Natural Inhibitors Targeting the Localization of Lipoprotein System in Vibrio parahaemolyticus
Abstract
1. Introduction
2. Results
2.1. Virtual Screening of Natural Compound Database
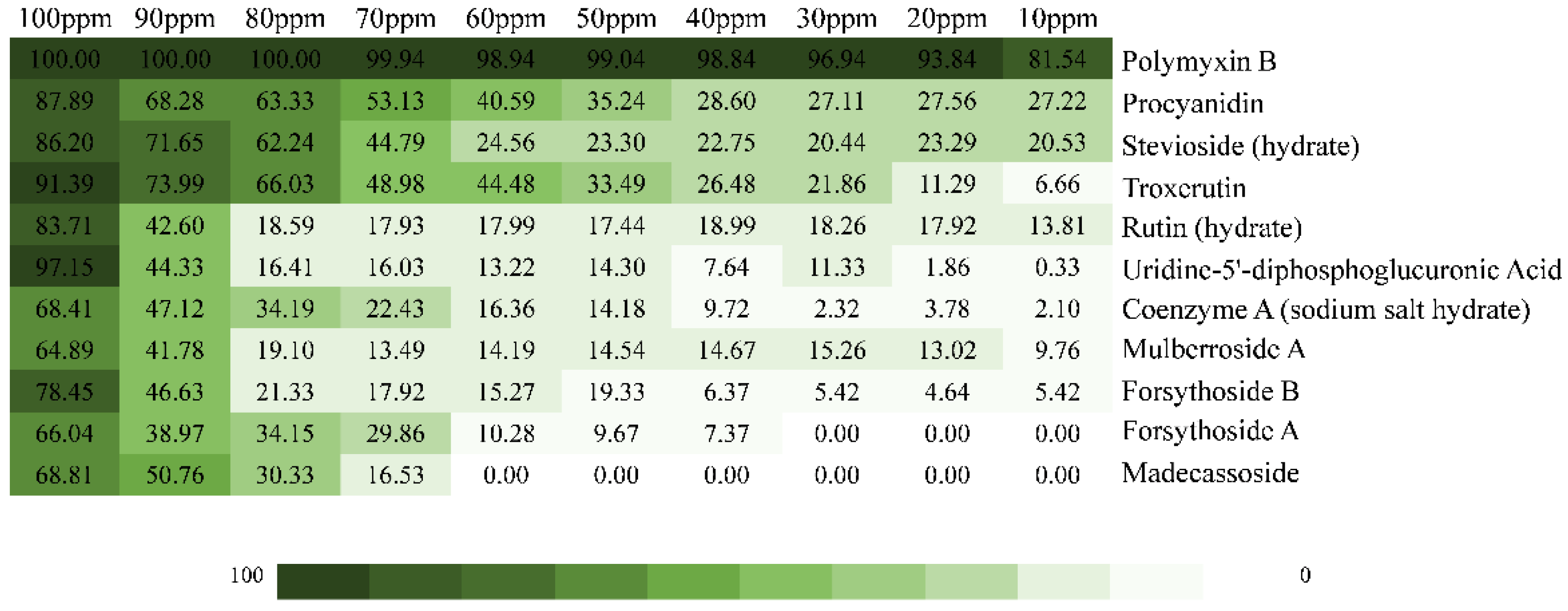
2.2. Inhibition Effect of the Top 10 Compounds on V. parahaemolyticus
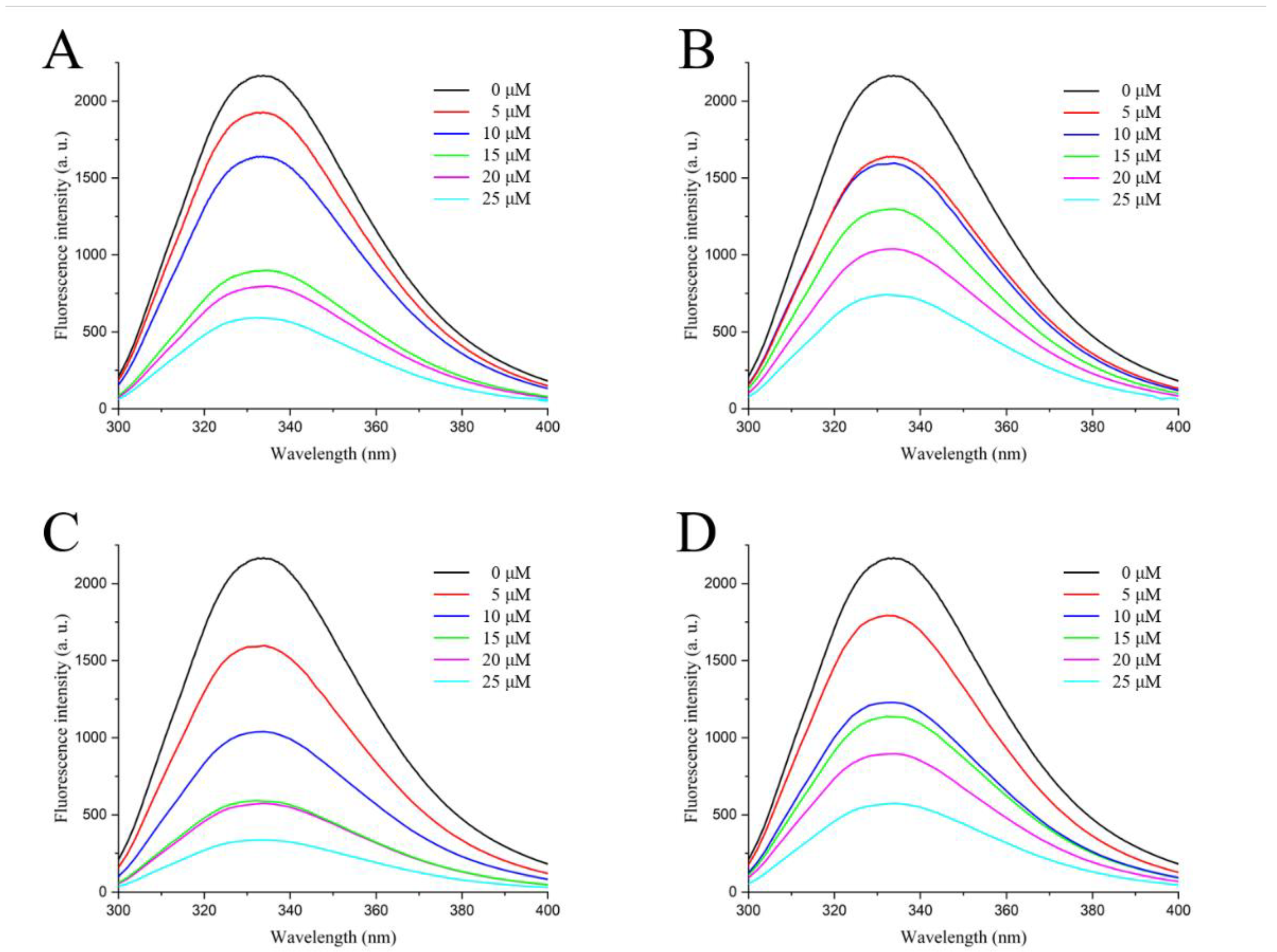
2.3. Fluorescence Spectroscopy of Active Compounds with LolB
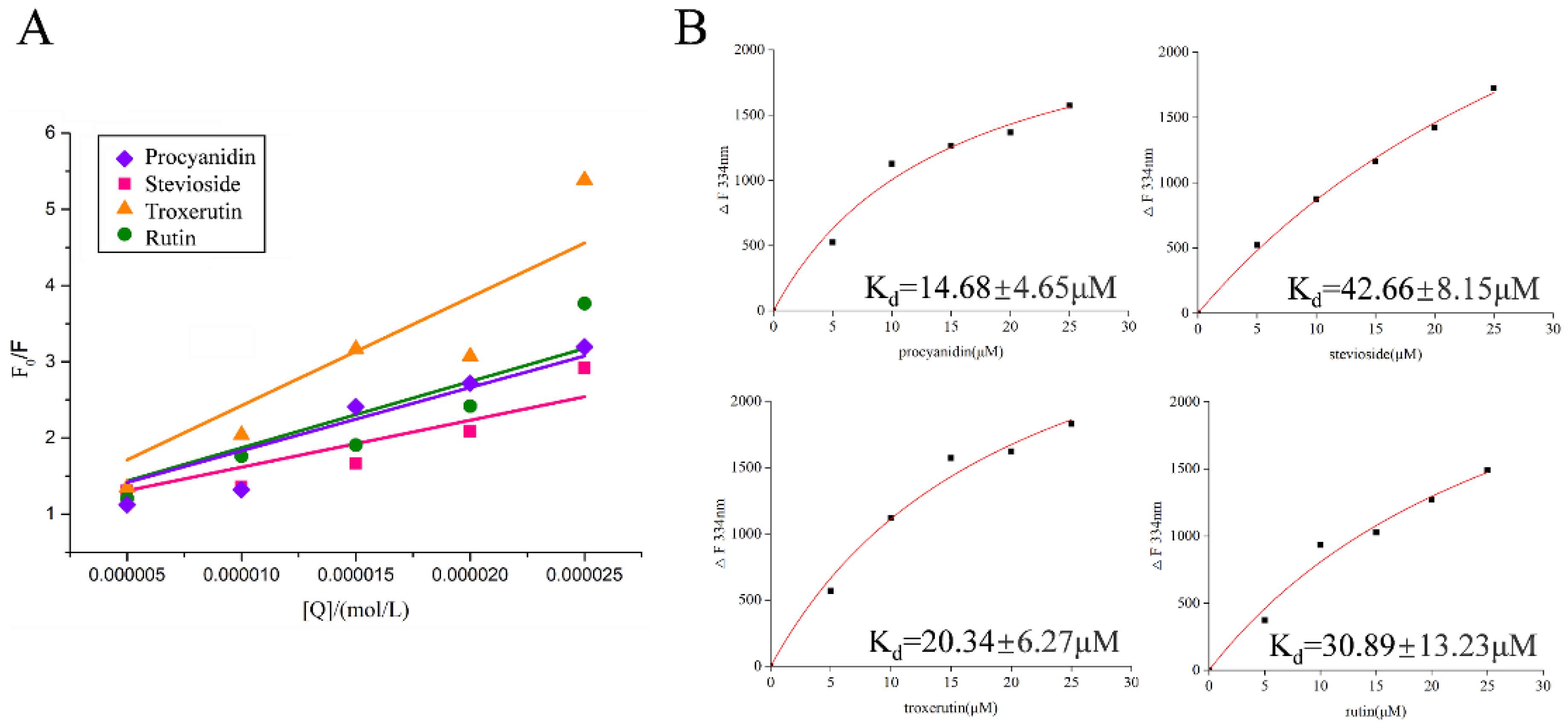
2.4. Fluorescence Quenching Mechanism of Active Compounds on LolB
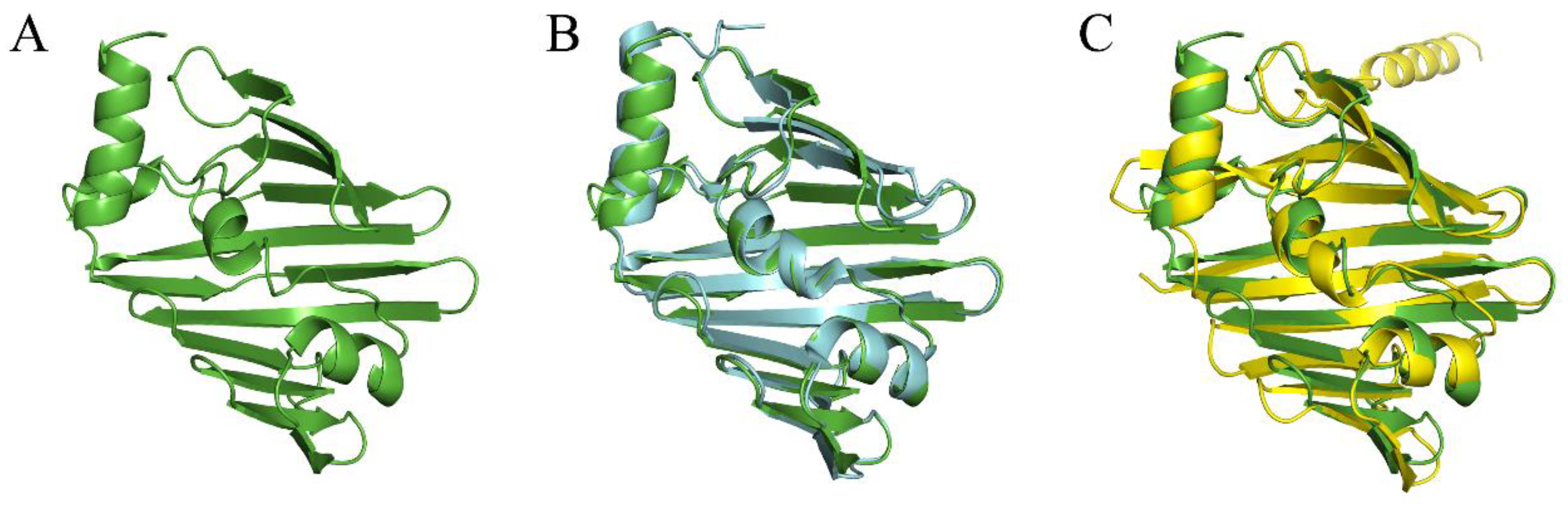
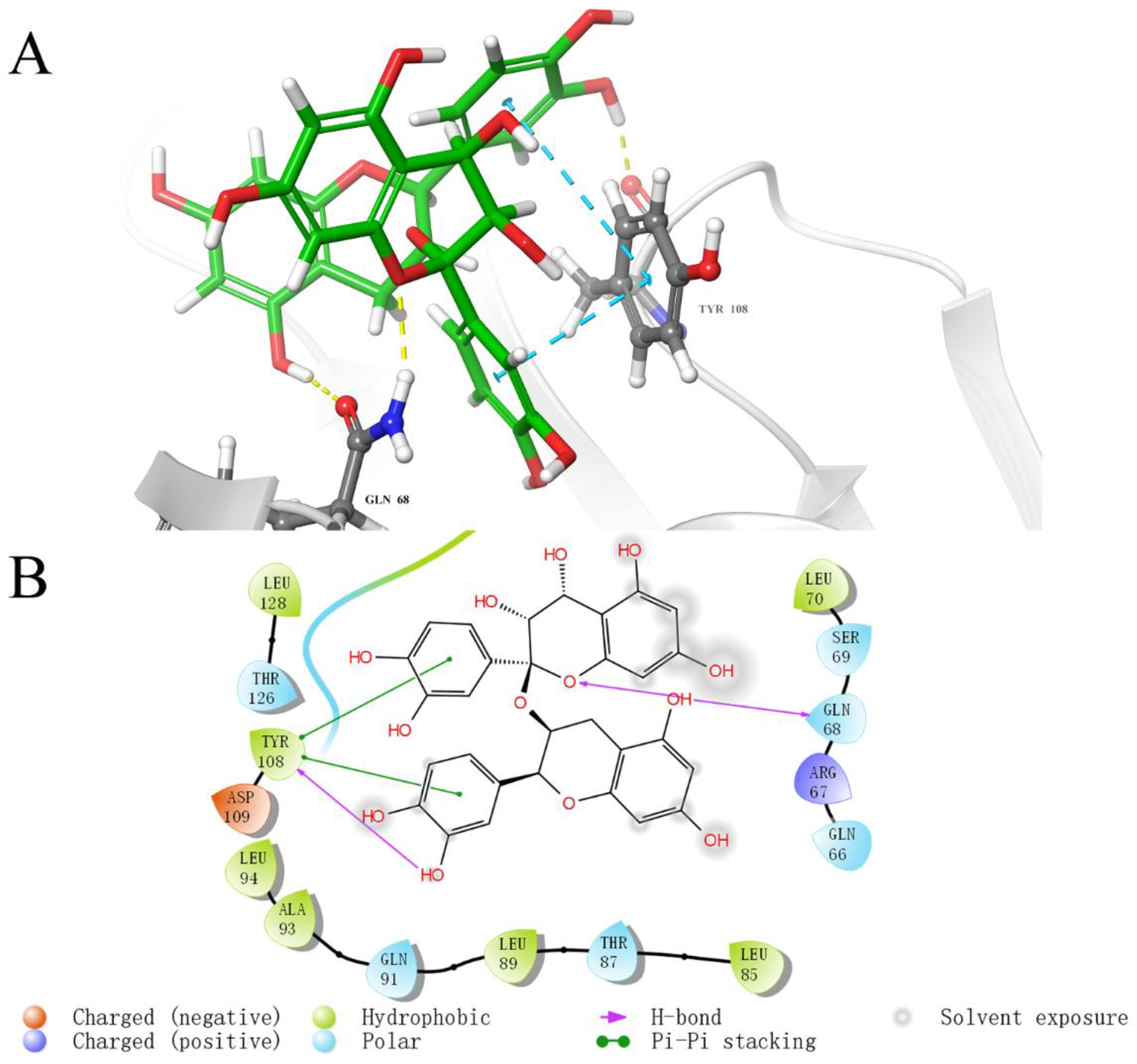
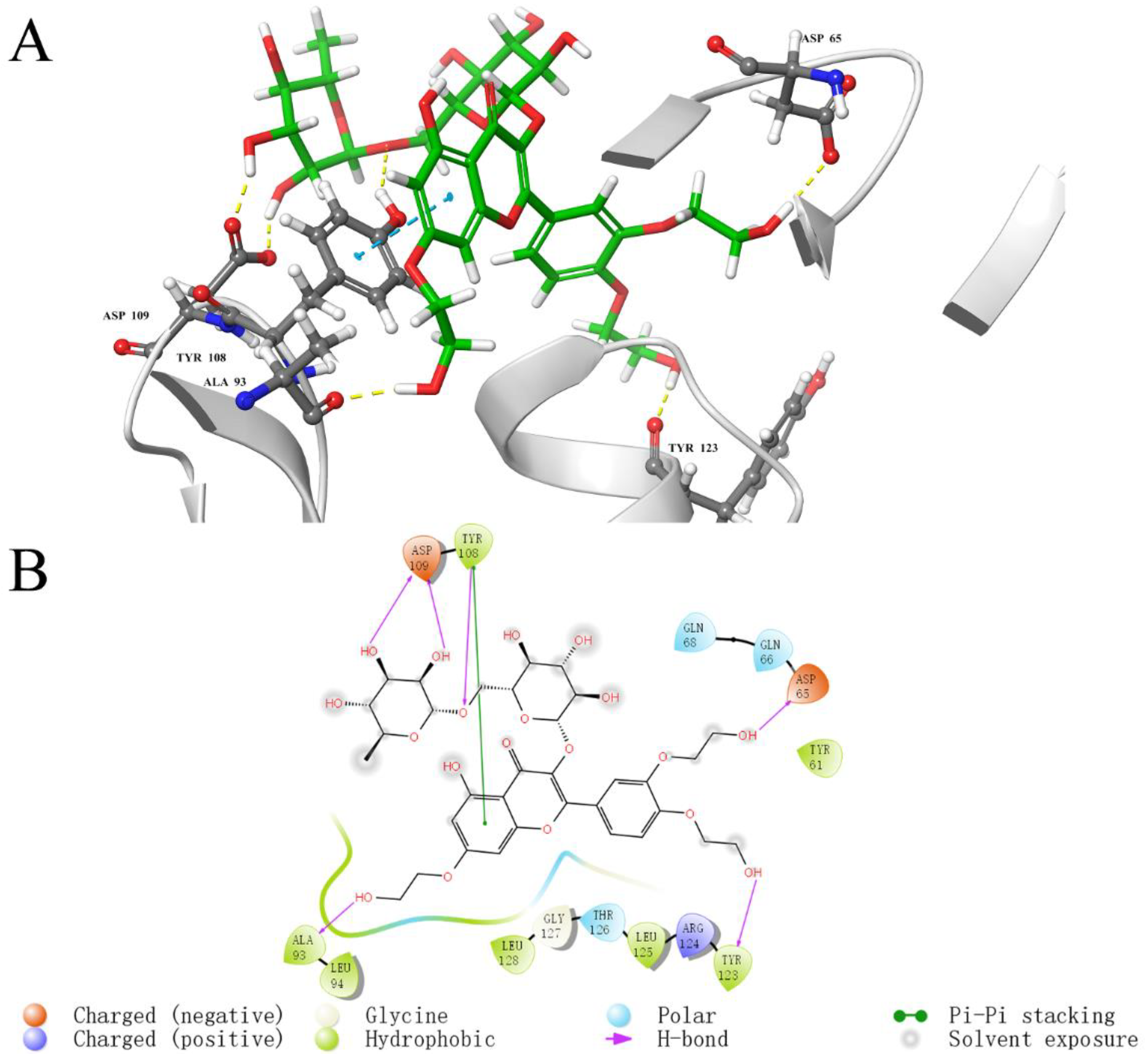
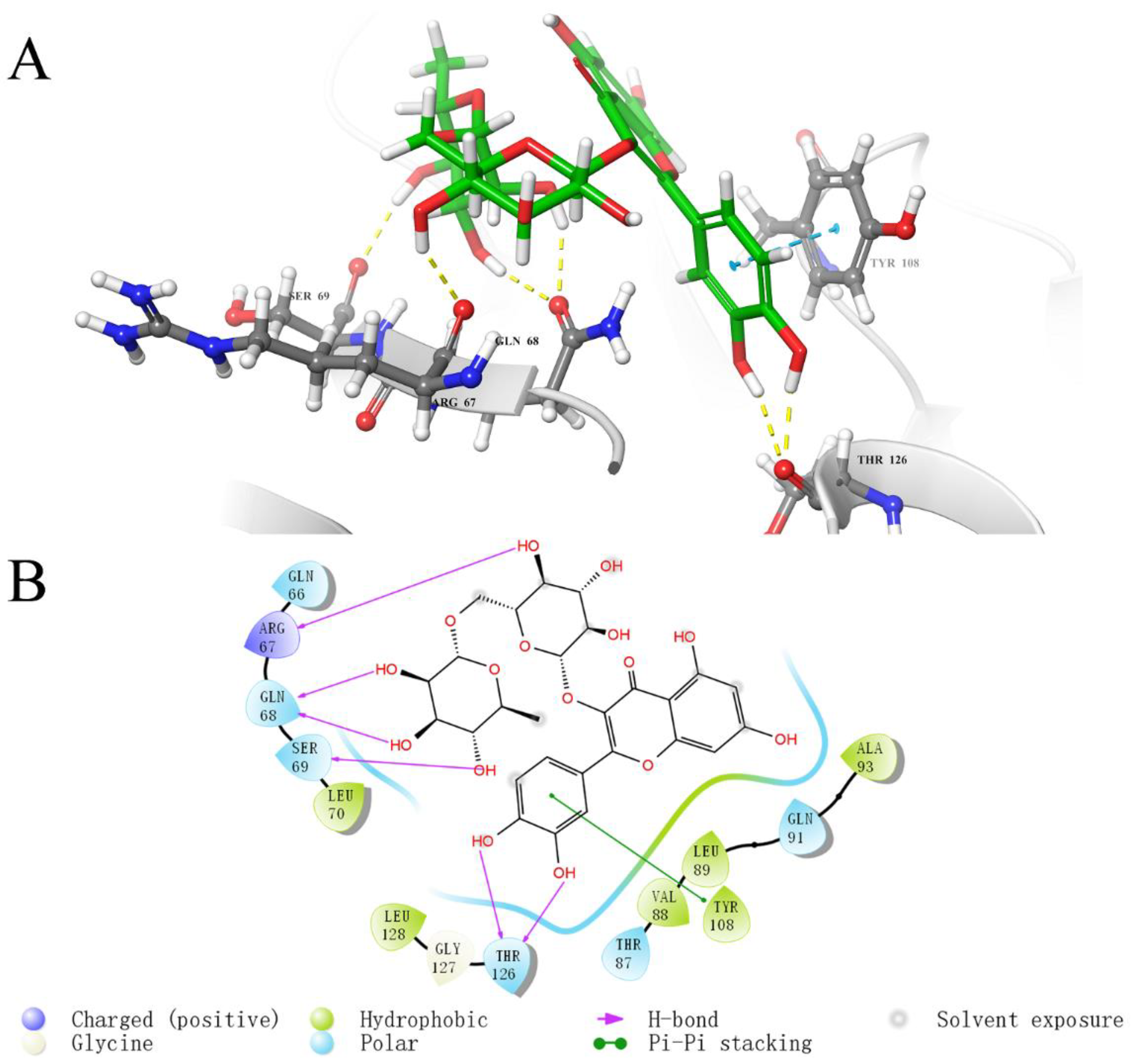
2.5. Analysis of Binding Sites between Active Compounds and LolB
3. Discussion
4. Materials and Methods
4.1. Homology Modeling
4.2. High-Throughput Virtual Screening
4.3. Determination of Inhibition on V. parahaemolyticus
4.4. Cloning, Expression and Purification of V. parahaemolyticus LolB Protein
4.4.1. Cloning of V. parahaemolyticus LolB
4.4.2. Expression and Purification of LolB Protein
4.5. Fluorescence Spectroscopy
4.6. Identification of Fluorescence Quenching Mechanism
4.7. Analysis of Molecular Docking
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Baker-Austin, C.; Oliver, J.D.; Alam, M.; Ali, A.; Waldor, M.K.; Qadri, F.; Martinez-Urtaza, J. Vibrio spp. infections. Nat. Rev. Dis. Prim. 2018, 4, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.-T.; Chen, I.-T.; Yang, Y.-T.; Ko, T.-P.; Huang, Y.-T.; Huang, J.-Y.; Huang, M.-F.; Lin, S.-J.; Chen, C.-Y.; Lin, S.-S.; et al. The opportunistic marine pathogen Vibrio parahaemolyticus becomes virulent by acquiring a plasmid that expresses a deadly toxin. Proc. Natl. Acad. Sci. USA 2015, 112, 10798–10803. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Kinch, L.N.; Ray, A.; Dalia, A.B.; Cong, Q.; Nunan, L.M.; Camilli, A.; Grishin, N.V.; Salomon, D.; Orth, K. Acute Hepatopancreatic Necrosis Disease-Causing Vibrio parahaemolyticus Strains Maintain an Antibacterial Type VI Secretion System with Versatile Effector Repertoires. Appl. Environ. Microbiol. 2017, 83, e00737-17. [Google Scholar] [CrossRef] [PubMed]
