructs −1082 to +5 bp and −186 to +5 bp were previously described [,]. Mouse Insl3 promoter deletions to −973 bp, −791 bp, −591 bp, −151 bp, −141 bp, −131 bp, −120 bp, −111 bp, and −79 bp were obtained by PCR using the −1082 bp Insl3 promoter as a template, along the primers listed in Table 1. All promoter fragments were cloned into a modified pXP1 luciferase reporter plasmid [,].

Table 1.
Sequence of the primers used to generate the various mouse Insl3 promoter deletion constructs.
Various trinucleotide and dinucleotide mutant constructs in the context of the −1082 bp reporter were generated using the QuikChange XL mutagenesis kit (Agilent Technologies Canada, Mississauga, ON, Canada), as recommended by the manufacturer, along with the oligonucleotides listed in Table 2 (only the sequence of the sense oligonucleotide is shown) where the mutations are in lowercase.

Table 2.
Sequence of the oligonucleotides used to generate the various mouse Insl3 promoter mutant constructs.
All the deletion and mutation reporter constructs were confirmed by sequencing (CHUQ Research Centre sequencing platform, Quebec City, QC, Canada). The following expression plasmids were obtained from different research groups: SF1/NR5A1 [], cJUN [], CREB [], C/EBPβ [], COUP-TFII/NR2F2 [], and NF-κB p50 [].
4.2. Cells Culture, Transfections, and Reporter Assays
Mouse MA-10 Leydig cells (ATCC, Manassas, VA, USA, Cat# CRL-3050, RRID:CVCL_D789) were grown in DMEM/F12 medium supplemented with 2.438 g/L sodium bicarbonate, 3.57 g/L HEPES, and 15% horse serum on gelatin-coated plates. Penicillin and streptomycin sulphate were added to the cell culture media to a final concentration of 50 mg/L, and cells were kept at 37 °C, 5% CO2 in a humidified incubator. MA-10 Leydig cells were validated by morphology and by quantifying steroidogenic output, as previously described [,,,,,,,]. MA-10 cells were transiently transfected using polyethylenimine hydrochloride (PEI) (Sigma-Aldrich Canada, Oakville, ON, Canada), as previously described [,,], or the calcium phosphate co-precipitation method, as described in [,,,]. Briefly, MA-10 cells were transfected 24 h after plating at a density of 100,000 cells/well, by using 0.5 μg of Insl3 promoter construct fused to the Firefly luciferase reporter gene, 20 ng of phRL-TK Renilla luciferase expression vector used as an internal control for transfection efficiency, and pSP64 as carrier DNA up to 1.5 μg/well. For transactivation assays, cells were transfected with 400 ng of the mouse Insl3 −1082/+5 bp reporter vector along with 100 ng of an empty expression vector (pcDNA3.1 as control), or expression vectors for the various transcription factors (50 ng) individually (completed to 100 ng with the empty pcDNA3.1 expression vector to keep the total amount of expression vector to 100 ng), or the combination of transcription factors (50 ng each). The culture media were renewed 3 h before and 16 h after the transfection. Two days after the transfection, MA-10 Leydig cells were harvested and luciferase activities were measured using the Dual Luciferase Assay System (Promega Corp, Madison, WI, USA) and the Luminoskan Ascent luminometer (Thermo Scientific, Milford, MA, USA). The data reported represent the average of at least four experiments, using different DNA preparations, and each performed in duplicate.
