Laser Capture Microdissection: A Gear for Pancreatic Cancer Research
Abstract
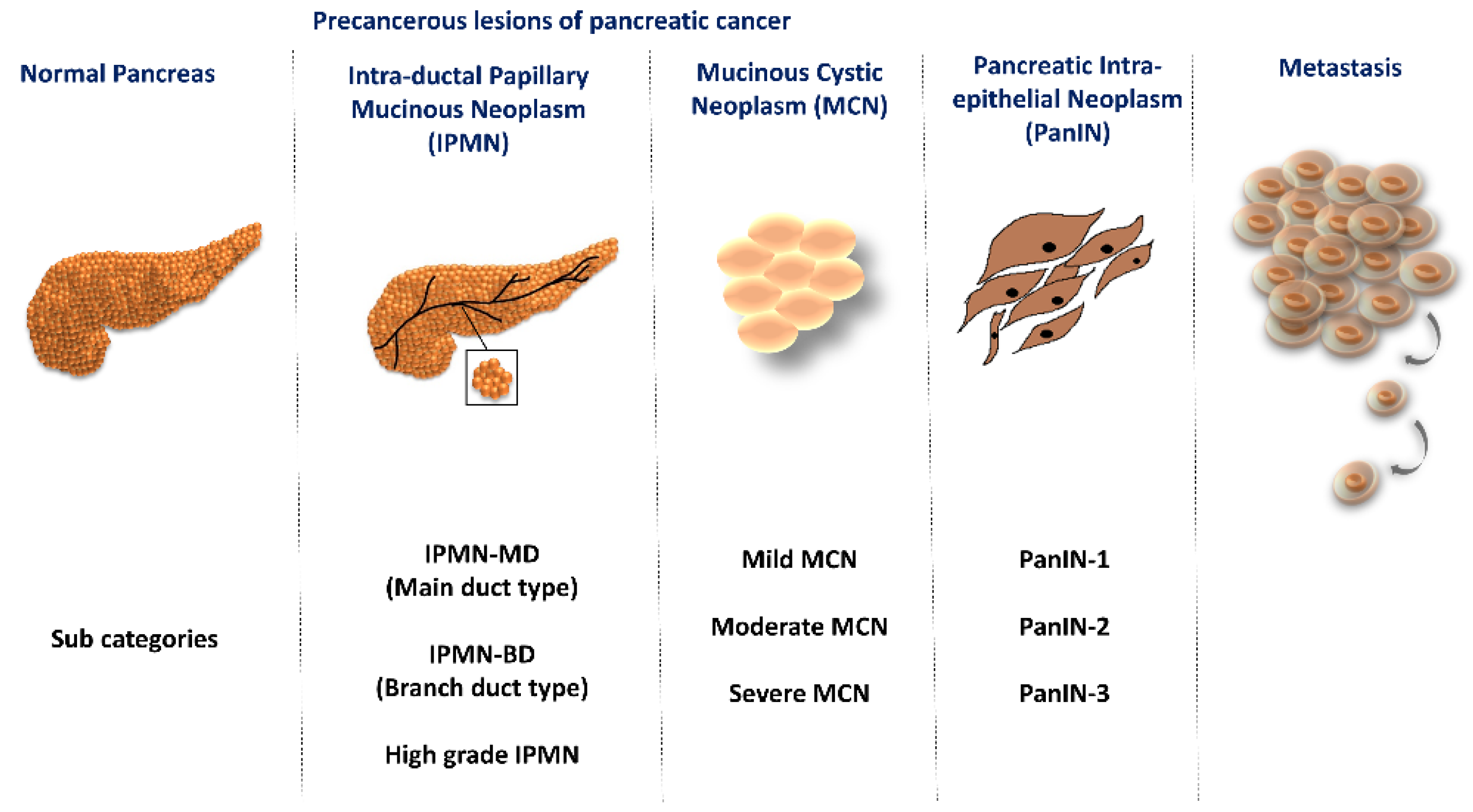
1. Introduction
2. Laser Capture Microdissection (LCM)

3. Impact of LCM on Pancreatic Cancer Research
3.1. Mutation Studies
3.2. Breakthrough of PC Subtypes and Their Relevance in Survival
3.3. Proteins, Pathways, and Cancer Management
| Author/Year | Finding | Sample Used | Techniques Used along with LCM | Reference |
|---|---|---|---|---|
| Emmert-buck et al., 1996 | Discovery of LCM technique | [32] | ||
| Crnogorac-Jurcevic et al., 2002 | Association of ABL2, NOTCH4, SOD1, XRCC1 with metastasis of PC | Fresh frozen tissue of PDAC and PC cell lines (ASPC1, Bxpc-3, CaPan1, CaPan2, HS766T, Mia PaCa-2, PANC-1, SU86.86) | Micro-array derived gene expression analysis, quantitative real-time PCR (qPCR), Tissue array, IHC | [52] |
| Shekouh et al., 2003 | Identification of DEGs in PDAC | Fresh frozen samples of PDAC and normal tissues | Isoelectric focusing, SDS-PAGE, silver staining, MALDI-TOF, IHC | [80] |
| Guweidhi et al., 2004 | role of 14-3-3sigma/stratifin in cell cycle regulation, and apoptosis | Fresh-frozen and PPFE samples of human PDAC and normal tissues | cDNA array, qPCR, southern blot, IHC, mutation analysis (sequencing), western blot, immunoprecipitation, FACS analysis | [84] |
| Kayed et al., 2005 | Role of FXYD3 in PC development | FFPE samples of PDAC and PC cell lines (ASPC-1, BxPc-3, CaPan-1, Colo-357, SU86.86, T3M4) | qPCR, DNA oligonucleotide microarray, IHC, northern blot, immunofluorescence | [67] |
| Wei et al., 2005 | The difference in KRAS mutation in the Chinese population | PDAC Samples | PCR and direct sequencing | [53] |
| Erkan et al., 2005 | Role of BNIP3 in chemoresistance resulting in poor prognosis and survival in PDAC | PDAC tissue samples and PC cell lines (ASPC-1, BxPc-3, CaPan-1, Colo-357, MiaPaCa-2, Panc-1, SU86.86, T3M4) | cDNA microarray, qPCR, IHC | [94] |
| Sato et al., 2005 | Down-regulation of CDKN1C in PC by an epigenetic mechanism | Fresh frozen IPMNs and normal tissues PC cell lines (AsPC1,BxPC3, CaPan1, CaPan2, CFPAC1, Hs766T, MiaPaCa2, Panc1), and xenografts | Microarray, semiquantitative reverse-transcription PCR, IHC, Methylation-specific PCR, and Bisulfite sequencing | [48] |
| Sitek et al., 2005 | Role of actin filament proteins in PanIN progression | Fresh frozen PanIN samples and PC cell lines (CFPAC, CAPAN, Hs766T, IMIMPC-2, SCPC-1, PATH-8988T) | 2-D electrophoresis (2-DE), fluorescence dye saturation labeling, MALDI-TOF | [81] |
| Fukushima et al., 2005 | Role of HC gp-39, lactoferrin, and HIP/PIP as potential predictive biomarker of PC | Fresh frozen tissues and serum samples | Oligonucleotide hybridization, IHC, qPCR, ELIS | [85] |
| Hwang et al., 2006 | Upregulation of PGK1 in PDAC and its potential role in therapeutic strategies or as a diagnostic biomarker | PDAC and normal tissues, serum samples | 2-DE, MALDI-TOF, ELISA, IHC, Western blot | [95] |
| Tzeng et al., 2007 | Conservation of EGFR in PC and its unavailability to act in the prognosis of PC | PDAC tissue samples, PC cell lines (S2-VP10 AND S2-103) | PCR, and sequencing | [64] |
| Kayed et al., 2007 | Role of BSP in cancer progression | PDAC and chronic pancreatitis (CP) tissue, PC cell lines (ASPC-1, BxPc-3, CAnPan-1, Colo-357, MiaPaCa-2, Panc-1, SU86.86, T3M4) | qPCR, cDNA array, IHC, Radioimmunoassay (RIA), FACS, Invitro invasion, scattering, and adhesion assays | [78] |
| Esposito et al., 2007 | Role of SPARC1 as a tumor suppressor gene in PC | Fresh frozen PDAC, PanIN tissue samples, and PC Cell lines (ASPC-1, BxPc-3, Capan1, colo-357, Su86.86, and T3M4) | FACS, in-vitro invasion assays, IHC | [79] |