- Letchumanan, V.; Chan, K.G.; Lee, L.-H. Vibrio parahaemolyticus: A review on the pathogenesis, prevalence, and advance molecular identification techniques. Front. Microbiol. 2014, 5, 705. [Google Scholar] [CrossRef] [PubMed]
- Mullard, A.; O’Neill, J. An audience with… Jim O’Neill. Nat. Rev. Drug Discov. 2016, 15, 526. [Google Scholar] [CrossRef]
- Okuda, S.; Tokuda, H. Lipoprotein Sorting in Bacteria. In Annual Review of Microbiology; Gottesman, S., Harwood, C.S., Eds.; Annual Reviews: San Mateo, CA, USA, 2011; Volume 65, pp. 239–259. [Google Scholar]
- Kovacs-Simon, A.; Titball, R.W.; Michell, S.L. Lipoproteins of Bacterial Pathogens. Infect. Immun. 2011, 79, 548–561. [Google Scholar] [CrossRef]
- Konovalova, A.; Kahne, D.E.; Silhavy, T.J. Outer Membrane Biogenesis. In Annual Review of Microbiology; Gottesman, S., Ed.; Annual Reviews: San Mateo, CA, USA, 2017; Volume 71, pp. 539–556. [Google Scholar]
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Tong, J.R.; Zhang, Z.H.; Huang, Z.H.; Zhang, X.; Shi, J.; Liu, H.Q.; Pan, Y.J.; Zhao, Y. Research Progress on Transmembrane Transport Mechanisms for the Main Components of the Outer Membrane in Gram-negative Bacteria. Prog. Biochem. Biophys. 2020, 47, 510–522. [Google Scholar] [CrossRef]
- Kebbi, Y.; Muhammad, A.I.; Sant’Ana, A.S.; Prado-Silva, L.D.; Liu, D.; Ding, T. Recent advances on the application of UV-LED technology for microbial inactivation: Progress and mechanism. Compr. Rev. Food Sci. Food Saf. 2020, 19, 3501–3527. [Google Scholar] [CrossRef]
- Rahman, S.; Khan, I.; Oh, D.-H. Electrolyzed Water as a Novel Sanitizer in the Food Industry: Current Trends and Future Perspectives. Compr. Rev. Food Sci. Food Saf. 2016, 15, 471–490. [Google Scholar] [CrossRef]
- Chen, B.; Huang, J.; Li, H.; Zeng, Q.-H.; Wang, J.J.; Liu, H.; Pan, Y.; Zhao, Y. Eradication of planktonic Vibrio parahaemolyticus and its sessile biofilm by curcumin-mediated photodynamic inactivation. Food Control 2020, 113, 107181. [Google Scholar] [CrossRef]