4.3. Preparation of Nuclear Extracts
Nuclear extracts from MA-10 Leydig cells were prepared using the method described by Schreiber [], with the following modifications: cells were rinsed with PBS-EDTA, harvested and pelleted by centrifugation for 45 s at 9000 RPM at 4 °C. The cells were resuspended in 700 µL of buffer A (HEPES 10 mM, EDTA 0.1 mM, EGTA 0.1 mM, DTT 1 mM, PMSF 0.5 mM, aprotinin 5 pg/mL, pepstatin 5 µg/mL, leupeptin 5 µg/mL) and incubated on ice for 15 min. Next, 50 µL of 10% Igepal was added and mixed vigorously for 10 s, followed by centrifugation for 30 sec at 13,000 RPM at 4 °C to pellet the nuclei. Nuclei were then incubated in 50 µL of buffer B (HEPES 20 mM, NaCl 400 mM, EDTA 1 mM, EGTA 1 mM, DTT 1 mM, PMSF 1 mM, aprotinin 5 µg/mL, pepstatin 5 pg/mL, leupeptin 5 µg/mL) with vigorous shaking for 45 min at 4 °C, followed by centrifugation for 5 min at 13,000 RPM at 4 °C. Protein concentrations were estimated using the standard Bradford assay (Bio-Rad Laboratories, Hercules, CA, USA). Nuclear proteins were stored at −80 °C until needed.
4.4. Electromobility Shift Assays
Electromobility shift assays (EMSAs) were performed as previously described [,,,,,,,,]. Briefly, 5 µg of nuclear extracts from MA-10 Leydig cells were incubated in 20 µL of 4 mM Tris-HCl (pH 8.0), 24 mM KCl, 0.4 mM EDTA (pH 8.0), 0.4 mM dithiothreitol, 5 mM MgCl2, 100 ng BSA, 10% glycerol, and 500 ng poly(dI-dC) for 1 h on ice. A 32P-labeled 42-bp double-stranded oligonucleotide containing the 35 bp (−186/−151 bp) region of the mouse Insl3 promoter was used as a probe (5′- GTT GGG GAG CGG CTC CTG GCA CAG CGC CGC ACC TGG GAG AGG -3′). Competition experiments were performed using 5× and 25× (molar excess) of unlabeled double-stranded oligonucleotides corresponding to the probe or harboring various mutations (shown in lowercase) in the 35-bp (−186/−151 bp) region of the mouse Insl3 promoter (Table 2). Competitions were also performed using oligonucleotides corresponding to consensus binding sites for different transcription factors (Table 3).

Table 3.
Sequence of oligonucleotides corresponding to consensus binding sites for different transcription factors used in EMSA assays.
4.5. Sequence Analysis
To identify binding sites for potential transcription factors, the 35-bp region of the mouse Insl3 promoter was analyzed using the bioinformatic tools TFbind (https://tfbind.hgc.jp/, last accessed 10 October 2022) [] and PROMO (PROMO version 3.0.2 using version 8.3 of TRANSFAC, last accessed 10 October 2022, http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3) [,]. Multiple sequence alignment was performed using the CLUSTAL Omega multiple sequence alignment tool (version 1.2.4, last accessed 12 October 2022, https://www.ebi.ac.uk/Tools/msa/clustalo/) [].
4.6. Statistical Analysis
Statistical analyses were carried out using Statistics Calculators (Statistics Kingdom, Melbourne, Australia, November 2017, https://www.statskingdom.com/kruskal-wallis-calculator.html, accessed on 7 October 2022). To identify significant differences between multiple groups, statistical analyses were carried out using a nonparametric Kruskal–Wallis one-way ANOVA on ranks followed by a Mann–Whitney U test to detect differences between pairs. For all statistical analyses, p < 0.05 was considered significant.
Author Contributions
X.C.G. performed the majority of the experiments along with K.J.P. and N.M.R. X.C.G., K.J.P. and J.J.T. analyzed and interpreted the data. J.J.T. conceived the study, coordinated and supervised the project, and wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by a grant from the Canadian Institutes of Health Research (CIHR) (funding reference number MOP-81387) to JJT.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data generated or analyzed during this study are included in this article.