| Soliman et al., 2007 | Importance of gene-environment interaction in cancerogenesis | FFPE samples of PDAC and normal tissues | PCR, DNA sequencing | [54] |
| Nakamura et al., 2007 | Importance of DEG studies in zonal heterogeneity of PDAC | Human PC cell line (L3.6pl), nude mice | Affymetrix HG-U133 plus 2.0 array, FISH | [73] |
| Hoffmann et al., 2008 | Overexpression of HIF1A during the hypoxic condition in PDAC and its correlation with PDGFA, VEGF, and FGF2 | PDAC FFPE samples | qPCR | [96] |
| Shi et al., 2009 | Involvement of acinar cells in the development of PanIN/PDAC | PanIN lesions | PCR, LigAmp analysis, IHC | [57] |
| Kubo et al., 2009 | Mutation of KRAS/BRAF in resequenced tyrosine kinase gene showing its importance in the downstream signaling pathway | PDAC samples and cell lines | WGA and sequencing | [87] |
| Collisson et al., 2011 | Subtypes of PDAC | PDAC FFPE samples and Cell lines (HPAC, Capan2, HPAF II, 6.03, CFPac1, MPanc96, 2.13, Panc1, MiaPaca2, 10.05, and Colo357) | IHC, microarray | [10] |
| Kayashima et al., 2011 | Stimulation of INSIG2 in PC during hypoxia condition | PC cell lines (SUIT-2, ASPC-1, BxPC-3, PANC-1, KP-1N, KP2, KP-3, MiaPaCa2, CaPan1, CaPan 2, CFPAC-1, SW1990, HS766T, H48N, NOR-P1, HDPE6-E6E7) and PanIN lesions | qPCR, microarray | [97] |
| Naidoo et al., 2012 | Protein composition of PDAC and lymph node metastasis | PDAC FFPE samples | Multidimensional Protein Identification Technology (MudPIT), IHC | [82] |
| Nakahara et al., 2012 | Role of miR-101 as a therapeutic target in IMPNs | FFPE samples, PC cell lines (PANC-1, PK8, PK9, PK-59, KLM-1, MIA PaCa2, PK-45P) | IHC, qPCR, knock-down of miR101 | [69] |
| Zhu et al., 2013 | A better understanding of tumor progression using proteomic analysis of PDAC samples | Fresh frozen PDAC and adjacent normal tissue | LC-MS/MS, Tissue microarray, IHC | [88] |
| Murphy et al., 2013 | Mutation of KRAS, TP53, and other somatic genes in PanIN-2 lesions and its role in PDAC progression | Frozen PDAC samples | Exome sequencing | [58] |
| Shan et al., 2014 | Downregulation of Cav-1 as a prognostic indicator in PC | Fresh frozen PDAC samples | IHC, reverse-transcriptase PCR, qPCR, FISH | [89] |
| Garcia-Carracedo et al., 2014 | PIK3CA mutation in pancreatic MCN | FFPE samples of MCN | IHC, direct sequencing | [51] |
| Sawai et al., 2015 | Role of AID in PDAC development | PPFE samples of PDAC tissues, transgenic mice | IHC, deep sequencing | [86] |
| Hasegawa et al., 2015 | Role of Sox4/Ezh2 in epigenetic mechanism and EMT pathway in PC patients | Fresh frozen PDAC samples | IHC, qPCR | [68] |
| Court et al., 2016 | Role of CTCs in molecular diagnostics of PC | PC cell lines (CFPAC-1, ASPC-1, Panc-1, BxPC-3, HPAF-II) and blood samples of pancreatobiliary cancer patients | WGA, KRAS PCR, Sanger sequencing | [90] |
| M.Ling et al., 2016 | Role of lncRNA H19 in PC tumorigenesis | Fresh frozen PDAC and normal tissues, PC cell lines (Colo-357, Capan1, MiaPaca-2, AsPC-1, BxPC-3, Panc-1, T3M4, SW1990) | qPCR, western blot, IHC | [91] |
| Fu et al., 2017 | Role of lncRNA HOTTIP in DFS of PC | Cell lines (PANC-1 and SW1990) | qPCR, western blot, FACS, IHC | [92] |
| Fang et al., 2017 | Showed PASC and PDAC originated from same progenitor cancer cells | FFPE samples of normal and tumor tissue | Whole-genome, whole-exome sequencing | [60] |
| Anug et al., 2018 | COMPASS trial | Fresh frozen PDAC samples and whole blood samples | WGS, RNA-seq, RNA-ISH | [72] |
| Maurer et al., 2019 | Molecular subtypes of PDAC | Fresh frozen PDAC samples | RNA sequencing | [71] |
| Nadella et al., 2019 | Role of gastrin in stimulating KRAS and in turn carcinogenesis | Gastrin Knockout mice | Reverse phase protein array, IHC, miRNA analysis | [66] |
