RNA-Binding Proteins as Epigenetic Regulators of Brain Functions and Their Involvement in Neurodegeneration
Abstract
:1. Introduction
2. Post-Transcriptional Regulation of Gene Expression in the Nervous System
2.1. Mechanisms of Controlled RNA Metabolism, Localization, and Translation
2.2. Intrinsically Disordered Regions (IDRs)
2.3. Brain Cell Asymmetry
2.4. RBPs as Regulators of mRNA Pre-Localization in Brain Cells
2.5. RBPs and Neuronal Plasticity
3. Role of RBPs in RNA Sorting to Extracellular Vesicles
3.1. Extracellular Vesicles (EVs): Origin and General Functions
3.2. Roles of EVs in the Nervous System
3.3. Specific Sorting of Molecules to Nascent EVs: Possible Role of RNA-Binding Proteins
4. RBPs and Neurological Diseases
4.1. Amyotrophic Lateral Sclerosis (ALS)
4.1.1. TDP-43
4.1.2. FUS
4.2. Multiple Sclerosis (MS)
4.3. Alzheimer’s Disease (AD)
5. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Derrigo, M.; Cestelli, A.; Savettieri, G.; Di Liegro, I. RNA-protein interactions in the control of stability and localization of messenger RNA (Review). Int. J. Mol. Med. 2000, 5, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Bolognani, F.; Perrone-Bizzozero, N.I. RNA-protein interactions and control of mRNA stability in neurons. J. Neurosci. Res. 2008, 86, 481–489. [Google Scholar] [CrossRef] [PubMed]
- Darnell, J.C.; Richter, J.D. Cytoplasmic RNA-binding proteins and the control of complex brain function. Cold Spring Harb. Perspect. Biol. 2012, 4, a012344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutten, S.; Sharangdhar, T.; Kiebler, M. Unmasking the messenger. RNA Biol. 2014, 11, 992–997. [Google Scholar] [CrossRef] [Green Version]
- Ravanidis, S.; Kattan, F.G.; Doxakis, E. Unraveling the Pathways to Neuronal Homeostasis and Disease: Mechanistic Insights into the Role of RNA-Binding Proteins and Associated Factors. Int. J. Mol. Sci. 2018, 19, 2280. [Google Scholar] [CrossRef] [Green Version]
- Mirisis, A.A.; Carew, T.J. The ELAV family of RNA-binding proteins in synaptic plasticity and long-term memory. Neurobiol. Learn. Mem. 2019, 161, 143–148. [Google Scholar] [CrossRef]
- Ohashi, R.; Shiina, N. Cataloguing and Selection of mRNAs Localized to Dendrites in Neurons and Regulated by RNA-Binding Proteins in RNA Granules. Biomolecules 2020, 10, 167. [Google Scholar] [CrossRef] [Green Version]
- Landinez-Macias, M.; Urwyler, O. The Fine Art of Writing a Message: RNA Metabolism in the Shaping and Remodeling of the Nervous System. Front. Mol. Neurosci. 2021, 14, 755686. [Google Scholar] [CrossRef]
- Guan, W.; Bellemin, S.; Bouchet, M.; Venkatasubramanian, L.; Guillermin, C.; Laurençon, A.; Kabir, C.; Darnas, A.; Godin, C.; Urdy, S.; et al. Post-transcriptional regulation of transcription factor codes in immature neurons drives neuronal diversity. Cell Rep. 2022, 39, 110992. [Google Scholar] [CrossRef]
- Parra, A.S.; Johnston, C.A. Emerging Roles of RNA-Binding Proteins in Neurodevelopment. J. Dev. Biol. 2022, 10, 23. [Google Scholar] [CrossRef]
- Woese, C.R. The Genetic Code; Harper & Row: Manhattan, NY, USA, 1967. [Google Scholar]
- Crick, F.H.C. The origin of the genetic code. J. Mol. Biol. 1968, 38, 367–379. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E. Evolution of the genetic apparatus. J. Mol. Biol. 1968, 38, 381–393. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, W. The RNA world. Nature 1986, 319, 618. [Google Scholar] [CrossRef]
- Joyce, G.F. The antiquity of RNA-based evolution. Nature 2002, 418, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Spirin, A.S. Omnipotent RNA. FEBS Lett. 2002, 530, 4–8. [Google Scholar] [CrossRef] [Green Version]
- Vlassov, A.V.; Kazakov, S.A.; Johnston, B.H.; Landweber, L.F. The RNA world on ice: A new scenario for the emergence of RNA information. J. Mol. Evol. 2005, 61, 264–273. [Google Scholar] [CrossRef]
- Taylor, W.R. Transcription and translation in an RNA world. Philos. Trans. Soc. B 2006, 361, 1751–1760. [Google Scholar] [CrossRef] [Green Version]
- Kruger, K.; Grabowski, P.J.; Zaug, A.J.; Sands, J.; Gottschling, D.E.; Cech, T.R. Self-splicing RNA: Autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell 1982, 31, 147–157. [Google Scholar] [CrossRef]
- Guerrier-Takada, C.; Gardiner, K.; Marsh, T.; Pace, N.; Altman, S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell 1983, 35, 849–857. [Google Scholar] [CrossRef]
- Bartel, D.P.; Szostak, J.W. Isolation of new ribozymes from a large pool of random sequences. Science 1993, 261, 1411–1418. [Google Scholar] [CrossRef]
- Kumar, R.K.; Yarus, M. RNA-catalyzed amino acid activation. Biochemistry 2001, 40, 6998–7004. [Google Scholar] [CrossRef] [PubMed]
- Hordijk, W. Autocatalytic sets and RNA secondary structure. J. Mol. Evol. 2017, 84, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, D.; Wang, Y. Protein-free ribosomal RNA scaffolds can assemble poly-lysine oligos from charged tRNA fragments. Biochem. Biophys. Res. Commun. 2021, 544, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Wolk, S.K.; Mayfield, W.S.; Gelinas, A.D.; Astling, D.; Guillot, J.; Brody, E.N.; Janjic, N.; Gold, L. Modified nucleotides may have enhanced early RNA catalysis. Proc. Natl. Acad. Sci. USA 2020, 117, 8236–8242. [Google Scholar] [CrossRef] [PubMed]
- Goldman, A.D.; Kacar, B. Cofactors are Remnants of Life’s Origin and Early Evolution. J. Mol. Evol. 2021, 89, 127–133. [Google Scholar] [CrossRef]
- Nam, J.-W.; Choi, S.-W.; You, B.-H. Incredible RNA: Dual Functions of Coding and Noncoding. Mol. Cells 2016, 39, 367–374. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.L. A Production of amino acids under possible primitive earth conditions. Science 1953, 117, 528–529. [Google Scholar] [CrossRef] [Green Version]
- Miller, S.L. Production of some organic compounds under possible primitive earth conditions. J. Am. Chem. Soc. 1955, 77, 2351–2361. [Google Scholar] [CrossRef]
- Miller, S.L. The Atmosphere of the Primitive Earth and the Prebiotic Synthesis of Aminoacids. In Cosmochemical Evolution and the Origins of Life; Springer: Dordrecht, The Netherlands, 1974; Volume 5, pp. 139–151. [Google Scholar]
- Ikehara, K. [GADV]-Protein world Hypothesis on the origin of life. Orig. Life Evol. Biosph. 2014, 44, 299–302. [Google Scholar] [CrossRef] [Green Version]
- Greenwald, J.; Kwiatkowski, W.; Riek, R. Peptide Amyloids in the origin of life. J. Mol. Biol. 2018, 430, 3735–3750. [Google Scholar] [CrossRef]
- Carny, O.; Gazit, E. A model for the role of short self-assembled peptides in the very early stages of the origin of life. FASEB J. 2005, 19, 1051–1055. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.P.; Banerji, A. Case for an RNA-prion world: A hypothesis based on conformational diversity. J. Biol. Phys. 2011, 37, 185–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maury, C.P.J. Amyloid and the origin of life: Self-replicating catalytic amyloids as prebiotic informational and protometabolic entities. Cell. Mol. Life Sci. 2018, 75, 1499–1507. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rufo, C.M.; Moroz, Y.S.; Moroz, O.V.; Stöhr, J.; Smith, T.A.; Hu, X.; DeGrado, W.F.; Korendovych, I.V. Short peptides self-assemble to produce catalytic amyloids. Nat. Chem. 2014, 6, 303–309. [Google Scholar] [CrossRef] [Green Version]
- Zozulia, O.; Dolan, M.A.; Korendovych, I.V. Catalytic peptide assemblies. Chem. Soc. Rev. 2018, 47, 3621–3639. [Google Scholar] [CrossRef]
- Kayed, R.; Sokolov, Y.; Edmonds, B.; McIntire, T.M.; Milton, S.C.; Hall, J.E.; Glabe, C.G. Permeabilization of lipid bilayers is a common conformation-dependent activity of soluble amyloid oligomers in protein misfolding diseases. J. Biol. Chem. 2004, 279, 46363–46366. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, K. Physicochemical interactions of amyloid beta-peptide with lipid bilayers. Biochim. Biophys. Acta 2007, 1768, 1935–1942. [Google Scholar] [CrossRef] [Green Version]
- Butterfield, S.M.; Lashuel, H.A. Amyloidogenic protein-membrane interactions: Mechanistic insight from model systems. Angew. Chem. Int. Ed. Engl. 2010, 49, 5628–5654. [Google Scholar] [CrossRef]
- Carny, O.; Gazit, E. Creating prebiotic sanctuary: Self-assembling supramolecular Peptide structures bind and stabilize RNA. Orig. Life Evol. Biosph. 2011, 41, 121–132. [Google Scholar] [CrossRef]
- Drobot, B.; Iglesias-Artola, J.M.; Le Vay, K.; Mayr, V.; Kar, M.; Kreysing, M.; Mutschler, H.; Tang, T.-Y.D. Compartmentalised RNA catalysis in membrane-free coacervate protocells. Nat. Commun. 2018, 9, 3643. [Google Scholar] [CrossRef]
- Olasagasti, F.; Rajamani, S. Lipid-assisted polymerization of nucleotides. Life 2019, 9, 83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cai, Y.; Wan, J. Competing Endogenous RNA Regulations in Neurodegenerative Disorders: Current Challenges and Emerging Insights. Front. Mol. Neurosci. 2018, 11, 370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salmena, L.; Poliseno, L.; Tay, Y.; Kats, L.; Pandolfi, P.P. A ceRNA hypothesis: The Rosetta Stone of a hidden RNA language? Cell 2011, 146, 353–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guil, S.; Esteller, M. RNA-RNA interactions in gene regulation: The coding and noncoding players. Trends Biochem. Sci. 2015, 40, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Fabian, M.R.; Sonenberg, N. The mechanics of miRNA-mediated gene silencing: A look under the hood of miRISC. Nat. Struct. Mol. Biol. 2012, 19, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Sioud, M. RNA Interference: Story and Mechanisms. In Methods in Molecular Biology; Humana: Louisville, KY, USA, 2021; Volume 2282, pp. 1–15. [Google Scholar] [CrossRef]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microrna sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Li, X.; Yang, L.; Chen, L.L. The biogenesis, functions, and challenges of circular rnas. Mol. Cell 2018, 71, 428–442. [Google Scholar] [CrossRef] [Green Version]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding rnas. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [Green Version]
- Kageyama, Y.; Kondo, T.; Hashimoto, Y. Coding vs non-coding: Translatability of short ORFs found in putative non-coding transcripts. Biochimie 2011, 93, 1981–1986. [Google Scholar] [CrossRef]
- Ule, J.; Blencowe, B.J. Alternative Splicing Regulatory Networks: Functions, Mechanisms, and Evolution. Mol. Cell 2019, 76, 329–345. [Google Scholar] [CrossRef]
- Marima, R.; Francies, F.Z.; Hull, R.; Molefi, T.; Oyomno, M.; Khanyile, R.; Mbatha, S.; Mabongo, M.; Owen Bates, D.; Dlamini, Z. MicroRNA and Alternative mRNA Splicing Events in Cancer Drug Response/Resistance: Potent Therapeutic Targets. Biomedicines 2021, 9, 1818. [Google Scholar] [CrossRef] [PubMed]
- Wei, L.; Lai, E.C. Regulation of the Alternative Neural Transcriptome by ELAV/Hu RNA Binding Proteins. Front. Genet. 2022, 13, 848626. [Google Scholar] [CrossRef] [PubMed]
- Vallejos Baier, R.; Picao-Osorio, J.; Alonso, C.R. Molecular Regulation of Alternative Polyadenylation (APA) within the Drosophila Nervous System. J. Mol. Biol. 2017, 429, 3290–3300. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Manley, J.L. Alternative polyadenylation of mRNA precursors. Nat. Rev. Mol. Cell Biol. 2017, 18, 18–30. [Google Scholar] [CrossRef]