- Zhang, Z.; Huang, Z.; Tong, J.; Wu, Q.; Pan, Y.; Malakar, P.K.; Zhao, Y. An outlook for food sterilization technology: Targeting the outer membrane of foodborne gram-negative pathogenic bacteria. Curr. Opin. Food Sci. 2021, 42, 15–22. [Google Scholar] [CrossRef]
- Lehman, K.M.; Grabowicz, M. Countering Gram-Negative Antibiotic Resistance: Recent Progress in Disrupting the Outer Membrane with Novel Therapeutics. Antibiotics 2019, 8, 163. [Google Scholar] [CrossRef]
- Bechelane-Maia, E.H.; Assis, L.C.; Alves de Oliveira, T.; Marques da Silva, A.; Gutterres Taranto, A. Structure-based virtual screening: From classical to artificial intelligence. Front. Chem. 2020, 8, 343. [Google Scholar] [CrossRef]
- Hou, T.J.; Xu, X.J. Recent Development and Application of Virtual Screening in Drug Discovery: An Overview. Curr. Pharm. Des. 2004, 10, 1011–1033. [Google Scholar] [CrossRef]
- Green, D.V. Virtual screening of chemical libraries for drug discovery. Expert Opin. Drug Discov. 2008, 3, 1011–1026. [Google Scholar] [CrossRef]
- Takeda, K.; Miyatake, H.; Yokota, N.; Matsuyama, S.; Tokuda, H.; Miki, K. Crystal structures of bacterial lipoprotein localization factors, LolA and LolB. EMBO J. 2003, 22, 3199–3209. [Google Scholar] [CrossRef]
- Papadopoulou, A.; Green, R.J.; Frazier, R.A. Interaction of Flavonoids with Bovine Serum Albumin: A Fluorescence Quenching Study. J. Agric. Food Chem. 2004, 53, 158–163. [Google Scholar] [CrossRef]
- Panat, N.A.; Maurya, D.K.; Ghaskadbi, S.S.; Sandur, S.K. Troxerutin, a plant flavonoid, protects cells against oxidative stress-induced cell death through radical scavenging mechanism. Food Chem. 2016, 194, 32–45. [Google Scholar] [CrossRef]
- Elmahdi, S.; DaSilva, L.V.; Parveen, S. Antibiotic resistance of Vibrio parahaemolyticus and Vibrio vulnificus in various countries: A review. Food Microbiol. 2016, 57, 128–134. [Google Scholar] [CrossRef]