Acknowledgments
We would like to thank Dany Chalbos (cJUN), Marc Montminy (CREB), Ming-Jer Tsai (COUP-TFII), Keith Parker (SF1), Steven McKnight (C/EBPβ), and Richard Pope (NF-κB p50) for generously providing plasmids used in this study.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Forest, M.G. Role of androgens in fetal and pubertal development. Horm. Res. 1983, 18, 69–83. [Google Scholar] [CrossRef]
- Nef, S.; Parada, L.F. Cryptorchidism in mice mutant for Insl3. Nat. Genet. 1999, 22, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Zimmermann, S.; Steding, G.; Emmen, J.M.; Brinkmann, A.O.; Nayernia, K.; Holstein, A.F.; Engel, W.; Adham, I.M. Targeted disruption of the Insl3 gene causes bilateral cryptorchidism. Mol. Endocrinol. 1999, 13, 681–691. [Google Scholar] [CrossRef] [PubMed]
- Ferlin, A.; Pepe, A.; Gianesello, L.; Garolla, A.; Feng, S.; Giannini, S.; Zaccolo, M.; Facciolli, A.; Morello, R.; Agoulnik, A.I.; et al. Mutations in the insulin-like factor 3 receptor are associated with osteoporosis. J. Bone Miner. Res. 2008, 23, 683–693. [Google Scholar] [CrossRef]
- Kawamura, K.; Kumagai, J.; Sudo, S.; Chun, S.Y.; Pisarska, M.; Morita, H.; Toppari, J.; Fu, P.; Wade, J.D.; Bathgate, R.A.; et al. Paracrine regulation of mammalian oocyte maturation and male germ cell survival. Proc. Natl. Acad. Sci. USA 2004, 101, 7323–7328. [Google Scholar] [CrossRef]
- Minagawa, I.; Murata, Y.; Terada, K.; Shibata, M.; Park, E.Y.; Sasada, H.; Kohsaka, T. Evidence for the role of INSL3 on sperm production in boars by passive immunisation. Andrologia 2018, 50, e13010. [Google Scholar] [CrossRef]
- Ferlin, A.; De Toni, L.; Agoulnik, A.I.; Lunardon, G.; Armani, A.; Bortolanza, S.; Blaauw, B.; Sandri, M.; Foresta, C. Protective role of testicular hormone INSL3 from atrophy and weakness in skeletal muscle. Front. Endocrinol. 2018, 9, 562. [Google Scholar] [CrossRef]
- Sedaghat, K.; Shen, P.J.; Finkelstein, D.I.; Henderson, J.M.; Gundlach, A.L. Leucine-rich repeat-containing G-protein-coupled receptor 8 in the rat brain: Enrichment in thalamic neurons and their efferent projections. Neuroscience 2008, 156, 319–333. [Google Scholar] [CrossRef]
- Hampel, U.; Klonisch, T.; Sel, S.; Schulze, U.; Garreis, F.; Seitmann, H.; Zouboulis, C.C.; Paulsen, F.P. Insulin-like factor 3 promotes wound healing at the ocular surface. Endocrinology 2013, 154, 2034–2045. [Google Scholar] [CrossRef][Green Version]
- Foresta, C.; Bettella, A.; Vinanzi, C.; Dabrilli, P.; Meriggiola, M.C.; Garolla, A.; Ferlin, A. A novel circulating hormone of testis origin in humans. J. Clin. Endocrinol. Metab. 2004, 89, 5952–5958. [Google Scholar] [CrossRef]
- Ivell, R.; Wade, J.D.; Anand-Ivell, R. INSL3 as a biomarker of Leydig cell functionality. Biol. Reprod. 2013, 88, 147. [Google Scholar] [CrossRef] [PubMed]
- Koskimies, P.; Spiess, A.N.; Lahti, P.; Huhtaniemi, I.; Ivell, R. The mouse relaxin-like factor gene and its promoter are located within the 3′ region of the JAK3 genomic sequence. FEBS Lett. 1997, 419, 186–190. [Google Scholar] [CrossRef]
- Sadeghian, H.; Anand-Ivell, R.; Balvers, M.; Relan, V.; Ivell, R. Constitutive regulation of the Insl3 gene in rat Leydig cells. Mol. Cell Endocrinol. 2005, 241, 10–20. [Google Scholar] [CrossRef] [PubMed]