- Gruber, A.J.; Zavolan, M. Alternative cleavage and polyadenylation in health and disease. Nat. Rev. Genet. 2019, 20, 599–614. [Google Scholar] [CrossRef]
- Guvenek, A.; Shin, J.; De Filippis, L.; Zheng, D.; Wang, W.; Pang, Z.P.; Tian, B. Neuronal Cells Display Distinct Stability Controls of Alternative Polyadenylation mRNA Isoforms, Long Non-Coding RNAs, and Mitochondrial RNAs. Front. Genet. 2022, 13, 840369. [Google Scholar] [CrossRef]
- Mitschka, S.; Mayr, C. Context-specific regulation and function of mRNA alternative polyadenylation. Nat. Rev. Mol. Cell Biol. 2022, 23, 779–796. [Google Scholar] [CrossRef]
- Pitolli, C.; Marini, A.; Sette, C.; Pagliarini, V. Non-canonical splicing and its implications in brain physiology and cancer. Int. J. Mol. Sci. 2022, 23, 2811. [Google Scholar] [CrossRef]
- Glanzer, J.; Miyashiro, K.Y.; Sul, J.Y.; Barrett, L.; Belt, B.; Haydon, P.; Eberwine, J. RNA splicing capability of live neuronal dendrites. Proc. Natl. Acad. Sci. USA 2005, 102, 16859–16864. [Google Scholar] [CrossRef]
- Khaladkar, M.; Buckley, P.T.; Lee, M.T.; Francis, C.; Eghbal, M.M.; Chuong, T.; Suresh, S.; Kuhn, B.; Eberwine, J.; Kim, J. Subcellular RNA sequencing reveals broad presence of cytoplasmic intron-sequence retaining transcripts in mouse and rat neurons. PLoS ONE 2013, 8, e76194. [Google Scholar] [CrossRef] [Green Version]
- Buckley, P.T.; Khaladkar, M.; Kim, J.; Eberwine, J. Cytoplasmic intron retention, function, splicing, and the sentinel RNA hypothesis. Wiley Interdiscip. Rev. RNA 2014, 5, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Amort, T.; Rieder, D.; Wille, A.; Khokhlova-Cubberley, D.; Riml, C.; Trixl, L.; Jia, X.Y.; Micura, R.; Lusser, A. Distinct 5-methylcytosine profiles in poly(A) RNA from mouse embryonic stem cells and brain. Genome Biol. 2017, 18, 1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schieweck, R.; Ninkovic, J.; Kiebler, M.A. RNA-binding proteins balance brain function in health and disease. Physiol. Rev. 2021, 101, 1309–1370. [Google Scholar] [CrossRef] [PubMed]
- Weng, Y.L.; Wang, X.; An, R.; Cassin, J.; Vissers, C.; Liu, Y.; Liu, Y.; Xu, T.; Wang, X.; Wong, S.Z.; et al. Epitranscriptomic m6A regulation of axon regeneration in the adult mammalian nervous system. Neuron 2018, 97, 313–325.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, K.D.; Patil, D.P.; Zhou, J.; Zinoviev, A.; Skabkin, M.A.; Elemento, O.; Pestova, T.V.; Qian, S.B.; Jaffrey, S.R. 5’UTR M(6)A Promotes Cap-Independent Translation. Cell 2015, 163, 999–1010. [Google Scholar] [CrossRef] [Green Version]
- Saletore, Y.; Meyer, K.; Korlach, J.; Vilfan, I.D.; Jaffrey, S.; Mason, C.E. The birth of the Epitranscriptome: Deciphering the function of RNA modifications. Genome Biol. 2012, 13, 175. [Google Scholar] [CrossRef] [Green Version]
- Savva, Y.A.; Rieder, L.E.; Reenan, R.A. The ADAR protein family. Genome Biol. 2012, 13, 252. [Google Scholar] [CrossRef] [Green Version]
- Lerner, T.; Papavasiliou, F.N.; Pecori, R. RNA Editors, Cofactors, and mRNA Targets: An Overview of the C-to-U RNA Editing Machinery and Its Implication in Human Disease. Genes 2018, 10, 13. [Google Scholar] [CrossRef]
- Karagianni, K.; Pettas, S.; Christoforidou, G.; Kanata, E.; Bekas, N.; Xanthopoulos, K.; Dafou, D.; Sklaviadis, T. A Systematic Review of Common and Brain-Disease-Specific RNA Editing Alterations Providing Novel Insights into Neurological and Neurodegenerative Disease Manifestations. Biomolecules 2022, 12, 465. [Google Scholar] [CrossRef]
- Di Liegro, C.M.; Schiera, G.; Di Liegro, I. Regulation of mRNA transport, localization and translation in the nervous system of mammals (Review). Int. J. Mol. Med. 2014, 33, 747–762. [Google Scholar] [CrossRef] [Green Version]
- Kiebler, M.A.; Bassell, G.J. Neuronal RNA granules: Movers and makers. Neuron 2006, 51, 685–690. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitchell, S.F.; Parker, R. Principles and properties of eukaryotic mRNPs. Mol. Cell. 2014, 54, 547–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lianoglou, S.; Garg, V.; Yang, J.L.; Leslie, C.S.; Mayr, C. Ubiquitously transcribed genes use alternative polyadenylation to achieve tissue-specific expression. Genes Dev. 2013, 27, 2380–2396. [Google Scholar] [CrossRef] [Green Version]
- Frye, M.; Harada, B.T.; Behm, M.; He, C. RNA modifications modulate gene expression during development. Science 2018, 361, 1346–1349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Chen, M.; Huang, H.; Zhu, J.; Song, H.; Zhu, J.; Park, J.; Ji, S.J. Dynamic m6A modification regulates local translation of mRNA in axons. Nucleic Acids Res. 2017, 46, 1412–1423. [Google Scholar] [CrossRef] [Green Version]
- Wassarman, D.A.; Steitz, J.A. Interactions of small nuclear RNAs with precursor messenger RNA during in vitro splicing. Science 1992, 257, 1918–1925. [Google Scholar] [CrossRef]
- Raj, B.; Blencowe, B.J. Alternative splicing in the mammalian nervous system: Recent insights into mechanisms and functional roles. Neuron 2015, 87, 14–27. [Google Scholar] [CrossRef] [Green Version]
- Baralle, F.E.; Giudice, J. Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 2017, 18, 437–451. [Google Scholar] [CrossRef]
- Jackson, T.C.; Kochanek, P.M. RNA Binding Motif 5 (RBM5) in the CNS-Moving Beyond Cancer to Harness RNA Splicing to Mitigate the Consequences of Brain Injury. Front. Mol. Neurosci. 2020, 13, 126. [Google Scholar] [CrossRef]
- Zheng, J.; Redmond, L.; Xu, C.; Kuang, J.; Liao, W. Alternative splicing in the variable domain of CaMKIIβ affects the level of F-actin association in developing neurons. Int. J. Clin. Exp. Pathol. 2014, 7, 2963–2975. [Google Scholar] [PubMed]
- Schreiner, D.; Nguyen, T.M.; Russo, G.; Heber, S.; Patrignani, A.; Ahrne, E.; Scheiffele, P. Targeted combinatorial alternative splicing generates brain region-specific repertoires of neurexins. Neuron 2014, 84, 386–398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhof, T.C. Synaptic Neurexin Complexes: A Molecular Code for the Logic of Neural Circuits. Cell 2017, 171, 745–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez, A.M.; Traunmüller, L.; Scheiffele, P. Neurexins: Molecular codes for shaping neuronal synapses. Nat. Rev. Neurosci. 2021, 22, 137–151. [Google Scholar] [CrossRef]
- Zhang, G.; Neubert, T.A.; Jordan, B.A. RNA binding proteins accumulate at the postsynaptic density with synaptic activity. J. Neurosci. 2012, 32, 599–609. [Google Scholar] [CrossRef] [Green Version]
- Fatscher, T.; Boehm, V.; Gehring, N.H. Mechanism, factors, and physiological role of nonsense-mediated mRNA decay. Cell. Mol. Life Sci. 2015, 72, 4523–4544. [Google Scholar] [CrossRef]
- Lewis, B.P.; Green, R.E.; Brenner, S.E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. USA 2003, 100, 189–192. [Google Scholar] [CrossRef] [Green Version]
- Ule, J.; Stefani, G.; Mele, A.; Ruggiu, M.; Wang, X.; Taneri, B.; Gaasterland, T.; Blencowe, B.J.; Darnell, R.B. An RNA map predicting nova-dependent splicing regulation. Nature 2006, 444, 580–586. [Google Scholar] [CrossRef]
- Tyagarajan, S.K.; Fritschy, J.M. Gephyrin: A master regulator of neuronal function? Nat. Rev. Neurosci. 2014, 15, 141–156. [Google Scholar] [CrossRef]
- Speese, S.D.; Ashley, J.; Jokhi, V.; Nunnari, J.; Barria, R.; Li, Y.; Ataman, B.; Koon, A.; Chang, Y.T.; Li, Q.; et al. Nuclear envelope budding enables large ribonucleoprotein particle export during synaptic Wnt signaling. Cell 2012, 149, 832–846. [Google Scholar] [CrossRef] [Green Version]
- Batish, M.; Bogaard, P.V.D.; Kramer, F.R.; Tyagi, S. Neuronal mrnas travel singly into dendrites. Proc. Natl. Acad. Sci. USA 2012, 109, 4645–4650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buxbaum, A.R.; Wu, B.; Singer, R.H. Single b-actin mRNA detection in neurons reveals a mechanism for its translatability. Science 2014, 343, 419–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, K.C.; Ephrussi, A. mRNA localization: Gene expression in the spatial dimension. Cell 2009, 136, 719–730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hüttelmaier, S.; Zenklusen, D.; Lederer, M.; Dictenberg, J.; Lorenz, M.; Meng, X.; Bassell, G.J.; Condeelis, J.; Singer, R.H. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature 2005, 438, 512–515. [Google Scholar] [CrossRef]
- Fritzsche, R.; Karra, D.; Bennett, K.L.; Ang, F.Y.; Heraud-Farlow, J.E.; Tolino, M.; Doyle, M.; Bauer, K.E.; Thomas, S.; Planyavsky, M.; et al. Interactome of two diverse RNA granules links mRNA localization to translational repression in neurons. Cell Rep. 2013, 5, 1749–1762. [Google Scholar] [CrossRef]
- Song, T.; Zheng, Y.; Wang, Y.; Katz, Z.; Liu, X.; Chen, S.; Singer, R.H.; Gu, W. Specific interaction of KIF11 with ZBP1 regulates the transport of β-actin mRNA and cell motility. J. Cell Sci. 2015, 128, 1001–1010. [Google Scholar] [CrossRef] [Green Version]
- Anderson, B.R.; Chopra, P.; Suhl, J.A.; Warren, S.T.; Bassell, G.J. Identification of consensus binding sites clarifies fmrp binding determinants. Nucleic Acids Res. 2016, 44, 6649–6659. [Google Scholar] [CrossRef] [Green Version]
- Mazroui, R.; Huot, M.E.; Tremblay, S.; Filion, C.; Labelle, Y.; Khandjian, E.W. Trapping of messenger RNA by fragile × mental retardation protein into cytoplasmic granules induces translation repression. Hum. Mol. Genet. 2002, 11, 3007–3017. [Google Scholar] [CrossRef]
- Darnell, J.C.; Van Driesche, S.J.; Zhang, C.; Hung, K.Y.; Mele, A.; Fraser, C.E.; Stone, E.F.; Chen, C.; Fak, J.J.; Chi, S.W.; et al. FMRP stalls ribosomal translocation on mrnas linked to synaptic function and autism. Cell 2011, 146, 247–261. [Google Scholar] [CrossRef]
- Napoli, I.; Mercaldo, V.; Boyl, P.P.; Eleuteri, B.; Zalfa, F.; De Rubeis, S.; Di Marino, D.; Mohr, E.; Massimi, M.; Falconi, M.; et al. The fragile X syndrome protein represses activity-dependent translation through cyfip1, a new 4e-bp. Cell 2008, 134, 1042–1054. [Google Scholar] [CrossRef] [Green Version]
- Pilaz, L.J.; Silver, D.L. Post-transcriptional regulation in corticogenesis: How RNA-binding proteins help build the brain. Wiley Interdiscip. Rev. RNA 2015, 6, 501–515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plante, I.; Davidovic, L.; Ouellet, D.L.; Gobeil, L.-A.; Tremblay, S.; Khandjian, E.W.; Provost, P. Dicer-derived micrornas are utilized by the fragile X mental retardation protein for assembly on target rNAS. J. Biomed. Biotechnol. 2006, 2006, 64347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muddashetty, R.S.; Nalavadi, V.C.; Gross, C.; Yao, X.; Xing, L.; Laur, O.; Warren, S.T.; Bassell, G.J. Reversible inhibition of psd-95 mRNA translation by mir-125a, fmrp phosphorylation, and mglur signaling. Mol. Cell 2011, 42, 673–688. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Tang, W.; Zhang, L.R.; Zhang, C.Y. Fmrp regulates mir196a-mediated repression of hoxb8 via interaction with the ago2 mid domain. Mol. Biosyst. 2014, 10, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of synaptic structure and function by fmrp-associated micrornas mir-125b and mir-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Jeong, J.H.; Nam, Y.J.; Kim, S.Y.; Kim, E.G.; Jeong, J.; Kim, H.K. The transport of staufen2-containing ribonucleoprotein complexes involves kinesin motor protein and is modulated by mitogen-activated protein kinase pathway. J. Neurochem. 2007, 102, 2073–2084. [Google Scholar] [CrossRef]