| Hiroshima et al., 2019 | Impact of FN1-ITGA3 on prognosis of PDAC | Fresh frozen tissue of PDAC | LC-MS/MS | [93] |
| Robin et al., 2020 | Prognostic role of stratifin | PDAC FFPE samples | Gene expression analysis, IHC, ELISA | [83] |
| Birnbaum et al., 2021 | Transcriptomic analysis of PDAC samples to identify molecular subtypes of PDAC | Fresh frozen PDAC samples | RNA-seq, RNA-ISH | [34] |
| Kalloger et al., 2021 | Prognostic roles of genes expressed in stroma and epithelium of PDAC | PDAC FFPE samples | mRNA quantification | [70] |
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Rawla, P.; Sunkara, T.; Gaduputi, V. Epidemiology of Pancreatic Cancer: Global Trends, Etiology and Risk Factors. World J. Oncol. 2019, 10, 10. [Google Scholar] [CrossRef] [PubMed]
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic Cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.P. Pancreatic Cancer Epidemiology: Understanding the Role of Lifestyle and Inherited Risk Factors. Nat. Rev. Gastroenterol. Hepatol. 2021, 18, 493–502. [Google Scholar] [CrossRef] [PubMed]
- Goral, V. Pancreatic Cancer: Pathogenesis and Diagnosis. Asian Pac. J. Cancer Prev. 2015, 16, 5619–5624. [Google Scholar] [CrossRef] [PubMed]
- Vincent, A.; Herman, J.; Schulick, R.; Hruban, R.H.; Goggins, M. Pancreatic Cancer. Lancet 2011, 378, 607–620. [Google Scholar] [CrossRef] [PubMed]
- Sun, J. Pancreatic Neuroendocrine Tumors. Intractable Rare Dis. Res. 2017, 6, 21–28. [Google Scholar] [CrossRef]
- Maharjan, C.K.; Ear, P.H.; Tran, C.G.; Howe, J.R.; Chandrasekharan, C.; Quelle, D.E. Pancreatic Neuroendocrine Tumors: Molecular Mechanisms and Therapeutic Targets. Cancers 2021, 13, 5117. [Google Scholar] [CrossRef]
- Thomas, D.; Radhakrishnan, P. Tumor-Stromal Crosstalk in Pancreatic Cancer and Tissue Fibrosis. Mol. Cancer 2019, 18, 14. [Google Scholar] [CrossRef]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual Microdissection Identifies Distinct Tumor- and Stroma-Specific Subtypes of Pancreatic Ductal Adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of Pancreatic Ductal Adenocarcinoma and Their Differing Responses to Therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.M.; Gingras, M.C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic Analyses Identify Molecular Subtypes of Pancreatic Cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Puleo, F.; Nicolle, R.; Blum, Y.; Cros, J.; Marisa, L.; Demetter, P.; Quertinmont, E.; Svrcek, M.; Elarouci, N.; Iovanna, J.; et al. Stratification of Pancreatic Ductal Adenocarcinomas Based on Tumor and Microenvironment Features. Gastroenterology 2018, 155, 1999–2013.e3. [Google Scholar] [CrossRef] [PubMed]
- Collisson, E.A.; Bailey, P.; Chang, D.K.; Biankin, A.V. Molecular Subtypes of Pancreatic Cancer. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 207–220. [Google Scholar] [CrossRef] [PubMed]
- Grant, T.J.; Hua, K.; Singh, A. Molecular Pathogenesis of Pancreatic Cancer. In Progress in Molecular Biology and Translational Science; Elsevier B.V.: Amsterdam, The Netherlands, 2016; Volume 144, pp. 241–275. [Google Scholar] [CrossRef]
- Zhang, L.; Sanagapalli, S.; Stoita, A. Challenges in Diagnosis of Pancreatic Cancer. World J. Gastroenterol. 2018, 24, 2047. [Google Scholar] [CrossRef] [PubMed]
- Sarantis, P.; Koustas, E.; Papadimitropoulou, A.; Papavassiliou, A.G.; Karamouzis, M.V. Pancreatic Ductal Adenocarcinoma: Treatment Hurdles, Tumor Microenvironment and Immunotherapy. World J. Gastrointest. Oncol. 2020, 12, 173. [Google Scholar] [CrossRef]
- Ballehaninna, U.K.; Chamberlain, R.S. Biomarkers for Pancreatic Cancer: Promising New Markers and Options beyond CA 19-9. Tumor Biol. 2013, 34, 3279–3292. [Google Scholar] [CrossRef]