- Jo, S.; Shin, C.; Shin, Y.; Kim, P.H.; Park, J.I.; Kim, M.; Park, B.; So, J.-S. Heavy metal and antibiotic co-resistance in Vibrio parahaemolyticus isolated from shellfish. Mar. Pollut. Bull. 2020, 156, 111246. [Google Scholar] [CrossRef] [PubMed]
- Choi, U.; Lee, C.-R. Antimicrobial Agents That Inhibit the Outer Membrane Assembly Machines of Gram-Negative Bacteria. J. Microbiol. Biotechnol. 2019, 29, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Nickerson, N.N.; Jao, C.C.; Xu, Y.; Quinn, J.; Skippington, E.; Alexander, M.K.; Miu, A.; Skelton, N.; Hankins, J.V.; Lopez, M.S.; et al. A Novel Inhibitor of the LolCDE ABC Transporter Essential for Lipoprotein Trafficking in Gram-Negative Bacteria. Antimicrob. Agents Chemother. 2018, 62, e02151-17. [Google Scholar] [CrossRef] [PubMed]
- Xia, J.; Feng, B.; Wen, G.; Xue, W.; Ma, G.; Zhang, H.; Wu, S. Bacterial Lipoprotein Biosynthetic Pathway as a Potential Target for Structure-based Design of Antibacterial Agents. Curr. Med. Chem. 2020, 27, 1132–1150. [Google Scholar] [CrossRef] [PubMed]
- Pathania, R.; Zlitni, S.; Barker, C.; Das, R.; Gerritsma, A.D.; Lebert, J.; Awuah, E.; Melacini, G.; Capretta, A.F.; Brown, E.D. Chemical genomics in Escherichia coli identifies an inhibitor of bacterial lipoprotein targeting. Nat. Chem. Biol. 2009, 5, 849–856. [Google Scholar] [CrossRef]
- Barker, C.A.; Allison, S.E.; Zlitni, S.; Nguyen, N.D.; Das, R.; Melacini, G.; Capretta, A.A.; Brown, E.D. Degradation of MAC13243 and studies of the interaction of resulting thiourea compounds with the lipoprotein targeting chaperone LolA. Bioorg. Med. Chem. Lett. 2013, 23, 2426–2431. [Google Scholar] [CrossRef]
- McLeod, S.M.; Fleming, P.R.; MacCormack, K.; McLaughlin, R.E.; Whiteaker, J.D.; Narita, S.-I.; Mori, M.; Tokuda, H.; Miller, A.A. Small-Molecule Inhibitors of Gram-Negative Lipoprotein Trafficking Discovered by Phenotypic Screening. J. Bacteriol. 2015, 197, 1075–1082. [Google Scholar] [CrossRef]