- Sharangdhar, T.; Sugimoto, Y.; Heraud-Farlow, J.; Fernández-Moya, S.M.; Ehses, J.; Ruiz de los Mozos, I.; Ule, J.; Kiebler, M.A. A retained intron in the 3’-UTR of Calm3 mRNA mediates its Staufen2- and activity-dependent localization to neuronal dendrites. EMBO Rep. 2017, 18, 1762–1774. [Google Scholar] [CrossRef]
- Saladino, P.; Di Liegro, C.M.; Proia, P.; Sala, A.; Schiera, G.; Lo Cicero, A.; Di Liegro, I. RNA-binding activity of the rat calmodulin-binding PEP-19 protein and of the long PEP-19 isoform. Int. J. Mol. Med. 2012, 29, 141–145. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.S.; Carson, J.H.; Barbarese, E.; Richter, J.D. Facilitation of dendritic mRNA transport by CPEB. Genes Dev. 2003, 17, 638–653. [Google Scholar] [CrossRef]
- Chiaruttini, C.; Vicario, A.; Li, Z.; Baj, G.; Braiuca, P.; Wu, Y.; Lee, F.S.; Gardossi, L.; Baraban, J.M.; Tongiorgi, E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (Val66Met) mutation. Proc. Natl. Acad. Sci. USA 2009, 106, 16481–16486. [Google Scholar] [CrossRef] [Green Version]
- Costa, R.O.; Martins, L.F.; Tahiri, E.; Duarte, C.B. Brain-derived neurotrophic factor-induced regulation of RNA metabolism in neuronal development and synaptic plasticity. Wiley Interdiscip. Rev. RNA 2022, 24, e1713. [Google Scholar] [CrossRef] [PubMed]
- Yoshimura, A.; Fujii, R.; Watanabe, Y.; Okabe, S.; Fukui, K.; Takumi, T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr. Biol. 2006, 16, 2345–2351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, M.; Kiebler, M.A. Mechanisms of dendritic mRNA transport and its role in synaptic tagging. EMBO J. 2011, 30, 3540–3552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreassi, C.; Riccio, A. To localize or not to localize: mRNA fate is in 3’UTR ends. Trends Cell Biol. 2009, 19, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Holt, C.E.; Bullock, S.L. Subcellular mRNA localization in animal cells and why it matters. Science 2009, 326, 1212–1216. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.; Gkogkas, C.G.; Sonenberg, N.; Holt, C.E. Remote control of gene function by local translation. Cell 2014, 157, 26–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafner, A.S.; Donlin-Asp, P.G.; Leitch, B.; Herzog, E.; Schuman, E.M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 2019, 364, eaau3644. [Google Scholar] [CrossRef]
- Glock, C.; Biever, A.; Tushev, G.; Nassim-Assir, B.; Kao, A.; Bartnik, I.; Tom Dieck, S.; Schuman, E.M. The translatome of neuronal cell bodies, dendrites, and axons. Proc. Natl. Acad. Sci. USA 2021, 118, e2113929118. [Google Scholar] [CrossRef]
- Keene, J.D.; Tenenbaum, S.A. Eukaryotic mRNPs may represent posttranscriptional operons. Mol. Cell 2002, 9, 1161–1167. [Google Scholar] [CrossRef]
- Holt, C.E.; Schuman, E.M. The central dogma decentralized: New perspectives on RNA function and local translation in neurons. Neuron 2013, 80, 648–657. [Google Scholar] [CrossRef] [Green Version]
- Wong, H.H.-W.; Lin, J.Q.; Ströhl, F.; Roque, C.G.; Cioni, J.-M.; Cagnetta, R.; Turner-Bridger, B.; Laine, R.F.; Harris, W.A.; Kaminski, C.F.; et al. RNA docking and local translation regulate site-specific axon remodeling in vivo. Neuron 2017, 95, 852–868.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leung, K.M.; van Horck, F.P.; Lin, A.C.; Allison, R.; Standart, N.; Holt, C.E. Asymmetrical-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 2006, 9, 1247–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rangaraju, V.; Tom Dieck, S.; Schuman, E.M. Local translation in neuronal compartments: How local is local? EMBO Rep. 2017, 18, 693–711. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fonkeu, Y.; Kraynyukova, N.; Hafner, A.S.; Kochen, L.; Sartori, F.; Schuman, E.M.; Tchumatchenko, T. How mRNA localization and protein synthesis sites influence dendritic protein distribution and dynamics. Neuron 2019, 103, 1109–1122.e7. [Google Scholar] [CrossRef]
- Spillane, M.; Ketschek, A.; Merianda, T.T.; Twiss, J.L.; Gallo, G. Mitochondria coordinate sites of axon branching through localized intra-axonal protein synthesis. Cell Rep. 2013, 5, 1564–1575. [Google Scholar] [CrossRef] [Green Version]
- Shigeoka, T.; Koppers, M.; Wong, H.H.; Lin, J.Q.; Cagnetta, R.; Dwivedy, A.; de Freitas Nascimento, J.; van Tartwijk, F.W.; Ströhl, F.; Cioni, J.M.; et al. On-site ribosome remodeling by locally synthesized ribosomal proteins in axons. Cell Rep. 2019, 29, 3605–3619.e10. [Google Scholar] [CrossRef] [Green Version]
- Shigeoka, T.; Jung, H.; Jung, J.; Turner-Bridger, B.; Ohk, J.; Lin, J.Q.; Amieux, P.S.; Holt, C.E. Dynamic axonal translation in developing and mature visual circuits. Cell 2016, 166, 181–192. [Google Scholar] [CrossRef] [Green Version]
- Laursen, L.S.; Chan, C.W.; Ffrench-Constant, C. Translation of myelin basic protein mRNA in oligodendrocytes is regulated by integrin activation and hnRNP-K. J. Cell Biol. 2011, 192, 797–811. [Google Scholar] [CrossRef] [Green Version]
- White, R.; Gonsior, C.; Kramer-Albers, E.M.; Stohr, N.; Huttelmaier, S.; Trotter, J. Activation of oligodendroglial Fyn kinase enhances translation of mRNAs transported in hnRNP A2-dependent RNA granules. J. Cell Biol. 2008, 181, 579–586. [Google Scholar] [CrossRef]
- Dell’Orco, M.; Oliver, R.J.; Perrone-Bizzozero, N. HuD Binds to and Regulates Circular RNAs Derived From Neuronal Development- and Synaptic Plasticity-Associated Genes. Front. Genet. 2020, 11, 790. [Google Scholar] [CrossRef]
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Samata, M.; Akhtar, A. Dosage compensation of the X chromosome: A complex epigenetic assignment involving chromatin regulators and long noncoding RNAs. Annu. Rev. Biochem. 2018, 87, 323–350. [Google Scholar] [CrossRef] [PubMed]
- Thakur, J.; Henikoff, S. Architectural RNA in chromatin organization. Biochem. Soc. Trans. 2020, 48, 1967–1978. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.C.; Fernandopulle, M.S.; Wang, G.; Choi, H.; Hao, L.; Drerup, C.M.; Patel, R.; Qamar, S.; Nixon-Abell, J.; Shen, Y.; et al. RNA Granules Hitchhike on Lysosomes for Long-Distance Transport, Using Annexin A11 as a Molecular Tether. Cell 2019, 179, 147–164.e20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abildgaard, M.H.; Brynjólfsdóttir, S.H.; Frankel, L.B. The Autophagy-RNA Interplay: Degradation and Beyond. Trends Biochem. Sci. 2020, 45, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Varadi, M.; Zsolyomi, F.; Guharoy, M.; Tompa, P. Functional Advantages of Conserved Intrinsic Disorder in RNA-Binding Proteins. PLoS ONE 2015, 10, e139731. [Google Scholar] [CrossRef]
- Järvelin, A.I.; Noerenberg, M.; Davis, I.; Castello, A. The new (dis)order in RNA regulation. Cell Commun. Signal. 2016, 14, 9. [Google Scholar] [CrossRef] [Green Version]
- Zagrovic, B.; Bartonek, L.; Polyansky, A.A. RNA-protein interactions in an unstructured context. FEBS Lett. 2018, 592, 2901–2916. [Google Scholar] [CrossRef] [Green Version]
- Sharma, R.; Raduly, Z.; Miskei, M.; Fuxreiter, M. Fuzzy complexes: Specific binding without complete folding. FEBS Lett. 2015, 589, 2533–2542. [Google Scholar] [CrossRef]
- Delaforge, E.; Kragelj, J.; Tengo, L.; Palencia, A.; Milles, S.; Bouvignies, G.; Salvi, N.; Blackledge, M.; Jensen, M.R. Deciphering the Dynamic Interaction Profile of an Intrinsically Disordered Protein by NMR Exchange Spectroscopy. J. Am. Chem. Soc. 2018, 140, 1148–1158. [Google Scholar] [CrossRef]
- Lin, Y.; Protter, D.S.W.; Rosen, M.K.; Parker, R. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell 2015, 60, 208–219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vogler, T.O.; Wheeler, J.R.; Nguyen, E.D.; Hughes, M.P.; Britson, K.A.; Lester, E.; Rao, B.; Betta, N.D.; Whitney, O.; Ewachiw, T.E.; et al. TDP-43 and RNA form amyloid-like myo-granules in regenerating muscle. Nature 2018, 563, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Tsang, B.; Arsenault, J.; Vernon, R.M.; Lin, H.; Sonenberg, N.; Wang, L.Y.; Bah, A.; Forman-Kay, J.D. Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. USA 2019, 116, 4218–4227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sudhakaran, I.P.; Ramaswami, M. Long-term memory consolidation: The role of RNA-binding proteins with prion-like domains. RNA Biol. 2017, 14, 568–586. [Google Scholar] [CrossRef] [PubMed]
- Calabretta, S.; Richard, S. Emerging roles of disordered sequences in RNA-binding proteins. Trends Biochem. Sci. 2015, 40, 662–672. [Google Scholar] [CrossRef] [PubMed]
- Hofweber, M.; Dormann, D. Friend or foe-Post-translational modifications as regulators of phase separation and RNP granule dynamics. J. Biol. Chem. 2019, 294, 7137–7150. [Google Scholar] [CrossRef] [Green Version]
- Loureiro, J.R.; Castro, A.F.; Figueiredo, A.S.; Silveira, I. Molecular Mechanisms in Pentanucleotide Repeat Diseases. Cells 2022, 11, 205. [Google Scholar] [CrossRef]
- Wang, J.; Choi, J.-M.; Holehouse, A.S.; Lee, H.O.; Zhang, X.; Jahnel, M.; Maharana, S.; Lemaitre, R.; Pozniakovsky, A.; Drechsel, D.; et al. A Molecular Grammar Governing the Driving Forces for Phase Separation of Prion-like RNA Binding Proteins. Cell 2018, 174, 688–699.e16. [Google Scholar] [CrossRef] [Green Version]
- Holehouse, A.S.; Ginell, G.M.; Griffith, D.; Boke, E. Clustering of Aromatic Residues in Prion-like Domains Can Tune the Formation, State, and Organization of Biomolecular Condensates. Biochemistry 2021, 60, 3566–3581. [Google Scholar] [CrossRef]
- Gu, X.; Schafer, N.P.; Wolynes, P.G. Vectorial channeling as a mechanism for translational control by functional prions and condensates. Proc. Natl. Acad. Sci. USA 2021, 118, e2115904118. [Google Scholar] [CrossRef]
- Chiang, W.C.; Lee, M.H.; Chen, T.C.; Huang, J.R. Interactions between the Intrinsically Disordered Regions of hnRNP-A2 and TDP-43 Accelerate TDP-43’s Conformational Transition. Int. J. Mol. Sci. 2020, 21, 5930. [Google Scholar] [CrossRef] [PubMed]
- Milovanovic, D.; Wu, Y.; Bian, X.; De Camilli, P. A liquid phase of synapsin and lipid vesicles. Science 2018, 361, 604–607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDonald, N.A.; Fetter, R.D.; Shen, K. Assembly of synaptic active zones requires phase separation of scaffold molecules. Nature 2020, 588, 454–458. [Google Scholar] [CrossRef]
- Fernández-Moya, S.M.; Ehses, J.; Bauer, K.E.; Schieweck, R.; Chakrabarti, A.M.; Lee, F.C.Y.; Illig, C.; Luscombe, N.M.; Harner, M.; Ule, J.; et al. RGS4 RNA Secondary Structure Mediates Staufen2 RNP Assembly in Neurons. Int. J. Mol. Sci. 2021, 22, 13021. [Google Scholar] [CrossRef] [PubMed]
- Garner, C.C.; Tucker, R.P.; Matus, A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature 1988, 336, 674–677. [Google Scholar] [CrossRef] [PubMed]
- Burgin, K.E.; Waxham, M.N.; Rickling, S.; Westgate, S.A.; Mobley, W.C.; Kelly, P.T. In situ hybridization histochemistry of Ca21/calmodulin-dependent protein kinase in developing rat brain. J. Neurosci. 1990, 10, 1788–1798. [Google Scholar] [CrossRef]