- Winter, J.M.; Yeo, C.J.; Brody, J.R. Diagnostic, Prognostic, and Predictive Biomarkers in Pancreatic Cancer. J. Surg. Oncol. 2013, 107, 15–22. [Google Scholar] [CrossRef]
- Daoud, A.Z.; Mulholland, E.J.; Cole, G.; McCarthy, H.O. MicroRNAs in Pancreatic Cancer: Biomarkers, Prognostic, and Therapeutic Modulators. BMC Cancer 2019, 19, 1130. [Google Scholar] [CrossRef]
- Mannarapu, M.; Dariya, B.; Bandapalli, O.R. Application of Single-Cell Sequencing Technologies in Pancreatic Cancer. Mol. Cell. Biochem. 2021, 476, 2429. [Google Scholar] [CrossRef]
- Gawad, C.; Koh, W.; Quake, S.R. Single-Cell Genome Sequencing: Current State of the Science. Nat. Rev. Genet. 2016, 17, 175–188. [Google Scholar] [CrossRef]
- Heath, J.R.; Ribas, A.; Mischel, P.S. Single Cell Analytic Tools for Drug Discovery and Development. Nat. Rev. Drug Discov. 2016, 15, 204. [Google Scholar] [CrossRef] [PubMed]
- Rantalainen, M. Application of Single-Cell Sequencing in Human Cancer. Brief. Funct. Genom. 2018, 17, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Hodne, K.; Weltzien, F.A. Single-Cell Isolation and Gene Analysis: Pitfalls and Possibilities. Int. J. Mol. Sci. 2015, 16, 25996. [Google Scholar] [CrossRef]
- Aslam, B.; Basit, M.; Atif Nisar, M.; Khurshid, M.; Rasool, M.H. Proteomics: Technologies and Their Applications. J. Chromatogr. Sci. 2017, 55, 182–196. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.; Schoendube, J.; Zimmermann, S.; Steeb, M.; Zengerle, R.; Koltay, P. Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 2015, 16, 16897–16919. [Google Scholar] [CrossRef] [PubMed]
- Mazutis, L.; Gilbert, J.; Ung, W.L.; Weitz, D.A.; Griffiths, A.D.; Heyman, J.A. Single-Cell Analysis and Sorting Using Droplet-Based Microfluidics. Nat. Protoc. 2013, 8, 870. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Ye, H.; Hu, Z. Microfluidic System for Cell Sorting. J. Phys. Conf. Ser. 2021, 2012, 12129. [Google Scholar] [CrossRef]
- Jayasinghe, S.N. Reimagining Flow Cytometric Cell Sorting. Adv. Biosyst. 2020, 4, 2000019. [Google Scholar] [CrossRef]
- Lei, Y.; Tang, R.; Xu, J.; Wang, W.; Zhang, B.; Liu, J.; Yu, X.; Shi, S. Applications of Single-Cell Sequencing in Cancer Research: Progress and Perspectives. J. Hematol. Oncol. 2021, 14, 91. [Google Scholar] [CrossRef]
- Fuller, A.P.; Palmer-Toy, D.; Erlander, M.G.; Sgroi, D.C. Laser Capture Microdissection and Advanced Molecular Analysis of Human Breast Cancer. J. Mammary Gland. Biol. Neoplasia 2003, 8, 335–345. [Google Scholar] [CrossRef]
- Emmert-Buck, M.R.; Bonner, R.F.; Smith, P.D.; Chuaqui, R.F.; Zhuang, Z.; Goldstein, S.R.; Weiss, R.A.; Liotta, L.A.; Emmert-Buck, M.R.; Chuaqui, R.F.; et al. Laser Capture Microdissection. Science 1996, 274, 998–1001. [Google Scholar] [CrossRef] [PubMed]
- Chung, S.H.; Shen, W. Laser Capture Microdissection: From Its Principle to Applications in Research on Neurodegeneration. Neural Regen. Res. 2015, 10, 897. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, D.J.; Begg, S.K.S.; Finetti, P.; Vanderburg, C.; Kulkarni, A.S.; Neyaz, A.; Hank, T.; Tai, E.; Deshpande, V.; Bertucci, F.; et al. Transcriptomic Analysis of Laser Capture Microdissected Tumors Reveals Cancer- and Stromal-Specific Molecular Subtypes of Pancreatic Ductal Adenocarcinoma. Clin. Cancer Res. 2021, 27, 2314–2325. [Google Scholar] [CrossRef] [PubMed]
- Espina, V.; Heiby, M.; Pierobon, M.; Liotta, L.A. Laser Capture Microdissection Technology. Expert Rev. Mol. Diagn. 2007, 7, 647–657. [Google Scholar] [CrossRef] [PubMed]
- Microdissection from Carl Zeiss; LCM User Protocols. LCM Laboratories. Available online: https://www.biotech.cornell.edu/sites/default/files/2020-06/Zeiss%20LCM%20Cell%20culture.pdf (accessed on 13 August 2022).