- Zhao, Y.; Liu, J.; Wu, Q.; Cao, H.; Ou, J.; Pan, Y.J. Nutritional and health efficacy of aquatic products for humans with the replenishment of terrestrial resources. Shuichan Xuebao 2021, 45, 1235–1247. [Google Scholar] [CrossRef]
- Bagchi, D.; Swaroop, A.; Preuss, H.G.; Bagchi, M. Free radical scavenging, antioxidant and cancer chemoprevention by grape seed proanthocyanidin: An overview. Mutat. Res. Mol. Mech. Mutagen. 2014, 768, 69–73. [Google Scholar] [CrossRef]
- Pinent, M.; Bladé, C.; Salvadó, M.J.; Blay, M.; Pujadas, G.; Fernández-Larrea, J.; Arola, L.; Ardèvol, A. Procyanidin Effects on Adipocyte-Related Pathologies. Crit. Rev. Food Sci. Nutr. 2006, 46, 543–550. [Google Scholar] [CrossRef]
- Dirir, A.M.; Daou, M.; Yousef, A.F.; Yousef, L.F. A review of alpha-glucosidase inhibitors from plants as potential candidates for the treatment of type-2 diabetes. Phytochem. Rev. 2021, 21, 1049–1079. [Google Scholar] [CrossRef]
- Lee, Y. Cancer Chemopreventive Potential of Procyanidin. Toxicol. Res. 2017, 33, 273–282. [Google Scholar] [CrossRef]
- Maroli, N.; Bhasuran, B.; Natarajan, J.; Kolandaivel, P. The potential role of procyanidin as a therapeutic agent against SARS-CoV-2: A text mining, molecular docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020, 40, 1230–1245. [Google Scholar] [CrossRef]
- Geuns, J.M. Stevioside. Phytochemistry 2003, 64, 913–921. [Google Scholar] [CrossRef]
- Chatsudthipong, V.; Muanprasat, C. Stevioside and related compounds: Therapeutic benefits beyond sweetness. Pharmacol. Ther. 2009, 121, 41–54. [Google Scholar] [CrossRef]
- Ahmadi, Z.; Mohammadinejad, R.; Roomiani, S.; Afshar, E.G.; Ashrafizadeh, M. Biological and Therapeutic Effects of Troxerutin: Molecular Signaling Pathways Come into View. J. Pharmacopunct. 2021, 24, 1–13. [Google Scholar] [CrossRef]
- Zamanian, M.; Bazmandegan, G.; Sureda, A.; Sobarzo-Sanchez, E.; Yousefi-Manesh, H.; Shirooie, S. The Protective Roles and Molecular Mechanisms of Troxerutin (Vitamin P4) for the Treatment of Chronic Diseases: A Mechanistic Review. Curr. Neuropharmacol. 2020, 19, 97–110. [Google Scholar] [CrossRef]