- Eberwine, J.; Miyashiro, K.; Kacharmina, J.E.; Job, C. Local translation of classes of mRNAs that are targeted to neuronal dendrites. Proc. Natl. Acad. Sci. USA 2001, 98, 7080–7085. [Google Scholar] [CrossRef] [Green Version]
- Grooms, S.Y.; Noh, K.M.; Regis, R.; Bassell, G.J.; Bryan, M.K.; Carroll, R.C.; Zukin, R.S. Activity bidirectionally regulates AMPA receptor mRNA abundance in dendrites of hippocampal neurons. J. Neurosci. 2006, 26, 8339–8351. [Google Scholar] [CrossRef] [Green Version]
- Mayford, M.; Baranes, D.; Podsypanina, K.; Kandel, E.R. The 3’-untranslated region of CaMKIIα is a cis-acting signal for the localization and translation of mRNA in dendrites. Proc. Natl. Acad. Sci. USA 1996, 93, 13250–13255. [Google Scholar] [CrossRef]
- Steward, O.; Wallace, C.S.; Lyford, G.L.; Worley, P.F. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron 1998, 21, 741–751. [Google Scholar] [CrossRef] [Green Version]
- Blichenberg, A.; Schwanke, B.; Rehbein, M.; Garner, C.C.; Richter, D.; Kindler, S. Identification of a cis-acting dendritic targeting element in MAP2 mRNAs. J. Neurosci. 1999, 19, 8818–8829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eom, T.; Antar, L.N.; Singer, R.H.; Bassell, G.J. Localization of a beta-actin messenger ribonucleoprotein complex with zipcode-binding protein modulates the density of dendritic filopodia and filopodial synapses. J. Neurosci. 2003, 23, 10433–10444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, J.J.; Gharami, K.; Liao, G.Y.; Woo, N.H.; Lau, A.G.; Vanevski, F.; Torre, E.R.; Jones, K.R.; Feng, Y.; Lu, B.; et al. Distinct role of long 3’ UTR BDNF mRNA in spine morphology and synaptic plasticity in hippocampal neurons. Cell 2008, 134, 175–187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tushev, G.; Glock, C.; Heumuller, M.; Biever, A.; Jovanovic, M.; Schuman, E.M. Alternative 3’UTRs modify the localization, regulatory potential, stability, and plasticity of mRNAs in neuronal compartments. Neuron 2018, 98, 495–511.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, M.; Kiebler, M.A. A zipcode unzipped. Genes Dev. 2012, 26, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Mikl, M.; Vendra, G.; Kiebler, M.A. Independent localization of MAP2, CaMKIIa and b-actin RNAs in low copy numbers. EMBO Rep. 2011, 12, 1077–1084. [Google Scholar] [CrossRef]
- Tübing, F.; Vendra, G.; Mikl, M.; Macchi, P.; Thomas, S.; Kiebler, M.A. Dendritically localized transcripts are sorted into distinct ribonucleoprotein particles that display fast directional motility along dendrites of hippocampal neurons. J. Neurosci. 2010, 30, 4160–4170. [Google Scholar] [CrossRef] [Green Version]
- Wilhelm, J.E.; Vale, R.D. RNA on the move: The mRNA localization pathway. J. Cell Biol. 1993, 123, 269–274. [Google Scholar] [CrossRef] [Green Version]
- Thomas, K.L.; Laroche, S.; Errington, M.L.; Bliss, T.V.; Hunt, S.P. Spatial and temporal changes in signal transduction pathways during LTP. Neuron 1994, 13, 737–745. [Google Scholar] [CrossRef]
- Wells, D.G.; Dong, X.; Quinlan, E.M.; Huang, Y.S.; Bear, M.F.; Richter, J.D.; Fallon, J.R. A role for the cytoplasmic polyadenylation element in NMDA receptor-regulated mRNA translation in neurons. J. Neurosci. 2001, 21, 9541–9548. [Google Scholar] [CrossRef] [Green Version]
- Bayer, K.U.; Schulman, H. CaM kinase: Still inspiring at 40. Neuron 2019, 103, 380–394. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.; Yasuda, M.; Coats, J.K.; Jones, Y.; Martone, M.E.; Mayford, M. Disruption of dendritic translation of CaMKIIα impairs stabilization of synaptic plasticity and memory consolidation. Neuron 2002, 36, 507–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cagnetta, R.; Frese, C.K.; Shigeoka, T.; Krijgsveld, J.; Holt, C.E. Rapid cue-specific remodeling of the nascent axonal proteome. Neuron 2018, 99, 29–46.e24. [Google Scholar] [CrossRef] [Green Version]
- Oe, S.; Yoneda, Y. Cytoplasmic polyadenylation element-like sequences are involved in dendritic targeting of BDNF mRNA in hippocampal neurons. FEBS Lett. 2010, 584, 3424–3430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baj, G.; Leone, E.; Chao, M.V.; Tongiorgi, E. Spatial segregation of BDNF transcripts enables BDNF to differentially shape distinct dendritic compartments. Proc. Natl. Acad. Sci. USA 2011, 108, 16813–16818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allen, M.; Bird, C.; Feng, W.; Liu, G.; Li, W.; Perrone-Bizzozero, N.I.; Feng, Y. HuD promotes BDNF expression in brain neurons via selective stabilization of the BDNF long 3’UTR mRNA. PLoS ONE 2013, 8, e55718. [Google Scholar] [CrossRef]
- Kim, S.; Kim, H.; Um, J.W. Synapse development organized by neuronal activity-regulated immediate-early genes. Exp. Mol. Med. 2018, 50, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Plath, N.; Ohana, O.; Dammermann, B.; Errington, M.L.; Schmitz, D.; Gross, C.; Mao, X.; Engelsberg, A.; Mahlke, C.; Welzl, H.; et al. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron 2006, 52, 437–444. [Google Scholar] [CrossRef] [Green Version]
- Giorgi, C.; Yeo, G.W.; Stone, M.E.; Katz, D.B.; Burge, C.; Turrigiano, G.; Moore, M.J. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell 2007, 130, 179–191. [Google Scholar] [CrossRef]
- Zalfa, F.; Giorgi, M.; Primerano, B.; Moro, A.; Di Penta, A.; Reis, S.; Oostra, B.; Bagni, C. The Fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 2003, 112, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Ashley, J.; Cordy, B.; Lucia, D.; Fradkin, L.G.; Budnik, V.; Thomson, T. Retrovirus-like Gag protein Arc1 binds RNA and traffics across synaptic boutons. Cell 2018, 172, 262–274.e11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Donnelly, C.J.; Park, M.; Spillane, M.; Yoo, S.; Pacheco, A.; Gomes, C.; Vuppalanchi, D.; McDonald, M.; Kim, H.H.; Merianda, T.T.; et al. Axonally synthesized β-actin and GAP-43 proteins support distinct modes of axonal growth. J. Neurosci. 2013, 33, 3311–3322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, M.; Huang, J.; Li, G.W.; Jiang, B.; Cheng, H.; Liu, X.; Jiang, X.; Zhang, X.; Yang, L.; Bao, L.; et al. Axon enriched linc RNA ALAE is required for axon elongation via regulation of local mRNA translation. Cell Rep. 2021, 35, 109053. [Google Scholar] [CrossRef] [PubMed]
- Batista, A.F.R.; Martinez, J.C.; Hengst, U. Intra-axonal synthesis of SNAP25 is required for the formation of presynaptic terminals. Cell Rep. 2017, 20, 3085–3098. [Google Scholar] [CrossRef] [Green Version]
- Taylor, A.M.; Wu, J.; Tai, H.C.; Schuman, E.M. Axonal translation of β-catenin regulates synaptic vesicle dynamics. J. Neurosci. 2013, 33, 5584–5589. [Google Scholar] [CrossRef] [Green Version]
- Klein, M.E.; Monday, H.; Jordan, B.A. Proteostasis and RNA Binding Proteins in Synaptic Plasticity and in the Pathogenesis of Neuropsychiatric Disorders. Neural Plast. 2016, 2, 3857934. [Google Scholar] [CrossRef] [Green Version]
- Cosker, K.E.; Fenstermacher, S.J.; Pazyra-Murphy, M.F.; Elliott, H.L.; Segal, R.A. The RNA-binding protein SFPQ orchestrates an RNA regulon to promote axon viability. Nat. Neurosci. 2016, 19, 690–696. [Google Scholar] [CrossRef]
- Cosker, K.E.; Pazyra-Murphy, M.F.; Fenstermacher, S.J.; Segal, R.A. Target-derived neurotrophins coordinate transcription and transport of Bclw to prevent axonal degeneration. J. Neurosci. 2013, 33, 5195–5207. [Google Scholar] [CrossRef] [Green Version]
- Yoon, B.C.; Jung, H.; Dwivedy, A.; O’Hare, C.M.; Zivraj, K.H.; Holt, C.E. Local translation of extranuclear lamin B promotes axon maintenance. Cell 2012, 148, 752–764. [Google Scholar] [CrossRef]
- Thapar, R.; Denmon, A.P. Signaling pathways that control mRNA turnover. Cell. Signal. 2013, 25, 1699–1710. [Google Scholar] [CrossRef] [Green Version]
- Fuller-Pace, F.V. RNA helicases: Modulators of RNA structure. Trends Cell Biol. 1994, 4, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Weeks, K.M. Protein-facilitated RNA folding. Curr. Opin. Struct. Biol. 1997, 7, 336–342. [Google Scholar] [CrossRef] [PubMed]
- Grassi, E.; Santoro, R.; Umbach, A.; Grosso, A.; Oliviero, S.; Neri, F.; Conti, L.; Ala, U.; Provero, P.; DiCunto, F.; et al. Choice of alternative polyadenylation sites, mediated by the RNA-binding protein Elavl3, plays a role in differentiation of inhibitory neuronal progenitors. Front. Cell. Neurosci. 2019, 12, 518. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoi, S.; Udagawa, T.; Fujioka, Y.; Honda, D.; Okado, H.; Watanabe, H.; Katsuno, M.; Ishigaki, S.; Sobue, G. 3’UTR length-dependent control of SynGAP isoform a2 mRNA by FUS and ELAV-like proteins promotes dendritic spine maturation and cognitive function. Cell Rep. 2017, 20, 3071–3084. [Google Scholar] [CrossRef] [Green Version]
- Oktaba, K.; Zhang, W.; Lotz, T.S.; Jun, D.J.; Lemke, S.B.; Ng, S.P.; Esposito, E.; Levine, M.; Hilgers, V. ELAV links paused Pol II to alternative polyadenylation in the drosophila nervous system. Mol. Cell. 2015, 57, 341–348. [Google Scholar] [CrossRef] [Green Version]
- Bauer, K.E.; Bargenda, N.; Schieweck, R.; Illig, C.; Segura, I.; Harner, M.; Kiebler, M.A. RNA supply drives physiological granule assembly in neurons. Nat. Commun. 2022, 13, 2781. [Google Scholar] [CrossRef]
- Hirokawa, N. mRNA transport in dendrites: RNA granules, motors, and tracks. J. Neurosci. 2006, 26, 7139–7142. [Google Scholar] [CrossRef] [Green Version]
- Kneussel, M.; Wagner, W. Myosin motors at neuronal synapses: Drivers of membrane transport and actin dynamics. Nat. Rev. Neurosci. 2013, 14, 233–247. [Google Scholar] [CrossRef]
- Abouward, R.; Schiavo, G. Walking the line: Mechanisms underlying directional mRNA transport and localisation in neurons and beyond. Cell. Mol. Life Sci. 2021, 78, 2665–2681. [Google Scholar] [CrossRef]
- Baumann, S.; Komissarov, A.; Gili, M.; Ruprecht, V.; Wieser, S.; Maurer, S.P. A reconstituted mammalian APC-kinesin complex selectively transports defined packages of axonal mRNAs. Sci. Adv. 2020, 6, eaaz1588. [Google Scholar] [CrossRef] [Green Version]
- Preitner, N.; Quan, J.; Nowakowski, D.W.; Hancock, M.L.; Shi, J.; Tcherkezian, J.; Young-Pearse, T.L.; Flanagan, J.G. APC is an RNA-binding protein, and its interactome provides a link to neural development and microtubule assembly. Cell 2014, 158, 368–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terenzio, M.; Koley, S.; Samra, N.; Rishal, I.; Zhao, Q.; Sahoo, P.K.; Urisman, A.; Marvaldi, L.; Oses-Prieto, J.A.; Forester, C.; et al. Locally translated mTOR controls axonal local translation in nerve injury. Science 2018, 359, 1416–1421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Driscoll, H.E.; Muraro, N.I.; He, M.; Baines, R.A. Pumilio-2 regulates translation of Nav1.6 to mediate homeostasis of membrane excitability. J. Neurosci. 2013, 33, 9644–9654. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martinez, J.C.; Randolph, L.K.; Iascone, D.M.; Pernice, H.F.; Polleux, F.; Hengst, U. Pum2 shapes the transcriptome in developing axons through retention of target mRNAs in the cell body. Neuron 2019, 104, 931–946.e5. [Google Scholar] [CrossRef]