- Maurer, H.C.; Olive, K.P. Laser Capture Microdissection on Frozen Sections for Extraction of High-Quality Nucleic Acids. In Methods in Molecular Biology; Humana Press Inc.: Totowa, NJ, USA, 2019; Volume 1882, pp. 253–259. [Google Scholar] [CrossRef]
- Wyatt Shields Iv, C.; Reyes, C.D.; López, G.P. Microfluidic Cell Sorting: A Review of the Advances in the Separation of Cells from Debulking to Rare Cell Isolation. Lab A Chip 2015, 15, 1230. [Google Scholar] [CrossRef]
- Adan, A.; Alizada, G.; Kiraz, Y.; Baran, Y.; Nalbant, A. Flow Cytometry: Basic Principles and Applications. Crit. Rev. Biotechnol. 2017, 37, 163–176. [Google Scholar] [CrossRef]
- Lawrie, L.C.; Curran, S. Laser Capture Microdissection and Colorectal Cancer Proteomics. Methods Mol. Biol. 2005, 293, 245–253. [Google Scholar] [CrossRef][Green Version]
- de Preter, K.; Vandesompele, J.; Heimann, P.; Kockx, M.M.; van Gele, M.; Hoebeeck, J.; de Smet, E.; Demarche, M.; Laureys, G.; van Roy, N.; et al. Application of Laser Capture Microdissection in Genetic Analysis of Neuroblastoma and Neuroblastoma Precursor Cells. Cancer Lett. 2003, 197, 53–61. [Google Scholar] [CrossRef]
- Rubin, M.A. Use of Laser Capture Microdissection, CDNA Microarrays, and Tissue Microarrays in Advancing Our Understanding of Prostate Cancer. J. Pathol. 2001, 195, 80–86. [Google Scholar] [CrossRef]
- Thennavan, A.; Sharma, M.; Chandrashekar, C.; Hunter, K.; Radhakrishnan, R. Exploring the Potential of Laser Capture Microdissection Technology in Integrated Oral Biosciences. Oral Dis. 2017, 23, 737–748. [Google Scholar] [CrossRef]
- Cheng, L.; Mann, S.A.; Lopez-Beltran, A.; Chovanec, M.; Santoni, M.; Wang, M.; Albany, C.; Adra, N.; Davidson, D.D.; Cimadamore, A.; et al. Molecular Characterization of Testicular Germ Cell Tumors Using Tissue Microdissection. Methods Mol. Biol. 2021, 2195, 31–47. [Google Scholar] [CrossRef] [PubMed]
- Liotta, L.A.; Pappalardo, P.A.; Carpino, A.; Haymond, A.; Howard, M.; Espina, V.; Wulfkuhle, J.; Petricoin, E. Laser Capture Proteomics: Spatial Tissue Molecular Profiling from the Bench to Personalized Medicine. Expert Rev. Proteom. 2021, 18, 845–861. [Google Scholar] [CrossRef] [PubMed]
- von Eggeling, F.; Hoffmann, F. Microdissection-An Essential Prerequisite for Spatial Cancer Omics. Proteomics 2020, 20, 2000077. [Google Scholar] [CrossRef] [PubMed]
- Fend, F.; Raffeld, M. Laser Capture Microdissection in Pathology. J. Clin. Pathol. 2000, 53, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Sato, N.; Matsubayashi, H.; Abe, T.; Fukushima, N.; Goggins, M. Epigenetic Down-Regulation of CDKN1C/P57KIP2 in Pancreatic Ductal Neoplasms Identified by Gene Expression Profiling. Clin. Cancer Res. 2005, 11, 4681–4688. [Google Scholar] [CrossRef]
- Matthaei, H.; Norris, A.L.; Tsiatis, A.C.; Olino, K.; Hong, S.M.; Dal Molin, M.; Goggins, M.G.; Canto, M.; Horton, K.M.; Jackson, K.D.; et al. Clinicopathological Characteristics and Molecular Analyses of Multifocal Intraductal Papillary Mucinous Neoplasms of the Pancreas. Ann. Surg. 2012, 255, 326. [Google Scholar] [CrossRef]
- Yonezawa, S.; Higashi, M.; Yamada, N.; Goto, M. Precursor Lesions of Pancreatic Cancer. Gut Liver 2008, 2, 137. [Google Scholar] [CrossRef]
- Garcia-Carracedo, D.; Chen, Z.M.; Qiu, W.; Huang, A.S.; Tang, S.M.; Hruban, R.H.; Su, G.H. PIK3CA Mutations in Mucinous Cystic Neoplasms of the Pancreas. Pancreas 2014, 43, 245. [Google Scholar] [CrossRef][Green Version]
- Crnogorac-Jurcevic, T.; Efthimiou, E.; Nielsen, T.; Loader, J.; Terris, B.; Stamp, G.; Baron, A.; Scarpa, A.; Lemoine, N.R. Expression Profiling of Microdissected Pancreatic Adenocarcinomas. Oncogene 2002, 21, 4587–4594. [Google Scholar] [CrossRef]
- Liu, T.; Wei, S.; Liang, Z.; Gao, J.; Wu, S.; Zhu, H.; Liu, H. Patterns of K-Ras Codon 12 and 13 Mutations Found in Pancreatic Adenocarcinoma of 30 Chinese Patients by Microdissection, PCR and Direct Sequencing. J. Gastroenterol. Hepatol. 2005, 20, 67–72. [Google Scholar] [CrossRef]
- Soliman, A.S.; Lo, A.-C.; Banerjee, M.; El-Ghawalby, N.; Khaled, H.M.; Bayoumi, S.; Seifeldin, I.A.; Abdel-Aziz, A.; Abbruzzese, J.L.; Greenson, J.K.; et al. Differences in K-Ras and P53 Gene Mutations among Pancreatic Adenocarcinomas Associated with Regional Environmental Pollution. Carcinogenesis 2007, 28, 1794–1799. [Google Scholar] [CrossRef] [PubMed]
- Izawa, T.; Obara, T.; Tanno, S.; Mizukami, Y.; Yanagawa, N.; Kohgo, Y. Clonality and Field Cancerization in Intraductal Papillary-Mucinous Tumors of the Pancreas. Cancer Interdiscip. Int. J. Am. Cancer Soc. 2001, 92, 1807–1817. [Google Scholar] [CrossRef]