- Ganeshpurkar, A.; Saluja, A.K. The Pharmacological Potential of Rutin. Saudi Pharm. J. 2016, 25, 149–164. [Google Scholar] [CrossRef]
- Farha, A.K.; Gan, R.-Y.; Li, H.-B.; Wu, D.-T.; Atanasov, A.G.; Gul, K.; Zhang, J.-R.; Yang, Q.-Q.; Corke, H. The anticancer potential of the dietary polyphenol rutin: Current status, challenges, and perspectives. Crit. Rev. Food Sci. Nutr. 2020, 62, 832–859. [Google Scholar] [CrossRef]
- Sharma, S.; Ali, A.; Ali, J.; Sahni, J.K.; Baboota, S. Rutin: Therapeutic potential and recent advances in drug delivery. Expert Opin. Investig. Drugs 2013, 22, 1063–1079. [Google Scholar] [CrossRef]
- Agrawal, P.K.; Agrawal, C.; Blunden, G. Rutin: A Potential Antiviral for Repurposing as a SARS-CoV-2 Main Protease (Mpro) Inhibitor. Nat. Prod. Commun. 2021, 16. [Google Scholar] [CrossRef]
- Gullón, B.; Lu-Chau, T.A.; Moreira, M.T.; Lema, J.M.; Eibes, G. Rutin: A review on extraction, identification and purification methods, biological activities and approaches to enhance its bioavailability. Trends Food Sci. Technol. 2017, 67, 220–235. [Google Scholar] [CrossRef]
- Okuda, S.; Tokuda, H. Model of mouth-to-mouth transfer of bacterial lipoproteins through inner membrane LolC, periplasmic LolA, and outer membrane LolB. Proc. Natl. Acad. Sci. USA 2009, 106, 5877–5882. [Google Scholar] [CrossRef] [PubMed]
- Tsukahara, J.; Mukaiyama, K.; Okuda, S.; Narita, S.-I.; Tokuda, H. Dissection of LolB function—Lipoprotein binding, membrane targeting and incorporation of lipoproteins into lipid bilayers. FEBS J. 2009, 276, 4496–4504. [Google Scholar] [CrossRef] [PubMed]
- Makino, K.; Oshima, K.; Kurokawa, K.; Yokoyama, K.; Uda, T.; Tagomori, K.; Iijima, Y.; Najima, M.; Nakano, M.; Yamashita, A.; et al. Genome sequence of Vibrio parahaemolyticus: A pathogenic mechanism distinct from that of V. cholerae. Lancet 2003, 361, 743–749. [Google Scholar] [CrossRef]