- Núñez, L.; Buxbaum, A.R.; Katz, Z.B.; Lopez-Jones, M.; Nwokafor, C.; Czaplinski, K.; Pan, F.; Rosenberg, J.; Monday, H.R.; Singer, R.H. Tagged actin mRNA dysregulation in IGF2BP1[Formula: See text] mice. Proc. Natl. Acad. Sci. USA 2022, 119, e2208465119. [Google Scholar] [CrossRef]
- Naeve, G.S.; Ramakrishnan, M.; Kramer, R.; Hevroni, D.; Citri, Y.; Theill, L.E. Neuritin: A gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc. Natl. Acad. Sci. USA 1997, 94, 2648–2653. [Google Scholar] [CrossRef] [Green Version]
- Sahoo, P.K.; Lee, S.J.; Jaiswal, P.B.; Alber, S.; Kar, A.N.; Miller-Randolph, S.; Taylor, E.E.; Smith, T.; Singh, B.; Ho, T.S.; et al. Axonal G3BP1 stress granule protein limits axonal mRNA translation and nerve regeneration. Nat. Commun. 2018, 9, 3358. [Google Scholar] [CrossRef] [Green Version]
- Calamita, P.; Gatti, G.; Miluzio, A.; Scagliola, A.; Biffo, S. Translating the game: Ribosomes as active players. Front. Genet. 2018, 9, 533. [Google Scholar] [CrossRef] [Green Version]
- Ceci, M.; Fazi, F.; Romano, N. The role of RNA-binding and ribosomal proteins as specific RNA translation regulators in cellular differentiation and carcinogenesis. Biochim. Biophys. Acta Mol. Basis. Dis. 2021, 1867, 166046. [Google Scholar] [CrossRef]
- Shi, Z.; Fujii, K.; Kovary, K.M.; Genuth, N.R.; Rost, H.L.; Teruel, M.N.; Barna, M. Heterogeneous ribosomes preferentially translate distinct subpools of mRNAs genome-wide. Mol. Cell 2017, 67, 71–83.e77. [Google Scholar] [CrossRef] [Green Version]
- Kondrashov, N.; Pusic, A.; Stumpf, C.R.; Shimizu, K.; Hsieh, A.C.; Ishijima, J.; Shiroishi, T.; Barna, M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell 2011, 145, 383–397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mazumder, B.; Sampath, P.; Seshadri, V.; Maitra, R.K.; DiCorleto, P.E.; Fox, P.L. Regulated release of L13a from the 60S ribosomal subunit as a mechanism of transcript-specific translational control. Cell 2003, 115, 187–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schiera, G.; Di Liegro, C.M.; Di Liegro, I. Cell-to-Cell Communication in Learning and Memory: From Neuro- and Glio-Transmission to Information Exchange Mediated by Extracellular Vesicles. Int. J. Mol. Sci. 2020, 21, 266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thelen, M.P.; Kye, M.J. The Role of Rna Binding Proteins for Local mRNA Translation: Implications in Neurological Disorders. Front. Mol. Biosci. 2019, 6, 161. [Google Scholar] [CrossRef] [Green Version]
- Joo, Y.; Benavides, D.R. Local Protein Translation and RNA Processing of Synaptic Proteins in Autism Spectrum Disorder. Int. J. Mol. Sci. 2021, 22, 2811. [Google Scholar] [CrossRef]
- Kim, W.; Kim, D.Y.; Lee, K.H. RNA-Binding Proteins and the Complex Pathophysiology of ALS. Int. J. Mol. Sci. 2021, 22, 2598. [Google Scholar] [CrossRef]
- Dent, E.W. Of microtubules and memory: Implications for microtubule dynamics in dendrites and spines. Mol. Biol. Cell 2017, 28, 1–8. [Google Scholar] [CrossRef]
- Nakayama, T.; Sawada, T. Involvement of microtubule integrity in memory impairment caused by colchicine. Pharmacol. Biochem. Behav. 2002, 71, 119–138. [Google Scholar] [CrossRef]
- Lynch, G.; Rex, C.S.; Chen, L.Y.; Gall, C.M. The substrates of memory: Defects, treatments, and enhancement. Eur. J. Pharmacol. 2008, 585, 2–13. [Google Scholar] [CrossRef]
- Priel, A.; Tuszynski, J.A.; Woolf, N.J. Neural cytoskeleton capabilities for learning and memory. J. Biol. Phys. 2010, 36, 3–21. [Google Scholar] [CrossRef] [Green Version]
- Lamprecht, R. The actin cytoskeleton in memory formation. Prog. Neurobiol. 2014, 117, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Rao, A.; Steward, O. Evidence that protein constituents of postsynaptic membrane specilization are locally synthesized: Analysis of proteins synthesized within synaptosomes. J. Neurosci. 1991, 11, 2881–2895. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giuditta, A.; Chun, J.T.; Eyman, M.; Cefaliello, C.; Bruno, A.P.; Crispino, M. Local gene expression in axons and nerve endings: The glia-neuron unit. Physiol. Rev. 2008, 88, 515–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rey, A.A.; Karaulanov, E.; Sharopov, S.; Arab, K.; Schäfer, A.; Gierl, M.; Guggenhuber, S.; Brandes, C.; Pennella, L.; Gruhn, W.H.; et al. Gadd45α modulates aversive learning through post-transcriptional regulation of memory-related mRNAs. EMBO Rep. 2019, 20, e46022. [Google Scholar] [CrossRef]
- Marion, R.M.; Fortes, P.; Beloso, A.; Dotti, C.; Ortin, J. A human sequence homologue of staufen is an rna-binding protein that is associated with polysomes and localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999, 19, 2212–2219. [Google Scholar] [CrossRef] [Green Version]
- Wickham, L.; Duchaine, T.; Luo, M.; Nabi, I.R.; DesGroseillers, L. Mammalian staufen is a double-stranded-RNA- and tubulin-binding protein which localizes to the rough endoplasmic reticulum. Mol. Cell. Biol. 1999, 19, 2220–2230. [Google Scholar] [CrossRef] [Green Version]
- Duchaine, T.F.; Hemraj, I.; Furic, L.; Deitinghoff, A.; Kiebler, M.A.; DesGroseillers, L. Staufen2 isoforms localize to the somatodendritic domain of neurons and interact with different organelles. J. Cell Sci. 2002, 115, 3285–3295. [Google Scholar] [CrossRef]
- Lebeau, G.; Maher-Laporte, M.; Topolnik, L.; Laurent, C.E.; Sossin, W.; Desgroseillers, L.; Lacaille, J.C. Staufen1 regulation of protein synthesis-dependent long-term potentiation and synaptic function in hippocampal pyramidal cells. Mol. Cell. Biol. 2008, 28, 2896–2907. [Google Scholar] [CrossRef] [Green Version]
- Berger, S.M.; Fernández-Lamo, I.; Schönig, K.; Fernández Moya, S.M.; Ehses, J.; Schieweck, R.; Clementi, S.; Enkel, T.; Grothe, S.; von Bohlen Und Halbach, O.; et al. Forebrain-specific, conditional silencing of Staufen2 alters synaptic plasticity, learning, and memory in rats. Genome Biol. 2017, 18, 222. [Google Scholar] [CrossRef]
- Vessey, J.P.; Macchi, P.; Stein, J.M.; Mikl, M.; Hawker, K.N.; Vogelsang, P.; Wieczorek, K.; Vendra, G.; Riefler, J.; Tubing, F.; et al. A loss of function allele for murine staufen1 leads to impairment of dendritic staufen1-rnp delivery and dendritic spine morphogenesis. Proc. Natl. Acad. Sci. USA 2008, 105, 16374–16379. [Google Scholar] [CrossRef] [Green Version]
- Kiebler, M.A.; Hemraj, I.; Verkade, P.; Kohrmann, M.; Fortes, P.; Marion, R.M.; Ortin, J.; Dotti, C.G. The mammalian staufen protein localizes to the somatodendritic domain of cultured hippocampal neurons: Implications for its involvement in mRNA transport. J. Neurosci. 1999, 19, 288–297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kohrmann, M.; Luo, M.; Kaether, C.; DesGroseillers, L.; Dotti, C.G.; Kiebler, M.A. Microtubule-dependent recruitment of staufen-green fluorescent protein into large rna-containing granules and subsequent dendritic transport in living hippocampal neurons. Mol. Biol. Cell 1999, 10, 2945–2953. [Google Scholar] [CrossRef] [PubMed]
- Sears, J.C.; Broadie, K. FMRP-PKA Activity Negative Feedback Regulates RNA Binding-Dependent Fibrillation in Brain Learning and Memory Circuitry. Cell Rep. 2020, 33, 108266. [Google Scholar] [CrossRef] [PubMed]
- Perrone-Bizzozero, N.; Bolognani, F. Role of HuD and other RNA-Binding proteins in neural development and plasticity. J. Neurosci. Res. 2002, 68, 121–126. [Google Scholar] [CrossRef]
- Oliver, R.J.; Brigman, J.L.; Bolognani, F.; Allan, A.M.; Neisewander, J.L.; Perrone-Bizzozero, N.I. Neuronal RNA-binding protein HuD regulates addiction-related gene expression and behaviour. Genes Brain Behav. 2018, 17, e12454. [Google Scholar] [CrossRef]
- Shi, H.; Zhang, X.; Weng, Y.L.; Lu, Z.; Liu, Y.; Lu, Z.; Li, J.; Hao, P.; Zhang, Y.; Zhang, F.; et al. m6A facilitates hippocampus-dependent learning and memory through YTHDF1. Nature 2018, 563, 249–253. [Google Scholar] [CrossRef]
- Qu, W.R.; Sun, Q.H.; Liu, Q.Q.; Jin, H.J.; Cui, R.J.; Yang, W.; Song, B.; Li, B.J. Role of CPEB3 protein in learning and memory: New insights from synaptic plasticity. Aging 2020, 12, 15169–15182. [Google Scholar] [CrossRef]
- Rayman, J.B.; Kandel, E.R. Functional Prions in the Brain. Cold Spring Harb. Perspect. Biol. 2017, 9, a023671. [Google Scholar] [CrossRef] [Green Version]
- Vijayakumar, J.; Perrois, C.; Heim, M.; Bousset, L.; Alberti, S.; Besse, F. The prion-like domain of drosophila imp promotes axonal transport of RNP granules in vivo. Nat. Commun. 2019, 10, 2593. [Google Scholar] [CrossRef]
- Atkins, C.M.; Davare, M.A.; Oh, M.C.; Derkach, V.; Soderling, T.R. Bidirectional regulation of cytoplasmic polyadenylation element-binding protein phosphorylation by Ca2+/calmodulin-dependent protein kinase II and protein phosphatase 1 during hippocampal long-term potentiation. J. Neurosci. 2005, 25, 5604–5610. [Google Scholar] [CrossRef] [Green Version]
- Valadi, H.; Ekström, K.; Bossios, A.; Sjöstrand, M.; Lee, J.J.; Lötvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mashburn-Warren, L.M.; Whiteley, M. Special delivery: Vesicle tracking in prokaryotes. Mol. Microbiol. 2006, 61, 839–846. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes-vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Schiera, G.; Di Liegro, C.M.; Di Liegro, I. Extracellular Membrane Vesicles as Vehicles for Brain Cell-to-Cell Interaction in Physiological as well as Pathological Conditions. Biomed. Res. Int. 2015, 2015, 152926. [Google Scholar] [CrossRef] [Green Version]
- Mateescu, B.; Kowal, E.J.K.; van Balkom, B.W.M.; Bartel, S.; Bhattacharyya, S.N.; Buzás, E.I.; Buck, A.H.; de Candia, P.; Chow, F.W.N.; Das, S.; et al. Obstacles and opportunities in the functional analysis of extracellular vesicle RNA—An ISEV position paper. J. Extracell. Vesicles 2017, 6, 1286095. [Google Scholar] [CrossRef] [Green Version]
- Meldolesi, J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol. 2018, 28, R435–R444. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.J. Microbe-Host Communication by Small RNAs in Extracellular Vesicles: Vehicles for Transkingdom RNA Transportation. Int. J. Mol. Sci. 2019, 20, 1487. [Google Scholar] [CrossRef] [Green Version]
- Woith, E.; Fuhrmann, G.; Melzig, M.F. Extracellular Vesicles-Connecting Kingdoms. Int. J. Mol. Sci. 2019, 20, 5695. [Google Scholar] [CrossRef]
- Hansen, E.P.; Fromm, B.; Andersen, S.D.; Marcilla, A.; Andersen, K.L.; Borup, A.; Williams, A.R.; Jex, A.R.; Gasser, R.B.; Young, N.D.; et al. Exploration of extracellular vesicles from Ascaris suum provides evidence of parasite-host cross talk. J. Extracell. Vesicles 2019, 8, 1578116. [Google Scholar] [CrossRef] [Green Version]
- Mardahl, M.; Borup, A.; Nejsum, P. A new level of complexity in parasite-host interaction: The role of extracellular vesicles. Adv. Parasitol. 2019, 104, 39–112. [Google Scholar] [CrossRef] [PubMed]
- Bayat, F.; Afshar, A.; Baghban, N. Algal Cells-Derived Extracellular Vesicles: A Review with Special Emphasis on Their Antimicrobial Effects. Front. Microbiol. 2021, 12, 785716. [Google Scholar] [CrossRef] [PubMed]
- Couch, Y.; Buzàs, E.I.; Di Vizio, D.; Gho, Y.S.; Harrison, P.; Hill, A.F.; Lötvall, J.; Raposo, G.; Stahl, P.D.; Théry, C.; et al. A brief history of nearly EV-erything—The rise and rise of extracellular vesicles. J. Extracell. Vesicles 2021, 10, e12144. [Google Scholar] [CrossRef]