- Spiess, A.N.; Balvers, M.; Tena-Sempere, M.; Huhtaniemi, I.; Parry, L.; Ivell, R. Structure and expression of the rat relaxin-like factor (RLF) gene. Mol. Reprod. Dev. 1999, 54, 319–325. [Google Scholar] [CrossRef]
- Zimmermann, S.; Schottler, P.; Engel, W.; Adham, I.M. Mouse Leydig insulin-like (Ley I-L) gene: Structure and expression during testis and ovary development. Mol. Reprod. Dev. 1997, 47, 30–38. [Google Scholar] [CrossRef]
- Burkhardt, E.; Adham, I.M.; Brosig, B.; Gastmann, A.; Mattei, M.G.; Engel, W. Structural organization of the porcine and human genes coding for a Leydig cell-specific insulin-like peptide (LEY I-L) and chromosomal localization of the human gene (INSL3). Genomics 1994, 20, 13–19. [Google Scholar] [CrossRef]
- Truong, A.; Bogatcheva, N.V.; Schelling, C.; Dolf, G.; Agoulnik, A.I. Isolation and expression analysis of the canine insulin-like factor 3 gene. Biol. Reprod. 2003, 69, 1658–1664. [Google Scholar] [CrossRef]
- Tremblay, M.A.; Mendoza-Villarroel, R.E.; Robert, N.M.; Bergeron, F.; Tremblay, J.J. KLF6 cooperates with NUR77 and SF1 to activate the human INSL3 promoter in mouse MA-10 leydig cells. J. Mol. Endocrinol. 2016, 56, 163–173. [Google Scholar] [CrossRef][Green Version]
- Koskimies, P.; Levallet, J.; Sipila, P.; Huhtaniemi, I.; Poutanen, M. Murine relaxin-like factor promoter: Functional characterization and regulation by transcription factors steroidogenic factor 1 and DAX-1. Endocrinology 2002, 143, 909–919. [Google Scholar] [CrossRef]
- Zimmermann, S.; Schwarzler, A.; Buth, S.; Engel, W.; Adham, I.M. Transcription of the Leydig insulin-like gene is mediated by steroidogenic factor-1. Mol. Endocrinol. 1998, 12, 706–713. [Google Scholar] [CrossRef]
- Tremblay, J.J.; Robert, N.M. Role of nuclear receptors in INSL3 gene transcription in Leydig cells. Ann. N. Y. Acad. Sci. 2005, 1061, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Robert, N.M.; Martin, L.J.; Tremblay, J.J. The orphan nuclear receptor NR4A1 regulates Insulin-like 3 gene transcription in Leydig cells. Biol. Reprod. 2006, 74, 322–330. [Google Scholar] [CrossRef] [PubMed]
- Laguë, E.; Tremblay, J.J. Antagonistic effects of testosterone and the endocrine disruptor mono-(2-ethylhexyl) phthalate on INSL3 transcription in Leydig cells. Endocrinology 2008, 149, 4688–4694. [Google Scholar] [CrossRef]
- Tremblay, J.J.; Robert, N.M.; Laguë, E. Nuclear receptors, testosterone and post-translational modifications in human INSL3 promoter activity in testicular Leydig cells. Ann. N. Y. Acad. Sci. 2009, 1160, 205–212. [Google Scholar] [CrossRef]
- Mendoza-Villarroel, R.E.; Di-Luoffo, M.; Camire, E.; Giner, X.C.; Brousseau, C.; Tremblay, J.J. The Insl3 gene is a direct target for the orphan nuclear receptor, COUP-TFII, in Leydig cells. J. Mol. Endocrinol. 2014, 53, 43–55. [Google Scholar] [CrossRef]
- Di-Luoffo, M.; Pierre, K.J.; Robert, N.M.; Girard, M.J.; Tremblay, J.J. The nuclear receptors SF1 and COUP-TFII cooperate on the Insl3 promoter in Leydig cells. Reproduction 2022, 164, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Strong, M.E.; Burd, M.A.; Peterson, D.G. Evaluation of the MA-10 cell line as a model of insl3 regulation and Leydig cell function. Anim. Reprod. Sci. 2019, 208, 106116. [Google Scholar] [CrossRef]