- Guyton, A.C.; Halls, J.E. Textbook of Medical Physiology; Elsevier B.V.: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Shi, C.; Hong, S.M.; Lim, P.; Kamiyama, H.; Khan, M.; Anders, R.A.; Goggins, M.; Hruban, R.H.; Eshleman, J.R. KRAS2 Mutations in Human Pancreatic Acinar-Ductal Metaplastic Lesions Are Limited to Those with PanIN: Implications for the Human Pancreatic Cancer Cell of Origin. Mol. Cancer Res. 2009, 7, 230–236. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.J.; Hart, S.N.; Lima, J.F.; Kipp, B.R.; Klebig, M.; Winters, J.L.; Szabo, C.; Zhang, L.; Eckloff, B.W.; Petersen, G.M.; et al. Genetic Alterations Associated with Progression from Pancreatic Intraepithelial Neoplasia to Invasive Pancreatic Tumor. Gastroenterology 2013, 145, 1098–1109.e1. [Google Scholar] [CrossRef]
- Baslan, T.; Morris, J.P.; Zhao, Z.; Reyes, J.; Ho, Y.J.; Tsanov, K.M.; Bermeo, J.; Tian, S.; Zhang, S.; Askan, G.; et al. Ordered and Deterministic Cancer Genome Evolution after P53 Loss. Nature 2022, 608, 795–802. [Google Scholar] [CrossRef]
- Fang, Y.; Su, Z.; Xie, J.; Xue, R.; Ma, Q.; Li, Y.; Zhao, Y.; Song, Z.; Lu, X.; Li, H.; et al. Genomic Signatures of Pancreatic Adenosquamous Carcinoma (PASC). J. Pathol. 2017, 243, 155–159. [Google Scholar] [CrossRef]
- Takano, A.; Hirotsu, Y.; Amemiya, K.; Nakagomi, H.; Naoki, O.I.; Toshio, O.; Hitoshi, M.; Masao, O. Genetic Basis of a Common Tumor Origin in the Development of Pancreatic Mixed Acinar-neuroendocrine-ductal Carcinoma: A Case Report. Oncol. Lett. 2017, 12, 4428–4432. [Google Scholar] [CrossRef][Green Version]
- Bai, Q.; Zhang, X.; Zhu, X.; Wang, L.; Huang, D.; Cai, X.; Zhou, X.; Wang, J.; Sheng, W. Pancreatic Carcinosarcoma with the Same KRAS Gene Mutation in Both Carcinomatous and Sarcomatous Components: Molecular Evidence for Monoclonal Origin of the Tumour. Histopathology 2016, 69, 393–405. [Google Scholar] [CrossRef]
- Qian, W.; Chen, K.; Qin, T.; Xiao, Y.; Li, J.; Yue, Y.; Zhou, C.; Ma, J.; Duan, W.; Lei, J.; et al. The EGFR-HSF1 Axis Accelerates the Tumorigenesis of Pancreatic Cancer. J. Exp. Clin. Cancer Res. 2021, 40, 25. [Google Scholar] [CrossRef]
- Tzeng, C.W.D.; Frolov, A.; Frolova, N.; Jhala, N.C.; Howard, J.H.; Buchsbaum, D.J.; Vickers, S.M.; Heslin, M.J.; Arnoletti, J.P. Epidermal Growth Factor Receptor (EGFR) Is Highly Conserved in Pancreatic Cancer. Surgery 2007, 141, 464–469. [Google Scholar] [CrossRef]
- Fujii, K.; Miyashita, K.; Yamada, Y.; Eguchi, T.; Taguchi, K.; Oda, Y.; Oda, S.; Yoshida, M.A.; Tanaka, M.; Tsuneyoshi, M. Simulation-Based Analyses Reveal Stable Microsatellite Sequences in Human Pancreatic Cancer. Cancer Genet. Cytogenet. 2009, 189, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Nadella, S.; Burks, J.; Huber, M.; Wang, J.; Cao, H.; Kallakury, B.; Tucker, R.D.; Boca, S.M.; Jermusyck, A.; Collins, I.; et al. Endogenous Gastrin Collaborates With Mutant KRAS in Pancreatic Carcinogenesis. Pancreas 2019, 48, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Kayed, H.; Org Kleeff, J.; Kolb, A.; Ketterer, K.; Keleg, S.; Felix, K.; Giese, T.; Penzel, R.; Zentgraf, H.; Büchler, M.W.; et al. FXYD3 Is Overexpressed in Pancreatic Ductal Adenocarcinoma and Influences Pancreatic Cancer Cell Growth. Int. J. Cancer 2005, 118, 43–54. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, S.; Nagano, H.; Konno, M.; Eguchi, H.; Tomokuni, A.; Tomimaru, Y.; Asaoka, T.; Wada, H.; Hama, N.; Kawamoto, K.; et al. A Crucial Epithelial to Mesenchymal Transition Regulator, Sox4/Ezh2 Axis Is Closely Related to the Clinical Outcome in Pancreatic Cancer Patients. Int. J. Oncol. 2016, 48, 145–152. [Google Scholar] [CrossRef] [PubMed]
- Nakahara, O.; Takamori, H.; Iwatsuki, M.; Baba, Y.; Sakamoto, Y.; Tanaka, H.; Chikamoto, A.; Horino, K.; Beppu, T.; Kanemitsu, K.; et al. Carcinogenesis of Intraductal Papillary Mucinous Neoplasm of the Pancreas: Loss of MicroRNA-101 Promotes Overexpression of Histone Methyltransferase EZH2. Ann. Surg. Oncol. 2012, 19, 565–571. [Google Scholar] [CrossRef]
- Kalloger, S.E.; Karasinska, J.M.; Keung, M.S.; Thompson, D.L.; Ho, J.; Chow, C.; Gao, D.; Topham, J.T.; Warren, C.; Wong, H.-L.; et al. Stroma vs Epithelium-Enhanced Prognostics through Histologic Stratification in Pancreatic Ductal Adenocarcinoma. Int. J. Cancer 2021, 148, 481–491. [Google Scholar] [CrossRef] [PubMed]