- Huang, X.; Lan, Y.; Shen, J.; Chen, Z.; Xie, Z. Extracellular Vesicles in Bone Homeostasis: Emerging Mediators of Osteoimmune Interactions and Promising Therapeutic Targets. Int. J. Biol. Sci. 2022, 18, 4088–4100. [Google Scholar] [CrossRef] [PubMed]
- Szuszkiewicz, J.; Myszczynski, K.; Reliszko, Z.P.; Heifetz, Y.; Kaczmarek, M.M. Early steps of embryo implantation are regulated by exchange of extracellular vesicles between the embryo and the endometrium. FASEB J. 2022, 36, e22450. [Google Scholar] [CrossRef]
- Xie, M.; Li, C.; She, Z.; Wu, F.; Mao, J.; Hun, M.; Luo, S.; Wan, W.; Tian, J.; Wen, C. Human umbilical cord mesenchymal stem cells derived extracellular vesicles regulate acquired immune response of lupus mouse in vitro. Sci. Rep. 2022, 12, 13101. [Google Scholar] [CrossRef]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 24641. [Google Scholar] [CrossRef] [Green Version]
- Feng, D.; Zhao, W.L.; Ye, Y.Y.; Bai, X.C.; Liu, R.Q.; Chang, L.F.; Zhou, Q.; Sui, S.F. Cellular internalization of exosomes occurs through phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef]
- Prada, I.; Meldolesi, J. Binding and Fusion of Extracellular Vesicles to the Plasma Membrane of Their Cell Targets. Int. J. Mol. Sci. 2016, 17, 1296. [Google Scholar] [CrossRef]
- Matsumoto, J.; Stewart, T.; Banks, W.A.; Zhang, J. The Transport Mechanism of Extracellular Vesicles at the Blood-Brain Barrier. Curr. Pharm. Des. 2017, 23, 6206–6214. [Google Scholar] [CrossRef]
- Schiera, G.; Di Liegro, C.M.; Puleo, V.; Colletta, O.; Fricano, A.; Cancemi, P.; Di Cara, G.; Di Liegro, I. Extracellular vesicles shed by melanoma cells contain a modified form of H1.0 linker histone and H1.0 mRNA-binding proteins. Int. J. Oncol. 2016, 49, 1807–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bang, C.; Batkai, S.; Dangwal, S.; Gupta, S.K.; Foinquinos, A.; Holzmann, A.; Just, A.; Remke, J.; Zimmer, K.; Zeug, A.; et al. Cardiac fibroblast-derived microRNA passenger strandenriched exosomes mediate cardiomyocyte hypertrophy. J. Clin. Investig. 2014, 124, 2136–2146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Janas, T.; Janas, M.M.; Sapon´, K.; Janas, T. Mechanisms of RNA loading into exosomes. FEBS Lett. 2015, 589, 1391–1398. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, C.; Hu, G.; Gao, L.; Hackfort, B.T.; Zucker, I.H. Extracellular vesicular MicroRNA-27a* contributes to cardiac hypertrophy in chronic heart failure. J. Mol. Cell. Cardiol. 2020, 143, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Doyle, L.M.; Wang, M.Z. Overview of Extracellular Vesicles, Their Origin, Composition, Purpose, and Methods for Exosome Isolation and Analysis. Cells 2019, 8, 727. [Google Scholar] [CrossRef] [Green Version]
- Stahl, P.D.; Raposo, G. Extracellular Vesicles: Exosomes and Microvesicles, Integrators of Homeostasis. Physiology 2019, 34, 169–177. [Google Scholar] [CrossRef]
- Witwer, K.W.; Théry, C. Extracellular vesicles or exosomes? On primacy, precision, and popularity influencing a choice of nomenclature. J. Extracell. Vesicles 2019, 8, 1648167. [Google Scholar] [CrossRef]
- Notarangelo, M.; Ferrara, D.; Potrich, C.; Lunelli, L.; Vanzetti, L.; Provenzani, A.; Basso, M.; Quattrone, A.; D’Agostino, V.G. Rapid Nickel-based Isolation of Extracellular Vesicles from Different Biological Fluids. Bio Protoc. 2020, 10, e3512. [Google Scholar] [CrossRef]
- Zhao, Z.; Wijerathne, H.; Godwin, A.K.; Soper, S.A. Isolation and analysis methods of extracellular vesicles (EVs). Extracell. Vesicles Circ. Nucleic Acids 2021, 2, 80–103. [Google Scholar] [CrossRef]
- Arifin, D.R.; Witwer, K.W.; Bulte, J.W.M. Non-Invasive imaging of extracellular vesicles: Quo vaditis in vivo? J. Extracell. Vesicles 2022, 11, e12241. [Google Scholar] [CrossRef]
- De Sousa, K.P.; Rossi, I.; Abdullahi, M.; Ramirez, M.I.; Stratton, D.; Inal, J.M. Isolation and characterization of extracellular vesicles and future directions in diagnosis and therapy. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2022, e1835. [Google Scholar] [CrossRef] [PubMed]
- Yoshioka, Y.; Shimomura, M.; Saito, K.; Ishii, H.; Doki, Y.; Eguchi, H.; Nakatsura, T.; Itoi, T.; Kuroda, M.; Mori, M.; et al. Circulating cancer-associated extracellular vesicles as early detection and recurrence biomarkers for pancreatic cancer. Cancer Sci. 2022, 113, 3498–3509. [Google Scholar] [CrossRef] [PubMed]
- Schiera, G.; Proia, P.; Alberti, C.; Mineo, M.; Savettieri, G.; Di Liegro, I. Neurons produce FGF2 and VEGF and secrete them at least in part by shedding extracellular vesicles. J. Cell. Mol. Med. 2007, 11, 1384–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fauré, J.; Lachenal, G.; Court, M.; Hirrlinger, J.; Chatellard-Causse, C.; Blot, B.; Grange, J.; Schoehn, G.; Goldberg, Y.; Boyer, V.; et al. Exosomes are released by cultured cortical neurones. Mol. Cell. Neurosci. 2006, 31, 642–648. [Google Scholar] [CrossRef] [PubMed]
- Chivet, M.; Hemming, F.; Pernet-Gallay, K.; Fraboulet, S.; Sadoul, R. Emerging role of neuronal exosomes in the central nervous system. Front. Physiol. 2012, 3, 145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zappulli, V.; Friis, K.P.; Fitzpatrick, Z.; Maguire, C.A.; Breakefield, X.O. Extracellular vesicles and intercellular communication within the nervous system. J. Clin. Investig. 2016, 126, 1198–1207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agnati, L.F.; Fuxe, K.; Zoli, M.; Ozini, I.; Toano, G.; Ferraguti, F. A correlation analysis of the regional distribution of central enkephalin and beta endorphin immunoreactive terminals and of opiate receptors in adult and old male rats. Evidence for the existence of two main types of communication in the central nervous system: The volume transmission and the wiring transmission. Acta Physiol. Scand. 1986, 128, 201–207. [Google Scholar] [CrossRef]
- Zoli, M.; Jansson, A.; Sykova, E.; Agnati, L.F.; Fuxe, K. Volume transmission in the CNS and its relevance for neuropsychopharmacology. Trends Pharmacol. Sci. 1999, 20, 142–150. [Google Scholar] [CrossRef]
- De-Miguel, F.F.; Trueta, C. Synaptic and extrasynaptic secretion of serotonin. Cell. Mol. Neurobiol. 2005, 25, 297–312. [Google Scholar] [CrossRef]
- Proia, P.; Schiera, G.; Mineo, M.; Ingrassia, A.M.R.; Santoro, G.; Savettieri, G.; Di Liegro, I. Astrocytes shed extracellular vesicles that contain fibroblast growth factor-2 and vascular endothelial growth factor. Int. J. Mol. Med. 2008, 21, 63–67. [Google Scholar] [CrossRef] [Green Version]
- Sbai, O.; Ould-Yahoui, A.; Ferhat, L.; Gueye, Y.; Bernard, A.; Charrat, E.; Mehanna, A.; Risso, J.J.; Chauvin, J.P.; Fenouillet, E.; et al. Differential vesicular distribution and tracking of MMP-2, MMP-9 and their inhibitors in astrocytes. Glia 2010, 58, 344–366. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.R.; Robinson, M.B.; Gifondorwa, D.J.; Tytell, M.; Milligan, C.E. Regulation of heat shock protein 70 release in astrocytes: Role of signaling kinases. Dev. Neurobiol. 2007, 67, 1815–1829. [Google Scholar] [CrossRef] [PubMed]
- Pascua-Maestro, R.; González, E.; Lillo, C.; Ganfornina, M.D.; Falcón-Pérez, J.M.; Sanchez, D. Extracellular Vesicles Secreted by Astroglial Cells Transport Apolipoprotein D to Neurons and Mediate Neuronal Survival Upon Oxidative Stress. Front. Cell. Neurosci. 2019, 12, 526. [Google Scholar] [CrossRef] [PubMed]
- Gosselin, R.-D.; Meylan, P.; Decosterd, I. Extracellular microvesicles from astrocytes contain functional glutamate transporters: Regulation by protein kinase C and cell activation. Front. Cell. Neurosci. 2013, 7, 251. [Google Scholar] [CrossRef] [Green Version]
- Krämer-Albers, E.M.; Bretz, N.; Tenzer, S.; Winterstein, C.; Möbius, W.; Berger, H.; Nave, K.A.; Schild, H.; Trotter, J. Oligodendrocytes secrete exosomes containing major myelin and stress protective proteins: Trophic support for axons? Proteom. Clin. Appl. 2007, 1, 1446–1461. [Google Scholar] [CrossRef]
- Frühbeis, C.; Fröhlich, D.; Kuo, W.P.; Amphornrat, J.; Thilemann, S.; Saab, A.S.; Kirchho, F.; Möbius, W.; Goebbels, S.; Nave, K.A.; et al. Neurotransmitter-triggered transfer of exosomes mediates oligodendrocyte-neuron communication. PLoS Biol. 2013, 11, e1001604. [Google Scholar] [CrossRef] [Green Version]
- Court, F.A.; Hendriks, W.T.J.; MacGillavry, H.D.; Alvarez, J.; Van Minnen, J. Schwann cell to axon transfer of ribosomes: Toward a novel understanding of the role of glia in the nervous system. J. Neurosci. 2008, 28, 11024–11029. [Google Scholar] [CrossRef] [Green Version]
- Twiss, J.L.; Fainzilber, M. Ribosomes in axons–scrounging from the neighbors? Trends Cell Biol. 2009, 19, 236–243. [Google Scholar] [CrossRef]
- Lopez-Verrilli, M.A.; Picou, F.; Court, F.A. Schwann cell-derived exosomes enhance axonal regeneration in the peripheral nervous system. Glia 2013, 61, 1795–1806. [Google Scholar] [CrossRef]
- López-Leal, R.; Díaz-Viraqué, F.; Catalán, R.J.; Saquel, C.; Enright, A.; Iraola, G.; Court, F.A. Schwann cell reprogramming into repair cells increases miRNA-21 expression in exosomes promoting axonal growth. J. Cell Sci. 2020, 133, jcs239004. [Google Scholar] [CrossRef]
- Ahmad, S.; Srivastava, R.K.; Singh, P.; Naik, U.P.; Srivastava, A.K. Role of extracellular vesicles in glia-neuron intercellular communication. Front. Mol. Neurosci. 2022, 15, 844194. [Google Scholar] [CrossRef] [PubMed]
- Bianco, F.; Pravettoni, E.; Colombo, A.; Schenk, U.; Möller, T.; Matteoli, M.; Verderio, C. Astrocyte-derived ATP induce vesicle shedding and IL-1 beta release from microglia. J. Immunol. 2005, 174, 7268–7277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bianco, F.; Perrotta, C.; Novellino, L.; Francolini, M.; Riganti, L.; Menna, E.; Saglietti, L.; Schuchman, E.H.; Furlan, R.; Clementi, E.; et al. Acid sphingomyelinase activity triggers microparticle release from glial cells. EMBO J. 2009, 28, 1043–1054. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fevrier, B.; Vilette, D.; Archer, F.; Loew, D.; Faigle, W.; Vidal, M.; Laude, H.; Raposo, G. Cells release prions in association with exosomes. Proc. Natl. Acad. Sci. USA 2004, 101, 9683–9688. [Google Scholar] [CrossRef] [Green Version]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef] [Green Version]
- Vella, L.J.; Sharples, R.A.; Nisbet, R.M.; Cappai, R.; Hill, A.F. The roleof exosomes in the processing of proteins associated with neurodegenerative diseases. Eur. Biophys. J. 2008, 37, 323–332. [Google Scholar] [CrossRef]
- Emmanouilidou, E.; Melachroinou, K.; Roumeliotis, T.; Garbis, S.D.; Ntzouni, M.; Margaritis, L.H.; Stefanis, L.; Vekrellis, K. Cell-produced alpha-synuclein is secreted in a calcium-dependent manner by exosomes and impacts neuronal survival. J. Neurosci. 2010, 30, 6838–6851. [Google Scholar] [CrossRef] [Green Version]
- Novak, P.; Prcina, M.; Kontsekova, E. Tauons and prions: Infamous cousins? J. Alzheimer’s Dis. 2011, 26, 413–430. [Google Scholar] [CrossRef]
- Nath, S.; Agholme, L.; Kurudenkandy, F.R.; Granseth, B.; Marcusson, J.; Hallbeck, M. Spreading of neurodegenerative pathology via neuron-to-neuron transmission of β-amyloid. J. Neurosci. 2012, 32, 8767–8777. [Google Scholar] [CrossRef]