- Nguyen, H.T.; Najih, M.; Martin, L.J. The AP-1 family of transcription factors are important regulators of gene expression within Leydig cells. Endocrine 2021, 74, 498–507. [Google Scholar] [CrossRef]
- Esteban-Lopez, M.; Agoulnik, A.I. Diverse functions of insulin-like 3 peptide. J. Endocrinol. 2020, 247, R1–R12. [Google Scholar] [CrossRef]
- Ascoli, M. Characterization of several clonal lines of cultured Leydig tumor cells: Gonadotropin receptors and steroidogenic responses. Endocrinology 1981, 108, 88–95. [Google Scholar] [CrossRef] [PubMed]
- Trbovich, A.M.; Martinelle, N.; O’Neill, F.H.; Pearson, E.J.; Donahoe, P.K.; Sluss, P.M.; Teixeira, J. Steroidogenic activities in MA-10 Leydig cells are differentially altered by cAMP and Mullerian inhibiting substance. J. Steroid Biochem. Mol. Biol. 2004, 92, 199–208. [Google Scholar] [CrossRef]
- Ascoli, M. Immortalized Leydig cell lines as models for studying Leydig cell physiology. In The Leydig Cell in Health and Disease, 2nd ed.; Payne, A.H.H., Hardy, M.P., Eds.; Humana Press: Totowa, NJ, USA, 2007; pp. 373–381. [Google Scholar] [CrossRef]
- Clark, B.J.; Wells, J.; King, S.R.; Stocco, D.M. The purification, cloning, and expression of a novel luteinizing hormone-induced mitochondrial protein in MA-10 mouse Leydig tumor cells. Characterization of the steroidogenic acute regulatory protein (StAR). J. Biol. Chem. 1994, 269, 28314–28322. [Google Scholar] [CrossRef]
- King, S.R.; Ronen-Fuhrmann, T.; Timberg, R.; Clark, B.J.; Orly, J.; Stocco, D.M. Steroid production after in vitro transcription, translation, and mitochondrial processing of protein products of complementary deoxyribonucleic acid for steroidogenic acute regulatory protein. Endocrinology 1995, 136, 5165–5176. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J.; Boucher, N.; Brousseau, C.; Tremblay, J.J. The orphan nuclear receptor NUR77 regulates hormone-induced StAR transcription in Leydig cells through a cooperation with CaMKI. Mol. Endocrinol. 2008, 22, 2021–2037. [Google Scholar] [CrossRef]
- Engeli, R.T.; Furstenberger, C.; Kratschmar, D.V.; Odermatt, A. Currently available murine Leydig cell lines can be applied to study early steps of steroidogenesis but not testosterone synthesis. Heliyon 2018, 4, e00527. [Google Scholar] [CrossRef] [PubMed]
- Balvers, M.; Spiess, A.N.; Domagalski, R.; Hunt, N.; Kilic, E.; Mukhopadhyay, A.K.; Hanks, E.; Charlton, H.M.; Ivell, R. Relaxin-like factor expression as a marker of differentiation in the mouse testis and ovary. Endocrinology 1998, 139, 2960–2970. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Kobayashi, K.; Murayama, K.; Kawahara, K.; Shima, Y.; Suzuki, A.; Tani, K.; Takahashi, A. FEAT enhances INSL3 expression in testicular Leydig cells. Genes Cells 2018, 23, 952–962. [Google Scholar] [CrossRef] [PubMed]
- Laguë, E.; Tremblay, J.J. Estradiol represses Insulin-like 3 expression and promoter activity in MA-10 Leydig cells. Toxicology 2009, 258, 101–105. [Google Scholar] [CrossRef]
- Clem, B.F.; Hudson, E.A.; Clark, B.J. Cyclic adenosine 3′,5′-monophosphate (cAMP) enhances cAMP-responsive element binding (CREB) protein phosphorylation and phospho-CREB interaction with the mouse steroidogenic acute regulatory protein gene promoter. Endocrinology 2005, 146, 1348–1356. [Google Scholar] [CrossRef][Green Version]