- Mazaheri, S.; Nafian, D.F. Modeling of membrane protein serpentin receptor with SWISS MODEL software. Clin. Biochem. 2011, 44, S321. [Google Scholar] [CrossRef]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; De Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef]
- Guex, N.; Peitsch, M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis 1997, 18, 2714–2723. [Google Scholar] [CrossRef]
- Sivasothy, Y.; Sulaiman, S.F.; Ooi, K.L.; Ibrahim, H.; Awang, K. Antioxidant and antibacterial activities of flavonoids and curcuminoids from Zingiber spectabile Griff. Food Control 2012, 30, 714–720. [Google Scholar] [CrossRef]
- Yu, K.; Lu, F.; Li, Q.; Chen, H.; Lu, B.; Liu, J.; Li, Z.; Dai, F.; Wu, D.; Lan, G. In situ assembly of Ag nanoparticles (AgNPs) on porous silkworm cocoon-based wound film: Enhanced antimicrobial and wound healing activity. Sci. Rep. 2017, 7, 2107. [Google Scholar] [CrossRef]
- Zhang, Z.; Sablok, G. Estimate Codon Usage Bias Using Codon Usage Analyzer (CUA). Methods Mol. Biol. 2017, 1667, 139–148. [Google Scholar] [CrossRef]
- Taraska, J.W.; Puljung, M.C.; Olivier, N.B.; Flynn, E.G.; Zagotta, W.N. Mapping the structure and conformational movements of proteins with transition metal ion FRET. Nat. Methods 2009, 6, 532–537. [Google Scholar] [CrossRef]
- Sindrewicz, P.; Li, X.; Yates, E.A.; Turnbull, J.E.; Lian, L.-Y.; Yu, L.-G. Intrinsic tryptophan fluorescence spectroscopy reliably determines galectin-ligand interactions. Sci. Rep. 2019, 9, 11851. [Google Scholar] [CrossRef]
- Kalirajan, R.; Kulshrestha, V.; Sankar, S.; Jubie, S. Docking studies, synthesis, characterization of some novel oxazine substituted 9-anilinoacridine derivatives and evaluation for their antioxidant and anticancer activities as topoisomerase II inhibitors. Eur. J. Med. Chem. 2012, 56, 217–224. [Google Scholar] [CrossRef]

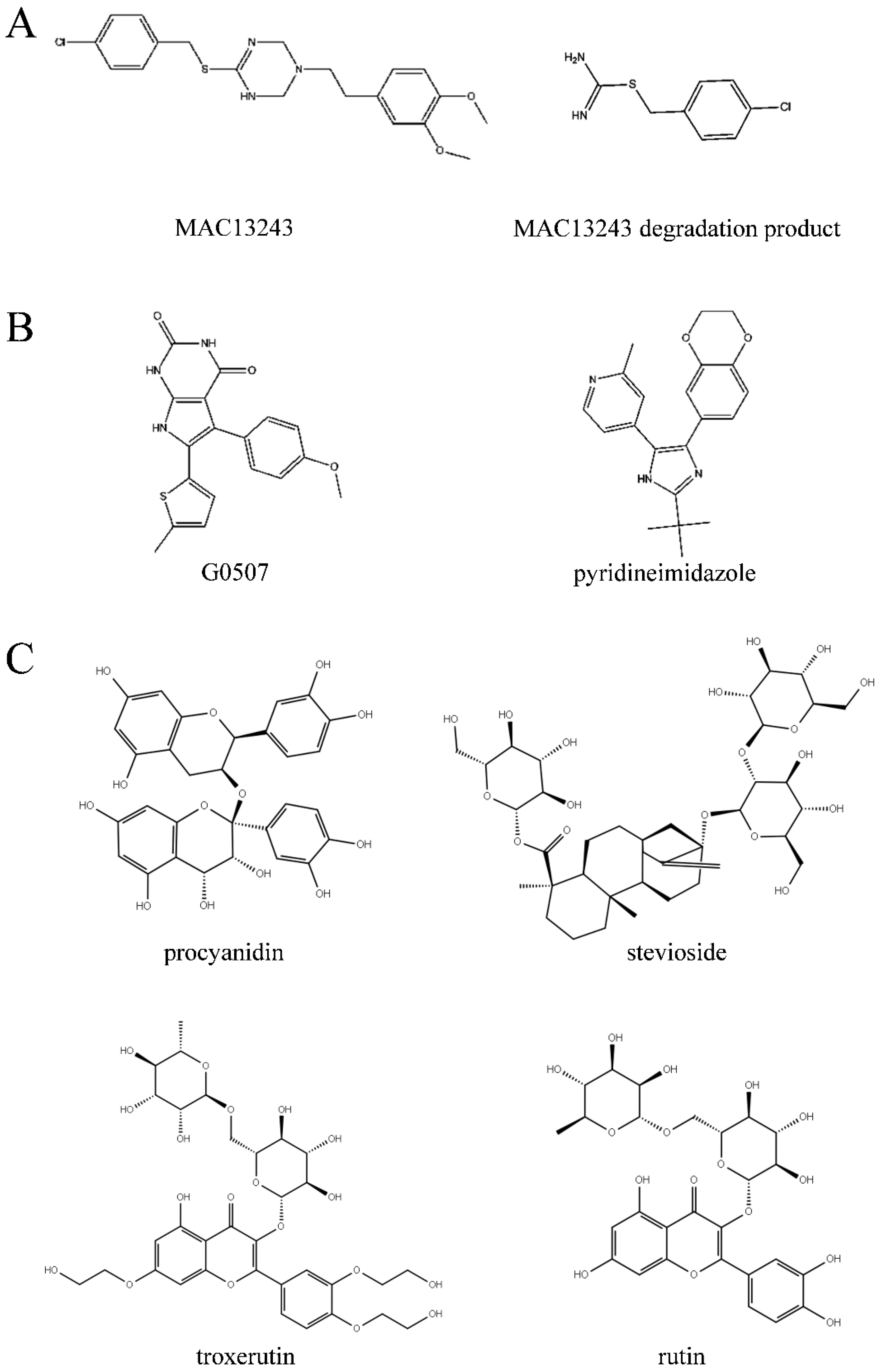

| Number | Glide Score | Compound | Formula | Mol wt |
|---|---|---|---|---|
| 1 | −12.023 | Stevioside (hydrate) | C38H62O19 | 822.895 |
| 2 | −9.858 | Forsythoside B | C34H44O19 | 756.707 |