- Budnik, V.; Ruiz-Cañada, C.; Wendler, F. Extracellular vesicles round off communication in the nervous system. Nat. Rev. Neurosci. 2016, 17, 160–172. [Google Scholar] [CrossRef] [Green Version]
- DeLeo, A.M.; Ikezu, T. Extracellular Vesicle Biology in Alzheimer’s Disease and Related Tauopathy. J. Neuroimmune Pharmacol. 2018, 13, 292–308. [Google Scholar] [CrossRef] [PubMed]
- De Gassart, A.; Geminard, C.; Fevrier, B.; Raposo, G.; Vidal, M. Lipid raft-associated protein sorting in exosomes. Blood 2003, 102, 4336–4344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinoshita, M.; Matsumori, N. Inimitable Impacts of Ceramides on Lipid Rafts Formed in Artificial and Natural Cell Membranes. Membranes 2022, 12, 727. [Google Scholar] [CrossRef] [PubMed]
- Elsherbini, A.; Qin, H.; Zhu, Z.; Tripathi, P.; Wang, G.; Crivelli, S.M.; Spassieva, S.D.; Bieberich, E. Extracellular Vesicles Containing Ceramide-Rich Platforms: “Mobile Raft” Isolation and Analysis. Methods Mol. Biol. 2021, 2187, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Crivelli, S.M.; Giovagnoni, C.; Zhu, Z.; Tripathi, P.; Elsherbini, A.; Quadri, Z.; Pu, J.; Zhang, L.; Ferko, B.; Berkes, D.; et al. Function of ceramide transfer protein for biogenesis and sphingolipid composition of extracellular vesicles. J. Extracell. Vesicles 2022, 11, e12233. [Google Scholar] [CrossRef] [PubMed]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brügger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Menck, K.; Sönmezer, C.; Worst, T.S.; Schulz, M.; Dihazi, G.H.; Streit, F.; Erdmann, G.; Kling, S.; Boutros, M.; Binder, C.; et al. Neutral sphingomyelinases control extracellular vesicles budding from the plasma membrane. J. Extracell. Vesicles 2017, 6, 1378056. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, F.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Qin, F.; Wang, W.; Ni, Y.; Gao, M.; Guo, M.; Sun, G. hnRNPA2B1-Mediated Extracellular Vesicles Sorting of miR-122-5p Potentially Promotes Lung Cancer Progression. Int. J. Mol. Sci. 2021, 22, 12866. [Google Scholar] [CrossRef]
- Lei, Y.; Guo, W.; Chen, B.; Chen, L.; Gong, J.; Li, W. Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol. Rep. 2018, 40, 3438–3446. [Google Scholar] [CrossRef] [Green Version]
- Kosaka, N.; Iguchi, H.; Yoshioka, Y.; Takeshita, F.; Matsuki, Y.; Ochiya, T. Secretory Mechanisms and Intercellular Transfer of MicroRNAs in Living Cells. J. Biol. Chem. 2010, 285, 17442–17452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, M.E.; Naude, P.J.W.; van der Westhuizen, F.H. Proteomics and metabolomics of HIV-associated neurocognitive disorders: A systematic review. J. Neurochem. 2021, 157, 429–449. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Hollinger, K.R.; Huang, Y.; Borjabad, A.; Kim, B.-H.; Arab, T.; Thomas, A.G.; Moniruzzaman, M.; Lovell, L.; Turchinovich, A.; et al. Neutral sphingomyelinase 2 inhibition attenuates extracellular vesicle release and improves neurobehavioral deficits in murine HIV. Neurobiol. Dis. 2022, 169, 105734. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.M.; Abdelmohsen, K.; Mustapic, M.; Kapogiannis, D.; Gorospe, M. RNA in extracellular vesicles. Wiley Interdiscip. Rev. RNA 2017, 8, e1413. [Google Scholar] [CrossRef]
- Anand, S.; Samuel, M.; Kumar, S.; Mathivanan, S. Ticket to a bubble ride: Cargo sorting into exosomes and extracellular vesicles. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 140203. [Google Scholar] [CrossRef]
- Groot, M.; Lee, H. Sorting Mechanisms for MicroRNAs into Extracellular Vesicles and Their Associated Diseases. Cells 2020, 9, 1044. [Google Scholar] [CrossRef]
- Kunadt, M.; Eckermann, K.; Stuendl, A.; Gong, J.; Russo, B.; Strauss, K.; Rai, S.; Kügler, S.; Falomir Lockhart, L.; Schwalbe, M.; et al. Extracellular vesicle sorting of α-Synuclein is regulated by sumoylation. Acta Neuropathol. 2015, 129, 695–713. [Google Scholar] [CrossRef] [Green Version]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.; et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.; Li, C.; Zhang, Y.; Zhang, D.; Otterbein, L.E.; Jin, Y. Caveolin-1 selectively regulates microRNA sorting into microvesicles after noxious stimuli. J. Exp. Med. 2019, 216, 2202–2220. [Google Scholar] [CrossRef] [Green Version]
- Monastyrskaya, K. Functional association between regulatory RNAs and the annexins. Int. J. Mol. Sci. 2018, 19, 591. [Google Scholar] [CrossRef] [Green Version]
- Fabbiano, F.; Corsi, J.; Gurrieri, E.; Trevisan, C.; Notarangelo, M.; D’Agostino, V.G. RNA packaging into extracellular vesicles: An orchestra of RNA-binding proteins? J. Extracell. Vesicles 2020, 10, e12043. [Google Scholar] [CrossRef] [PubMed]
- Leidal, A.M.; Huang, H.H.; Marsh, T.; Solvik, T.; Zhang, D.; Ye, J.; Kai, F.; Goldsmith, J.; Liu, J.Y.; Huang, Y.H.; et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 2020, 22, 187–199. [Google Scholar] [CrossRef] [PubMed]
- O’Brien, K.; Breyne, K.; Ughetto, S.; Laurent, L.C.; Breakefield, X.O. RNA delivery by extracellular vesicles in mammalian cells and its applications. Nat. Rev. Mol. Cell Biol. 2020, 21, 585–606. [Google Scholar] [CrossRef] [PubMed]
- Santangelo, L.; Giurato, G.; Cicchini, C.; Montaldo, C.; Mancone, C.; Tarallo, R.; Battistelli, C.; Alonzi, T.; Weisz, A.; Tripodi, M. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016, 17, 799–808. [Google Scholar] [CrossRef] [Green Version]
- Gao, T.; Liu, X.; He, B.; Nie, Z.; Zhu, C.; Zhang, P.; Wang, S. Exosomal lncRNA 91H is associated with poor development in colorectal cancer by modifying HNRNPK expression. Cancer Cell Int. 2018, 18, 11. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Temoche-Diaz, M.M.; Karfilis, K.V.; Ri, S.; Schekman, R. Y-box protein 1 is required to sort microRNAs into exosomes in cells and in a cell-free reaction. Elife 2016, 5, e19276. [Google Scholar] [CrossRef]
- Mukherjee, K.; Ghoshal, B.; Ghosh, S.; Chakrabarty, Y.; Shwetha, S.; Das, S.; Bhattacharyya, S.N. Reversible HuR-micro RNA binding controls extracellular export of miR-122 and augments stress response. EMBO Rep. 2016, 17, 1184–1203. [Google Scholar] [CrossRef]
- Bampton, A.; Gittings, L.M.; Fratta, P.; Lashley, T.; Gatt, A. The role of hnRNPs in frontotemporal dementia and amyotrophic lateral sclerosis. Acta Neuropathol. 2020, 140, 599–623. [Google Scholar] [CrossRef]
- Klingeborn, M.; Stamer, W.D.; Bowes Rickman, C. Polarized exosome release from the retinal pigmented epithelium. Adv. Exp. Med. Biol. 2018, 1074, 539–544. [Google Scholar] [CrossRef]
- Turchinovich, A.; Drapkina, O.; Tonevitsky, A. Transcriptome of Extracellular Vesicles: State-of-the-Art. Front. Immunol. 2019, 10, 202. [Google Scholar] [CrossRef] [Green Version]
- Ling, S.-C.; Polymenidou, M.; Cleveland, D.W. Converging mechanisms in ALS and FTD: Disrupted RNA and protein homeostasis. Neuron 2013, 79, 416–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polymenidou, M.; Lagier-Tourenne, C.; Hutt, K.R.; Huelga, S.C.; Moran, J.; Liang, T.Y.; Ling, S.C.; Sun, E.; Wancewicz, E.; Mazur, C.; et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nat. Neurosci. 2011, 14, 459–468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, A.; Bharathi, V.; Sivalingam, V.; Girdhar, A.; Patel, B.K. Molecular Mechanisms of TDP-43 Misfolding and Pathology in Amyotrophic Lateral Sclerosis. Front. Mol. Neurosci. 2019, 12, 25. [Google Scholar] [CrossRef] [PubMed]
- Neumann, M.; Sampathu, D.M.; Kwong, L.K.; Truax, A.C.; Micsenyi, M.C.; Chou, T.T.; Bruce, J.; Schuck, T.; Grossman, M.; Clark, C.M.; et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 2006, 314, 130–133. [Google Scholar] [CrossRef] [Green Version]
- Santamaria, N.; Alhothali, M.; Alfonso, M.H.; Breydo, L.; Uversky, V.N. Intrinsic disorder in proteins involved in amyotrophic lateral sclerosis. Cell. Mol. Life Sci. 2017, 74, 1297–1318. [Google Scholar] [CrossRef]
- Portz, B.; Lee, B.L.; Shorter, J. FUS and TDP-43 Phases in Health and Disease. Trends Biochem. Sci. 2021, 46, 550–563. [Google Scholar] [CrossRef]
- Udan, M.; Baloh, R.H. Implications of the prion-related Q/N domains in TDP-43 and FUS. Prion 2011, 5, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Yokoshi, M.; Okada, H.; Kawahara, Y. The cleavage pattern of TDP-43 determines its rate of clearance and cytotoxicity. Nat. Commun. 2015, 6, 6183. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.J.; Xu, Y.F.; Cook, C.; Gendron, T.F.; Roettges, P.; Link, C.D.; Lin, W.L.; Tong, J.; Castanedes-Casey, M.; Ash, P.; et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. USA 2009, 106, 7607–7612. [Google Scholar] [CrossRef]
- Zhang, Y.J.; Gendron, T.F.; Xu, Y.F.; Ko, L.W.; Yen, S.H.; Petrucelli, L. Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol. Neurodegener. 2010, 5, 33. [Google Scholar] [CrossRef] [Green Version]
- Berning, B.A.; Walker, A.K. The Pathobiology of TDP-43 C-Terminal Fragments in ALS and FTLD. Front. Neurosci. 2019, 13, 335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daoud, H.; Suhail, H.; Szuto, A.; Camu, W.; Salachas, F.; Meininger, V.; Bouchard, J.P.; Dupré, N.; Dion, P.A.; Rouleau, G.A. UBQLN2 mutations are rare in French and French-Canadian amyotrophic lateral sclerosis. Neurobiol. Aging 2012, 33, 2230.e1–2230.e5. [Google Scholar] [CrossRef] [PubMed]
- Sleigh, J.N.; Tosolini, A.P.; Gordon, D.; Devoy, A.; Fratta, P.; Fisher, E.M.C.; Talbot, K.; Schiavo, G. ALS mice carrying pathological mutant TDP-43, but not mutant FUS, display axonal transport defects in vivo. Cell Rep. 2020, 30, 3655–3662.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mann, J.R.; Gleixner, A.M.; Mauna, J.C.; Gomes, E.; DeChellis-Marks, M.R.; Needham, P.G.; Copley, K.E.; Hurtle, B.; Portz, B.; Pyles, N.J.; et al. RNA Binding Antagonizes Neurotoxic Phase Transitions of TDP-43. Neuron 2019, 102, 321–338.e8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maharana, S.; Wang, J.; Papadopoulos, D.K.; Richter, D.; Pozniakovsky, A.; Poser, I.; Bickle, M.; Rizk, S.; Guillén-Boixet, J.; Franzmann, T.M.; et al. RNA buffers the phase separation behavior of prion-like RNA binding proteins. Science 2018, 360, 918–921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iko, Y.; Kodama, T.S.; Kasai, N.; Oyama, T.; Morita, E.H.; Muto, T.; Okumura, M.; Fujii, R.; Takumi, T.; Tate, S.-I.; et al. Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 2004, 279, 44834–44840. [Google Scholar] [CrossRef] [Green Version]
- Sukhanova, M.V.; Singatulina, A.S.; Pastré, D.; Lavrik, O.I. Fused in Sarcoma (FUS) in DNA Repair: Tango with Poly(ADP-ribose) Polymerase 1 and Compartmentalisation of Damaged DNA. Int. J. Mol. Sci. 2020, 21, 7020. [Google Scholar] [CrossRef] [PubMed]
- Baade, I.; Hutten, S.; Sternburg, E.L.; Pörschke, M.; Hofweber, M.; Dormann, D.; Kehlenbach, R.H. The RNA-binding protein FUS is chaperoned and imported into the nucleus by a network of import receptors. J. Biol. Chem. 2021, 296, 100659. [Google Scholar] [CrossRef]
- Lagier-Tourenne, C.; Polymenidou, M.; Hutt, K.R.; Vu, A.Q.; Baughn, M.; Huelga, S.C.; Clutario, K.M.; Ling, S.C.; Liang, T.Y.; Mazur, C.; et al. Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat. Neurosci. 2012, 15, 1488–1497. [Google Scholar] [CrossRef]