- Manna, P.R.; Dyson, M.T.; Eubank, D.W.; Clark, B.J.; Lalli, E.; Sassone-Corsi, P.; Zeleznik, A.J.; Stocco, D.M. Regulation of steroidogenesis and the steroidogenic acute regulatory protein by a member of the cAMP response-element binding protein family. Mol. Endocrinol. 2002, 16, 184–199. [Google Scholar] [CrossRef]
- Manna, P.R.; Eubank, D.W.; Lalli, E.; Sassone-Corsi, P.; Stocco, D.M. Transcriptional regulation of the mouse steroidogenic acute regulatory protein gene by the cAMP response-element binding protein and steroidogenic factor 1. J. Mol. Endocrinol. 2003, 30, 381–397. [Google Scholar] [CrossRef] [PubMed]
- El-Asmar, B.; Giner, X.C.; Tremblay, J.J. Transcriptional cooperation between NF-κB and C/EBPβ regulates Nur77 transcription in Leydig cells. J. Mol. Endocrinol. 2009, 142, 131–138. [Google Scholar]
- Nalbant, D.; Williams, S.C.; Stocco, D.M.; Khan, S.A. Luteinizing hormone-dependent gene regulation in Leydig cells may be mediated by CCAAT/enhancer-binding protein-beta. Endocrinology 1998, 139, 272–279. [Google Scholar] [CrossRef][Green Version]
- Reinhart, A.J.; Williams, S.C.; Clark, B.J.; Stocco, D.M. SF-1 (steroidogenic factor-1) and C/EBP beta (CCAAT/enhancer binding protein-beta) cooperate to regulate the murine StAR (steroidogenic acute regulatory) promoter. Mol. Endocrinol. 1999, 13, 729–741. [Google Scholar]
- Tsai-Morris, C.H.; Geng, Y.; Xie, X.Z.; Buczko, E.; Dufau, M.L. Transcriptional protein binding domains governing basal expression of the rat luteinizing hormone receptor gene. J. Biol. Chem. 1994, 269, 15868–15875. [Google Scholar] [CrossRef]
- Tremblay, J.J.; Viger, R.S. Transcription factor GATA-4 enhances Müllerian inhibiting substance gene transcription through a direct interaction with the nuclear receptor SF-1. Mol. Endocrinol. 1999, 13, 1388–1401. [Google Scholar] [CrossRef]
- Tremblay, J.J.; Viger, R.S. GATA factors differentially activate multiple gonadal promoters through conserved GATA regulatory elements. Endocrinology 2001, 142, 977–986. [Google Scholar] [CrossRef]
- Lala, D.S.; Rice, D.A.; Parker, K.L. Steroidogenic factor I, a key regulator of steroidogenic enzyme expression, is the mouse homolog of fushi tarazu-factor I. Mol. Endocrinol. 1992, 6, 1249–1258. [Google Scholar] [CrossRef] [PubMed]
- Teyssier, C.; Belguise, K.; Galtier, F.; Chalbos, D. Characterization of the physical interaction between estrogen receptor alpha and JUN proteins. J. Biol. Chem. 2001, 276, 36361–36369. [Google Scholar] [CrossRef]
- Mayr, B.; Montminy, M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell Biol. 2001, 2, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Umek, R.M.; McKnight, S.L. Regulated expression of three C/EBP isoforms during adipose conversion of 3T3-L1 cells. Genes Dev. 1991, 5, 1538–1552. [Google Scholar] [CrossRef] [PubMed]
- Pereira, F.A.; Qiu, Y.; Zhou, G.; Tsai, M.J.; Tsai, S.Y. The orphan nuclear receptor COUP-TFII is required for angiogenesis and heart development. Genes Dev 1999, 13, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Sidiropoulos, P.; Song, G.; Pagliari, L.J.; Birrer, M.J.; Stein, B.; Anrather, J.; Pope, R.M. TNF-alpha gene expression in macrophages: Regulation by NF-kappa B is independent of c-Jun or C/EBP beta. J. Immunol. 2000, 164, 4277–4285. [Google Scholar] [CrossRef]