- Maurer, C.; Holmstrom, S.; He, J.; Laise, P.; Su, T.; Ahmed, A.; Hibshoosh, H.; Chabot, J.; Oberstein, P.; Sepulveda, A.; et al. Experimental Microdissection Enables Functional Harmonisation of Pancreatic Cancer Subtypes. Gut 2019, 68, 1034–1043. [Google Scholar] [CrossRef]
- Aung, K.L.; Fischer, S.E.; Denroche, R.E.; Jang, G.H.; Dodd, A.; Creighton, S.; Southwood, B.; Liang, S.B.; Chadwick, D.; Zhang, A.; et al. Genomics-Driven Precision Medicine for Advanced Pancreatic Cancer: Early Results from the COMPASS Trial. Clin. Cancer Res. 2018, 24, 1344. [Google Scholar] [CrossRef]
- Nakamura, T.; Kuwai, T.; Kitadai, Y.; Sasaki, T.; Fan, D.; Coombes, K.R.; Kim, S.-J.; Fidler, I.J. Zonal Heterogeneity for Gene Expression in Human Pancreatic Carcinoma. Cancer Res. 2007, 67, 7597–7604. [Google Scholar] [CrossRef]
- Aspinall-O’Dea, M.; Costello, E. The Pancreatic Cancer Proteome—Recent Advances and Future Promise. Proteom. Clin. Appl. 2007, 1, 1066–1079. [Google Scholar] [CrossRef]
- Abyadeh, M.; Meyfour, A.; Gupta, V.; Moghaddam, M.Z.; Fitzhenry, M.J.; Shahbazian, S.; Salekdeh, G.H.; Mirzaei, M. Molecular Sciences Recent Advances of Functional Proteomics in Gastrointestinal Cancers-a Path towards the Identification of Candidate Diagnostic, Prognostic, and Therapeutic Molecular Biomarkers. Int. J. Mol. Sci. 2020, 21, 8532. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Zhou, Q.; Guo, F.; Chen, M.; Tao, X.; Dong, D. S100 Proteins in Pancreatic Cancer: Current Knowledge and Future Perspectives. Front. Oncol. 2021, 11, 3429. [Google Scholar] [CrossRef] [PubMed]
- Kaleağasıoğlu, F.; Berger, M.R. SIBLINGs and SPARC Families: Their Emerging Roles in Pancreatic Cancer. World J. Gastroenterogy WJG 2014, 20, 14747. [Google Scholar] [CrossRef] [PubMed]
- Kayed, H.; Kleeff, J.; Keleg, S.; Felix, K.; Giese, T.; Berger, M.R.; Büchler, M.W.; Friess, H. Effects of Bone Sialoprotein on Pancreatic Cancer Cell Growth, Invasion and Metastasis. Cancer Lett. 2007, 245, 171–183. [Google Scholar] [CrossRef]
- Esposito, I.; Kayed, H.; Keleg, S.; Giese, T.; Sage, E.H.; Schirmacher, P.; Friess, H.; Kleeff, J. Tumor-Suppressor Function of SPARC-like Protein 1/Hevin in Pancreatic Cancer. Neoplasia 2007, 9, 8–17. [Google Scholar] [CrossRef]
- Shekouh, A.R.; Thompson, C.C.; Prime, W.; Campbell, F.; Hamlett, J.; Simon Herrington, C.; Lemoine, N.R.; Crnogorac-Jurcevic, T.; Buechler, M.W.; Friess, H.; et al. Application of Laser Capture Microdissectioncombined with Two-Dimensional Electrophoresis for the Discovery of Differentially Regulated Proteins in Pancreatic Ductal Adenocarcinoma. Proteom. Syst. Biol. 2003, 3, 1988–2001. [Google Scholar] [CrossRef]
- Sitek, B.; Lüttges, J.; Marcus, K.; Klöppel, G.; Schmiegel, W.; Meyer, H.E.; Hahn, S.A.; Stühler, K. Application of Fluorescence Difference Gel Electrophoresis Saturation Labelling for the Analysis of Microdissected Precursor Lesions of Pancreatic Ductal Adenocarcinoma. Proteomics 2005, 5, 2665–2679. [Google Scholar] [CrossRef]
- Library, W.O.; Naidoo, K.; Jones, R.; Dmitrovic, B.; Wijesuriya, N.; Kocher, H.; Hart, I.R.; Crnogorac-Jurcevic, T. Proteome of Formalin-Fixed Paraffin-Embedded Pancreatic Ductal Adenocarcinoma and Lymph Node Metastases. J. Pathol. 2012, 226, 756–763. [Google Scholar] [CrossRef]
- Robin, F.; Angenard, G.; Cano, L.; Courtin-Tanguy, L.; Gaignard, E.; Khene, Z.E.; Bergeat, D.; Clément, B.; Boudjema, K.; Coulouarn, C.; et al. Molecular Profiling of Stroma Highlights Stratifin as a Novel Biomarker of Poor Prognosis in Pancreatic Ductal Adenocarcinoma. Br. J. Cancer 2020, 123, 72. [Google Scholar] [CrossRef]
- Guweidhi, A.; Kleeff, J.; Giese, N.; el Fitori, J.; Ketterer, K.; Giese, T.; Büchler, M.W.; Korc, M.; Friess, H. Enhanced Expression of 14-3-3sigma in Pancreatic Cancer and Its Role in Cell Cycle Regulation and Apoptosis. Carcinogenesis 2004, 25, 1575–1585. [Google Scholar] [CrossRef]
- Fukushima, N.; Koopmann, J.; Sato, N.; Prasad, N.; Carvalho, R.; Leach, S.D.; Hruban, R.H.; Goggins, M. Gene Expression Alterations in the Non-Neoplastic Parenchyma Adjacent to Infiltrating Pancreatic Ductal Adenocarcinoma. Mod. Pathol. 2005, 18, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Sawai, Y.; Kodama, Y.; Shimizu, T.; Ota, Y.; Maruno, T.; Eso, Y.; Kurita, A.; Shiokawa, M.; Tsuji, Y.; Uza, N.; et al. Molecular and Cellular Pathobiology Activation-Induced Cytidine Deaminase Contributes to Pancreatic Tumorigenesis by Inducing Tumor-Related Gene Mutations. Cancer Res. 2015, 75, 3292–3301. [Google Scholar] [CrossRef] [PubMed]