- Mitchell, J.C.; McGoldrick, P.; Vance, C.; Hortobagyi, T.; Sreedharan, J.; Rogelj, B.; Tudor, E.L.; Smith, B.N.; Klasen, C.; Miller, C.C.; et al. Overexpression of human wild-type FUS causes progressive motor neuron degeneration in an age- and dose-dependent fashion. Acta Neuropathol. 2013, 125, 273–288. [Google Scholar] [CrossRef] [Green Version]
- Guerrero, E.N.; Wang, H.; Mitra, J.; Hegde, P.M.; Stowell, S.E.; Liachko, N.F.; Kraemer, B.C.; Garruto, R.M.; Rao, K.S.; Hegde, M.L. TDP-43/FUS in motor neuron disease: Complexity and challenges. Prog. Neurobiol. 2016, 145–146, 78–97. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dormann, D.; Rodde, R.; Edbauer, D.; Bentmann, E.; Fischer, I.; Hruscha, A.; Than, M.E.; Mackenzie, I.R.A.; Capell, A.; Schmid, B.; et al. ALS-associated fused in sarcoma (FUS) mutations disrupt Transportin-mediated nuclear import. EMBO J. 2010, 29, 2841–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelkovnikova, T.A.; Peters, O.M.; Deykin, A.V.; Connor-Robson, N.; Robinson, H.; Ustyugov, A.A.; Bachurin, S.O.; Ermolkevich, T.G.; Goldman, I.L.; Sadchikova, E.R.; et al. Fused in sarcoma (FUS) protein lacking nuclear localization signal (NLS) and major RNA binding motifs triggers proteinopathy and severe motor phenotype in transgenic mice. J. Biol. Chem. 2013, 288, 25266–25274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lassmann, H. Pathogenic Mechanisms Associated With Different Clinical Courses of Multiple Sclerosis. Front. Immunol. 2019, 9, 3116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friese, M.A.; Schattling, B.; Fugger, L. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat. Rev. Neurol. 2014, 10, 225–238. [Google Scholar] [CrossRef]
- Correale, J.; Gaitán, M.I.; Ysrraelit, M.C.; Fiol, M.P. Progressive multiple sclerosis: From pathogenic mechanisms to treatment. Brain 2017, 140, 527–546. [Google Scholar] [CrossRef] [Green Version]
- Vercellino, M.; Masera, S.; Lorenzatti, M.; Condello, C.; Merola, A.; Mattioda, A.; Tribolo, A.; Capello, E.; Mancardi, G.L.; Mutani, R.; et al. Demyelination, Inflammation, and Neurodegeneration in Multiple Sclerosis Deep Gray Matter. J. Neuropathol. Exp. Neurol. 2009, 68, 489–502. [Google Scholar] [CrossRef] [Green Version]
- Dutta, R.; McDonough, J.; Yin, X.; Peterson, J.; Chang, A.; Torres, T.; Gudz, T.; Macklin, W.B.; Lewis, D.A.; Fox, R.J.; et al. Mitochondrial dysfunction as a cause of axonal degeneration in multiple sclerosis patients. Ann. Neurol. 2006, 59, 478–489. [Google Scholar] [CrossRef]
- Su, K.G.; Banker, G.; Bourdette, D.; Forte, M. Axonal degeneration in multiple sclerosis: The mitochondrial hypothesis. Curr. Neurol. Neurosci. Rep. 2009, 9, 411–417. [Google Scholar] [CrossRef]
- Libner, C.D.; Salapa, H.E.; Levin, M.C. The Potential Contribution of Dysfunctional RNA-Binding Proteins to the Pathogenesis of Neurodegeneration in Multiple Sclerosis and Relevant Models. Int. J. Mol. Sci. 2020, 21, 4571. [Google Scholar] [CrossRef]
- Salapa, H.E.; Hutchinson, C.; Popescu, B.F.; Levin, M.C. Neuronal RNA-binding protein dysfunction in multiple sclerosis cortex. Ann. Clin. Transl. Neurol. 2020, 7, 1214–1224. [Google Scholar] [CrossRef] [PubMed]
- Salapa, H.E.; Libner, C.D.; Levin, M.C. Dysfunctional RNA-binding protein biology and neurodegeneration in experimental autoimmune encephalomyelitis in female mice. J. Neurosci. Res. 2020, 98, 704–717. [Google Scholar] [CrossRef] [PubMed]
- Masaki, K.; Sonobe, Y.; Ghadge, G.; Pytel, P.; Lépine, P.; Pernin, F.; Cui, Q.L.; Antel, J.P.; Zandee, S.; Prat, A.; et al. RNA-binding protein altered expression and mislocalization in MS. Neurol. Neuroimmunol. Neuroinflamm. 2020, 7, e704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Douglas, J.N.; Gardner, L.A.; Salapa, H.E.; Lalor, S.J.; Lee, S.; Segal, B.M.; Sawchenko, P.E.; Levin, M.C. Antibodies to the RNA-binding protein hnRNP A1 contribute to neurodegeneration in a model of central nervous system autoimmune inflammatory disease. J. Neuroinflamm. 2016, 13, 178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borgonetti, V.; Sanna, M.D.; Lucarini, L.; Galeotti, N. Targeting the RNA-binding protein HuR alleviates neuroinflammation in experimental autoimmune encephalomyelitis: Potential therapy for multiple sclerosis. Neurother. J. Am. Soc. Exp. Neuro Ther. 2020, 18, 412–429. [Google Scholar] [CrossRef]
- Chen, J.; Martindale, J.L.; Abdelmohsen, K.; Kumar, G.; Fortina, P.M.; Gorospe, M.; Rostami, A.; Yu, S. RNA-Binding Protein HuR Promotes Th17 Cell Differentiation and Can Be Targeted to Reduce Autoimmune Neuroinflammation. J. Immunol. 2020, 204, 2076–2087. [Google Scholar] [CrossRef]
- Chen, J.; Martindale, J.L.; Cramer, C.; Gorospe, M.; Atasoy, U.; Drew, P.D.; Yu, S. The RNA-binding protein HuR contributes to neuroinflammation by promoting C-C chemokine receptor 6 (CCR6) expression on Th17 cells. J. Biol. Chem. 2017, 292, 14532–14543. [Google Scholar] [CrossRef] [Green Version]
- Sanna, M.D.; Quattrone, A.; Galeotti, N. Silencing of the RNA-binding protein HuR attenuates hyperalgesia and motor disability in experimental autoimmune encephalomyelitis. Neuropharmacology 2017, 123, 116–125. [Google Scholar] [CrossRef]
- Pistono, C.; Monti, M.C.; Marchesi, N.; Boiocchi, C.; Campagnoli, L.I.M.; Morlotti, D.; Cuccia, M.; Govoni, S.; Montomoli, C.; Mallucci, G.; et al. Unraveling a new player in multiple sclerosis pathogenesis: The RNA-binding protein HuR. Mult. Scler. Relat. Disord. 2020, 41, 102048. [Google Scholar] [CrossRef]
- Khan, S.; Barve, K.H.; Kumar, M.S. Recent Advancements in Pathogenesis, Diagnostics and Treatment of Alzheimer’s Disease. Curr. Neuropharmacol. 2020, 18, 1106–1125. [Google Scholar] [CrossRef]
- Lester, E.; Ooi, F.K.; Bakkar, N.; Ayers, J.; Woerman, A.L.; Wheeler, J.; Bowser, R.; Carlson, G.A.; Prusiner, S.B.; Parker, R. Tau aggregates are RNA-protein assemblies that mislocalize multiple nuclear speckle components. Neuron 2021, 109, 1675–1691.e9. [Google Scholar] [CrossRef] [PubMed]
- Dixit, H.; Kumar, C.S.; Chaudhary, R.; Thaker, D.; Gadewal, N.; Dasgupta, D. Role of Phosphorylation and Hyperphosphorylation of Tau in Its Interaction with βα Dimeric Tubulin Studied from a Bioinformatics Perspective. Avicenna J. Med. Biotechnol. 2021, 13, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Combs, B.; Mueller, R.L.; Morfini, G.; Brady, S.T.; Kanaan, N.M. Tau and Axonal Transport Misregulation in Tauopathies. Adv. Exp. Med. Biol. 2019, 1184, 81–95. [Google Scholar] [CrossRef] [PubMed]
- Kampers, T.; Friedhoff, P.; Biernat, J.; Mandelkow, E.M.; Mandelkow, E. RNA stimulates aggregation of microtubule-associated protein tau into Alzheimer-like paired helical filaments. FEBS Lett. 1996, 399, 344–349. [Google Scholar] [CrossRef]
- Wang, X.; Wang, D.; Zhao, J.; Qu, M.; Zhou, X.; He, H.; He, R. The proline-rich domain and the microtubule binding domain of protein tau acting as RNA binding domains. Protein Pept. Lett. 2006, 13, 679–685. [Google Scholar] [CrossRef]
- Hochmair, J.; Exner, C.; Franck, M.; Dominguez-Baquero, A.; Diez, L.; Brognaro, H.; Kraushar, M.L.; Mielke, T.; Radbruch, H.; Kaniyappan, S.; et al. Molecular crowding and RNA synergize to promote phase separation, microtubule interaction, and seeding of Tau condensates. EMBO J. 2022, 41, e108882. [Google Scholar] [CrossRef]
- Sengupta, U.; Montalbano, M.; McAllen, S.; Minuesa, G.; Kharas, M.; Kayed, R. Formation of Toxic Oligomeric Assemblies of RNA-binding Protein: Musashi in Alzheimer’s disease. Acta Neuropathol. Commun. 2018, 6, 113. [Google Scholar] [CrossRef] [Green Version]
- Montalbano, M.; McAllen, S.; Puangmalai, N.; Sengupta, U.; Bhatt, N.; Johnson, O.D.; Kharas, M.G.; Kayed, R. RNA-binding proteins Musashi and tau soluble aggregates initiate nuclear dysfunction. Nat. Commun. 2020, 11, 4305. [Google Scholar] [CrossRef]
- Drummond, E.; Pires, G.; MacMurray, C.; Askenazi, M.; Nayak, S.; Bourdon, M.; Safar, J.; Ueberheide, B.; Wisniewski, T. Phosphorylated tau interactome in the human Alzheimer’s disease brain. Brain 2020, 143, 2803–2817. [Google Scholar] [CrossRef]
- Tsoi, P.S.; Quan, M.D.; Choi, K.J.; Dao, K.M.; Ferreon, J.C.; Ferreon, A.C.M. Electrostatic modulation of hnRNPA1 low-complexity domain liquid-liquid phase separation and aggregation. Protein Sci. 2021, 30, 1408–1417. [Google Scholar] [CrossRef]
- Filippova, N.; Yang, X.; Ananthan, S.; Sorochinsky, A.; Hackney, J.R.; Gentry, Z.; Bae, S.; King, P.; Nabors, L.B. Hu antigen R (HuR) multimerization contributes to glioma disease progression. J. Biol. Chem. 2017, 292, 16999–17010. [Google Scholar] [CrossRef] [PubMed]
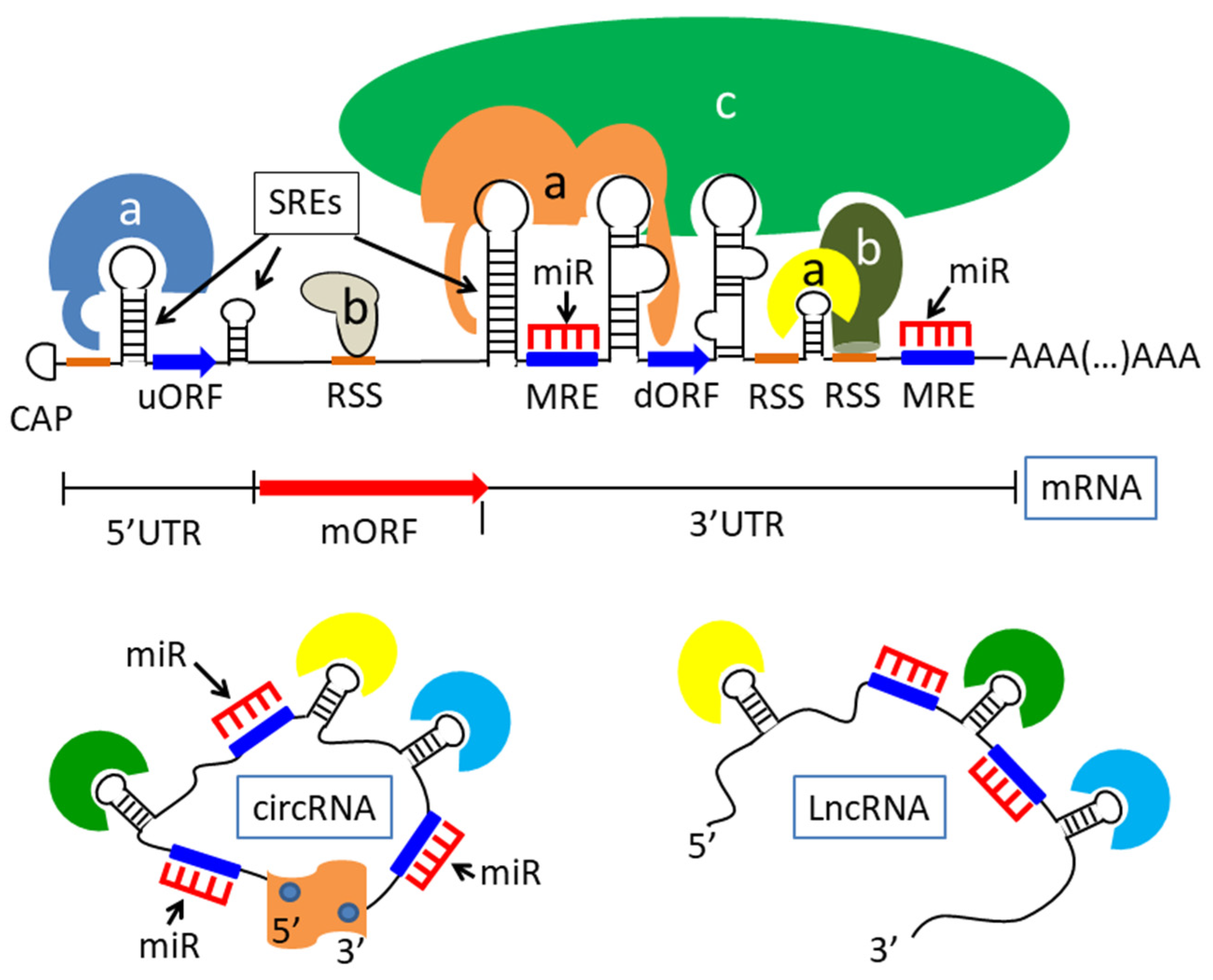
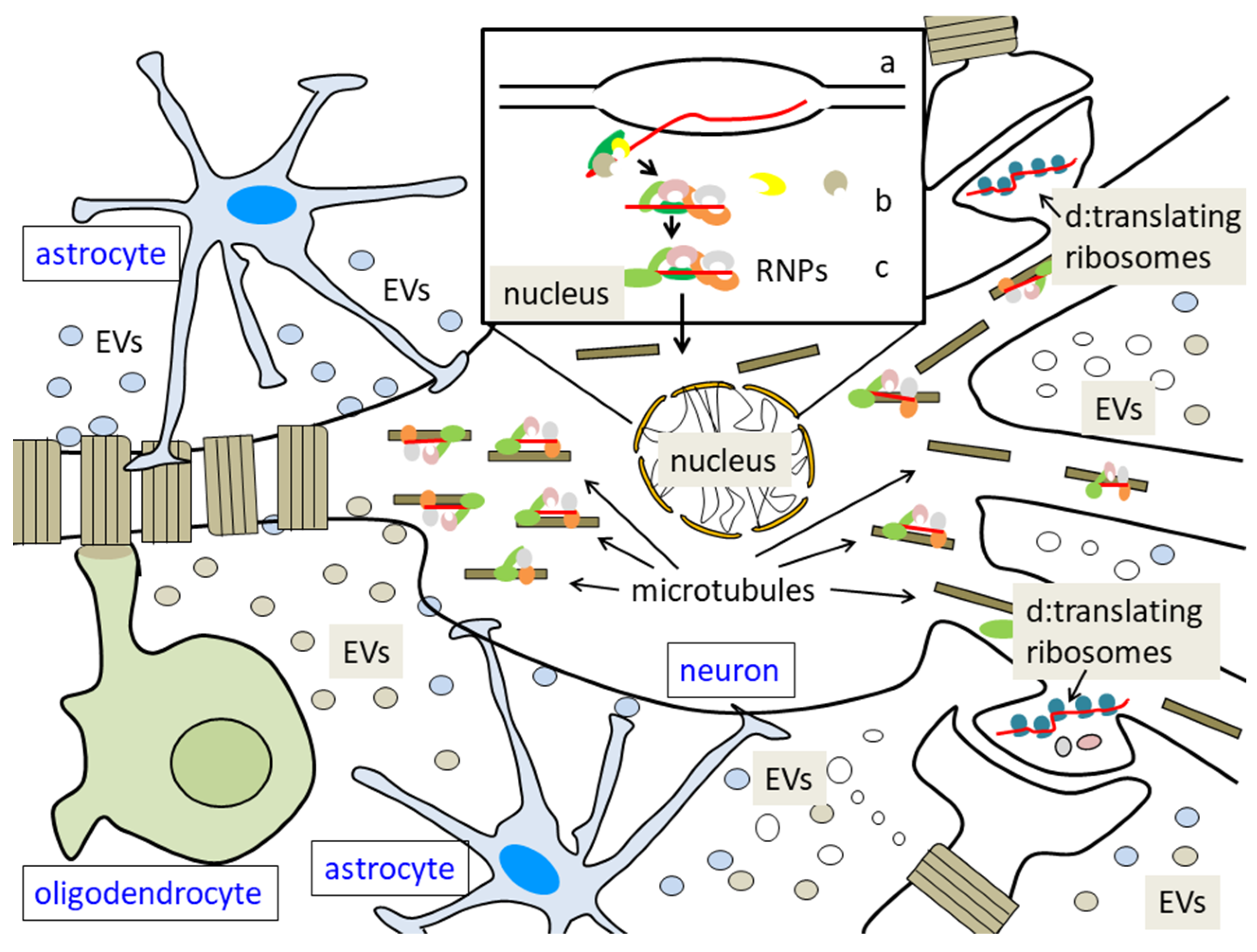
| Neurological Disease | RBPs | Predominant Localization in Normal Conditions | Suggested Functions | Tendency to Form Aggregates |
|---|---|---|---|---|
| Amyotrophic Lateral Sclerosis (SLA) | TDP-43 [334,335,336,337,338,339,340,341,342,343,344,345,346,347,348] FUS [349,350,351,352,353,354,355,356] | Nuclear (neurons) Nuclear (neurons) | -splicing -granule formation -RNA transport and protection [334,335,336,337,346] Processing of different RNAs (among which miRNAs) [352,354] | yes [336,337,338,339,340,341,342,346] RNA binding seems to be protective for aggregation [348] yes [356] |
| Multiple Sclerosis (MS) | TDP-43 [364,365,366] HnRNPA1 [364,365,367] HuR | Nuclear (neurons) Nuclear (neurons) Cytoplasm (microglial cells) [368] | see above -different aspects of RNA metabolism [331] -miRNA sorting to EVs [318] | see above yes [383] observed in gliomas [384] |
| Alzheimer’s Disease (AD) | Tau | neuronal axons | Main function: regulation of microtubules dynamics [374] Hypothesized functions: -RNA binding [378,379], with a possible effect on splicing [374] | yes Tau has been reported to form complexes with some RBPs, such as Musashi [381] and HnRNPA1 [382] NOTE: RNA can cause conversion in vitro of soluble Tau into paired helical filaments [377] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Liegro, C.M.; Schiera, G.; Schirò, G.; Di Liegro, I. RNA-Binding Proteins as Epigenetic Regulators of Brain Functions and Their Involvement in Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 14622. https://doi.org/10.3390/ijms232314622
Di Liegro CM, Schiera G, Schirò G, Di Liegro I. RNA-Binding Proteins as Epigenetic Regulators of Brain Functions and Their Involvement in Neurodegeneration. International Journal of Molecular Sciences. 2022; 23(23):14622. https://doi.org/10.3390/ijms232314622
Chicago/Turabian StyleDi Liegro, Carlo Maria, Gabriella Schiera, Giuseppe Schirò, and Italia Di Liegro. 2022. "RNA-Binding Proteins as Epigenetic Regulators of Brain Functions and Their Involvement in Neurodegeneration" International Journal of Molecular Sciences 23, no. 23: 14622. https://doi.org/10.3390/ijms232314622
APA StyleDi Liegro, C. M., Schiera, G., Schirò, G., & Di Liegro, I. (2022). RNA-Binding Proteins as Epigenetic Regulators of Brain Functions and Their Involvement in Neurodegeneration. International Journal of Molecular Sciences, 23(23), 14622. https://doi.org/10.3390/ijms232314622