- Abdou, H.S.; Bergeron, F.; Tremblay, J.J. A cell-autonomous molecular cascade initiated by AMP-activated protein kinase represses steroidogenesis. Mol. Cell Biol. 2014, 34, 4257–4271. [Google Scholar] [CrossRef] [PubMed]
- Abdou, H.S.; Villeneuve, G.; Tremblay, J.J. The calcium signaling pathway regulates leydig cell steroidogenesis through a transcriptional cascade involving the nuclear receptor NR4A1 and the steroidogenic acute regulatory protein. Endocrinology 2013, 154, 511–520. [Google Scholar] [CrossRef] [PubMed]
- Daems, C.; Di-Luoffo, M.; Paradis, E.; Tremblay, J.J. MEF2 cooperates with forskolin/cAMP and GATA4 to regulate Star gene expression in mouse MA-10 Leydig cells. Endocrinology 2015, 156, 2693–2703. [Google Scholar] [CrossRef]
- Enangue Njembele, A.N.; Bailey, J.L.; Tremblay, J.J. In vitro exposure of Leydig cells to an environmentally relevant mixture of organochlorines represses early steps of steroidogenesis. Biol. Reprod. 2014, 90, 118. [Google Scholar] [CrossRef]
- Enangue Njembele, A.N.; Demmouche, Z.B.; Bailey, J.L.; Tremblay, J.J. Mechanism of action of an environmentally relevant organochlorine mixture in repressing steroid hormone biosynthesis in Leydig cells. Int. J. Mol. Sci. 2022, 23, 3997. [Google Scholar] [CrossRef]
- Enangue Njembele, A.N.; Tremblay, J.J. Mechanisms of MEHP Inhibitory Action and Analysis of Potential Replacement Plasticizers on Leydig Cell Steroidogenesis. Int. J. Mol. Sci. 2021, 22, 11456. [Google Scholar] [CrossRef]
- Hebert-Mercier, P.O.; Bergeron, F.; Robert, N.M.; Mehanovic, S.; Pierre, K.J.; Mendoza-Villarroel, R.E.; de Mattos, K.; Brousseau, C.; Tremblay, J.J. Growth hormone-induced STAT5B regulates Star gene expression through a cooperation with cJUN in mouse MA-10 Leydig cells. Endocrinology 2022, 163, 1–13. [Google Scholar] [CrossRef]
- Mendoza-Villarroel, R.E.; Robert, N.M.; Martin, L.J.; Brousseau, C.; Tremblay, J.J. The nuclear receptor NR2F2 activates Star expression and steroidogenesis in mouse MA-10 and MLTC-1 Leydig cells. Biol. Reprod. 2014, 91, 26. [Google Scholar] [CrossRef] [PubMed]
- Pierre, K.J.; Tremblay, J.J. Differential response of transcription factors to activated kinases in steroidogenic and non-steroidogenic cells. Int. J. Mol. Sci. 2022, 23, 13153. [Google Scholar] [CrossRef] [PubMed]
- Mehanovic, S.; Mendoza-Villarroel, R.E.; Viger, R.S.; Tremblay, J.J. The nuclear receptor COUP-TFII regulates Amhr2 gene transcription via a GC-rich promoter element in mouse Leydig cells. J. Endocr. Soc. 2019, 3, 2236–2257. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J.; Boucher, N.; El-Asmar, B.; Tremblay, J.J. cAMP-induced expression of the orphan nuclear receptor Nur77 in testicular Leydig cells involves a CaMKI pathway. J. Androl. 2009, 30, 134–145. [Google Scholar] [CrossRef]
- Schreiber, E.; Matthias, P.; Muller, M.M.; Schaffner, W. Rapid detection of octamer binding proteins with ’mini-extracts’, prepared from a small number of cells. Nucleic Acids Res. 1989, 17, 6419. [Google Scholar] [CrossRef]
- Bergeron, F.; Bagu, E.T.; Tremblay, J.J. Transcription of platelet-derived growth factor receptor alpha in Leydig cells involves specificity protein 1 and 3. J. Mol. Endocrinol. 2011, 46, 125–138. [Google Scholar] [CrossRef]
- Daems, C.; Martin, L.J.; Brousseau, C.; Tremblay, J.J. MEF2 is restricted to the male gonad and regulates expression of the orphan nuclear receptor NR4A1. Mol. Endocrinol. 2014, 28, 886–898. [Google Scholar] [CrossRef]