| 3 | −9.907 | Coenzyme A (sodium salt hydrate) | C21H37N7NaO17P3S | 807.53 |
| 4 | −8.994 | Rutin (hydrate) | C27H36O19 | 664.566 |
| 5 | −8.789 | Madecassoside | C48H78O20 | 975.132 |
| 6 | −8.626 | Troxerutin | C33H42O19 | 742.68 |
| 7 | −8.307 | Procyanidin | C30H26O13 | 594.525 |
| 8 | −8.155 | Forsythoside A | C29H36O15 | 624.592 |
| 9 | −7.847 | Mulberroside A | C26H32O14 | 568.528 |
| 10 | −7.781 | Uridine-5’-diphosphoglucuronic Acid | C15H19N2Na3O18P2 | 646.23 |
| Compound | Ksv (104 L/mol) | Kq (1012 L/mol·s) | R2 |
|---|---|---|---|
| Procyanidin | 8.3054 | 8.3054 | 0.9817 |
| Stevioside (hydrate) | 6.1570 | 6.1570 | 0.9802 |
| Troxerutin | 14.2317 | 14.2317 | 0.9641 |
| Rutin (hydrate) | 8.7014 | 8.7014 | 0.9700 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, J.; Tong, J.; Wu, Q.; Liu, J.; Yuan, M.; Tian, C.; Xu, H.; Malakar, P.K.; Pan, Y.; Zhao, Y.; et al. Natural Inhibitors Targeting the Localization of Lipoprotein System in Vibrio parahaemolyticus. Int. J. Mol. Sci. 2022, 23, 14352. https://doi.org/10.3390/ijms232214352
Liu J, Tong J, Wu Q, Liu J, Yuan M, Tian C, Xu H, Malakar PK, Pan Y, Zhao Y, et al. Natural Inhibitors Targeting the Localization of Lipoprotein System in Vibrio parahaemolyticus. International Journal of Molecular Sciences. 2022; 23(22):14352. https://doi.org/10.3390/ijms232214352
Chicago/Turabian StyleLiu, Jiawen, Jinrong Tong, Qian Wu, Jing Liu, Mengqi Yuan, Cuifang Tian, Huan Xu, Pradeep K. Malakar, Yingjie Pan, Yong Zhao, and et al. 2022. "Natural Inhibitors Targeting the Localization of Lipoprotein System in Vibrio parahaemolyticus" International Journal of Molecular Sciences 23, no. 22: 14352. https://doi.org/10.3390/ijms232214352
APA StyleLiu, J., Tong, J., Wu, Q., Liu, J., Yuan, M., Tian, C., Xu, H., Malakar, P. K., Pan, Y., Zhao, Y., & Zhang, Z. (2022). Natural Inhibitors Targeting the Localization of Lipoprotein System in Vibrio parahaemolyticus. International Journal of Molecular Sciences, 23(22), 14352. https://doi.org/10.3390/ijms232214352