- Kubo, T.; Kuroda, Y.; Kokubu, A.; Hosoda, F.; Arai, Y.; Hiraoka, N.; Hirohashi, S.; Shibata, T. Resequencing Analysis of the Human Tyrosine Kinase Gene Family in Pancreatic Cancer. Pancreas 2009, 38, e200–e206. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Nie, S.; Wu, J.; Lubman, D.M. Target Proteomic Profiling of Frozen Pancreatic CD24+ Adenocarcinoma Tissues by Immuno-Laser Capture Microdissection and Nano-LC-MS/MS. J. Proteome Res. 2013, 12, 2791. [Google Scholar] [CrossRef] [PubMed]
- Shan, T.; Lu, H.; Ji, H.; Li, Y.; Guo, J.; Chen, X.; Wu, T. Loss of Stromal Caveolin-1 Expression: A Novel Tumor Microenvironment Biomarker That Can Predict Poor Clinical Outcomes for Pancreatic Cancer. PLoS ONE 2014, 9, e97239. [Google Scholar] [CrossRef] [PubMed]
- Court, C.M.; Ankeny, J.S.; Sho, S.; Hou, S.; Li, Q.; Hsieh, C.; Song, M.; Liao, X.; Rochefort, M.M.; Wainberg, Z.A.; et al. Reality of Single Circulating Tumor Cell Sequencing for Molecular Diagnostics in Pancreatic Cancer. J. Mol. Diagn. 2016, 18, 688. [Google Scholar] [CrossRef]
- Ma, L.; Tian, X.; Wang, F.; Zhang, Z.; Du, C.; Xie, X.; Kornmann, M.; Yang, Y. The Long Noncoding RNA H19 Promotes Cell Proliferation via E2F-1 in Pancreatic Ductal Adenocarcinoma. Cancer Biol. Ther. 2016, 17, 1051–1061. [Google Scholar] [CrossRef]
- Fu, Z.; Chen, C.; Zhou, Q.; Wang, Y.; Zhao, Y.; Zhao, X.; Li, W.; Zheng, S.; Ye, H.; Wang, L.; et al. LncRNA HOTTIP Modulates Cancer Stem Cell Properties in Human Pancreatic Cancer by Regulating HOXA9. Cancer Lett. 2017, 410, 68–81. [Google Scholar] [CrossRef]
- Hiroshima, Y.; Kasajima, R.; Kimura, Y.; Komura, D.; Ishikawa, S.; Ichikawa, Y.; Bouvet, M.; Yamamoto, N.; Oshima, T.; Morinaga, S.; et al. Novel Targets Identified by Integrated Cancer-Stromal Interactome Analysis of Pancreatic Adenocarcinoma. Cancer Lett. 2020, 469, 217–227. [Google Scholar] [CrossRef]
- Erkan, M.; Kleeff, J.; Esposito, I.; Giese, T.; Ketterer, K.; Büchler, M.W.; Giese, N.A.; Friess, H. Loss of BNIP3 Expression Is a Late Event in Pancreatic Cancer Contributing to Chemoresistance and Worsened Prognosis. Oncogene 2005, 24, 4421–4432. [Google Scholar] [CrossRef]
- Hwang, T.L.; Liang, Y.; Chien, K.Y.; Yu, J.S. Overexpression and Elevated Serum Levels of Phosphoglycerate Kinase 1 in Pancreatic Ductal Adenocarcinoma. Proteomics 2006, 6, 2259–2272. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.C.; Mori, R.; Vallbohmer, D.; Brabender, J.; Klein, E.; Drebber, U.; Baldus, S.E.; Cooc, J.; Azuma, M.; Metzger, R.; et al. High Expression of HIF1a Is a Predictor of Clinical Outcome in Patients with Pancreatic Ductal Adenocarcinomas and Correlated to PDGFA, VEGF, and BFGF. Neoplasia 2008, 10, 674–679. [Google Scholar] [CrossRef] [PubMed]
- Kayashima, T.; Nakata, K.; Ohuchida, K.; Ueda, J.; Shirahane, K.; Fujita, H.; Cui, L.; Mizumoto, K.; Tanaka, M. Insig2 Is Overexpressed in Pancreatic Cancer and Its Expression Is Induced by Hypoxia. Jpn. Cancer Assoc. 2011, 102, 1137–1143. [Google Scholar] [CrossRef] [PubMed]
- Tao, J.; Yang, G.; Zhou, W.; Qiu, J.; Chen, G.; Luo, W.; Zhao, F.; You, L.; Zheng, L.; Zhang, T.; et al. Targeting Hypoxic Tumor Microenvironment in Pancreatic Cancer. J. Hematol. Oncol. 2021, 14, 14. [Google Scholar] [CrossRef]
- Couvelard, A.; O’Toole, D.; Leek, R.; Turley, H.; Sauvanet, A.; Degott, C.; Ruszniewski, P.; Belghiti, J.; Harris, A.L.; Gatter, K.; et al. Expression of Hypoxia-Inducible Factors Is Correlated with the Presence of a Fibrotic Focus and Angiogenesis in Pancreatic Ductal Adenocarcinomas. Histopathology 2005, 46, 668–676. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rao, B.H.; Souček, P.; Hlaváč, V. Laser Capture Microdissection: A Gear for Pancreatic Cancer Research. Int. J. Mol. Sci. 2022, 23, 14566. https://doi.org/10.3390/ijms232314566
Rao BH, Souček P, Hlaváč V. Laser Capture Microdissection: A Gear for Pancreatic Cancer Research. International Journal of Molecular Sciences. 2022; 23(23):14566. https://doi.org/10.3390/ijms232314566
Chicago/Turabian StyleRao, Bhavana Hemantha, Pavel Souček, and Viktor Hlaváč. 2022. "Laser Capture Microdissection: A Gear for Pancreatic Cancer Research" International Journal of Molecular Sciences 23, no. 23: 14566. https://doi.org/10.3390/ijms232314566
APA StyleRao, B. H., Souček, P., & Hlaváč, V. (2022). Laser Capture Microdissection: A Gear for Pancreatic Cancer Research. International Journal of Molecular Sciences, 23(23), 14566. https://doi.org/10.3390/ijms232314566