- Di-Luoffo, M.; Daems, C.; Bergeron, F.; Tremblay, J.J. Novel targets for the transcription factors MEF2 in MA-10 Leydig cells. Biol. Reprod. 2015, 93, 9. [Google Scholar] [CrossRef]
- Garon, G.; Bergeron, F.; Brousseau, C.; Robert, N.M.; Tremblay, J.J. FOXA3 is expressed in multiple cell lineages in the mouse testis and regulates Pdgfra expression in Leydig cells. Endocrinology 2017, 158, 1886–1897. [Google Scholar] [CrossRef]
- Martin, L.J.; Tremblay, J.J. The human 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase type 2 promoter is a novel target for the immediate early orphan nuclear receptor NUR77 in steroidogenic cells. Endocrinology 2005, 146, 861–869. [Google Scholar] [CrossRef]
- Tu, S.; Teng, Y.C.; Yuan, C.; Wu, Y.T.; Chan, M.Y.; Cheng, A.N.; Lin, P.H.; Juan, L.J.; Tsai, M.D. The ARID domain of the H3K4 demethylase RBP2 binds to a DNA CCGCCC motif. Nat. Struct. Mol. Biol. 2008, 15, 419–421. [Google Scholar] [CrossRef]
- Persengiev, S.P.; Kondova, I.I.; Millette, C.F.; Kilpatrick, D.L. GLI family members are differentially expressed during the mitotic phase of spermatogenesis. Oncogene 1997, 14, 2259–2264. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ström, L.; Lundgren, M.; Severinson, E. Binding of Ikaros to germline Ig heavy chain gamma1 and epsilon promoters. Mol. Immunol. 2003, 39, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Giatzakis, C.; Papadopoulos, V. Differential utilization of the promoter of peripheral-type benzodiazepine receptor by steroidogenic versus nonsteroidogenic cell lines and the role of Sp1 and Sp3 in the regulation of basal activity. Endocrinology 2004, 145, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Yasuda, K.; Hirayoshi, K.; Hirata, H.; Kubota, H.; Hosokawa, N.; Nagata, K. The Kruppel-like factor ZF9 and proteins in the SP1 family regulate the expression of HSP47, a collagen-specific molecular chaperone. J. Biol. Chem. 2002, 277, 44613–44622. [Google Scholar] [CrossRef] [PubMed]
- Hewetson, A.; Hendrix, E.C.; Mansharamani, M.; Lee, V.H.; Chilton, B.S. Identification of the RUSH consensus-binding site by cyclic amplification and selection of targets: Demonstration that RUSH mediates the ability of prolactin to augment progesterone-dependent gene expression. Mol. Endocrinol. 2002, 16, 2101–2112. [Google Scholar] [CrossRef]
- Jethanandani, P.; Kramer, R.H. Alpha7 integrin expression is negatively regulated by deltaEF1 during skeletal myogenesis. J. Biol. Chem. 2005, 280, 36037–36046. [Google Scholar] [CrossRef]
- Hill, A.A.; Riley, P.R. Differential regulation of Hand1 homodimer and Hand1-E12 heterodimer activity by the cofactor FHL2. Mol. Cell. Biol. 2004, 24, 9835–9847. [Google Scholar] [CrossRef]
- Tsunoda, T.; Takagi, T. Estimating transcription factor bindability on DNA. Bioinformatics 1999, 15, 622–630. [Google Scholar] [CrossRef]
- Farre, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Rosello, L.; Alba, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653. [Google Scholar] [CrossRef]
- Messeguer, X.; Escudero, R.; Farre, D.; Nunez, O.; Martinez, J.; Alba, M.M. PROMO: Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 2002, 18, 333–334. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).