Identification and Expression Analysis of the NPF Genes in Cotton
Abstract
1. Introduction
2. Results
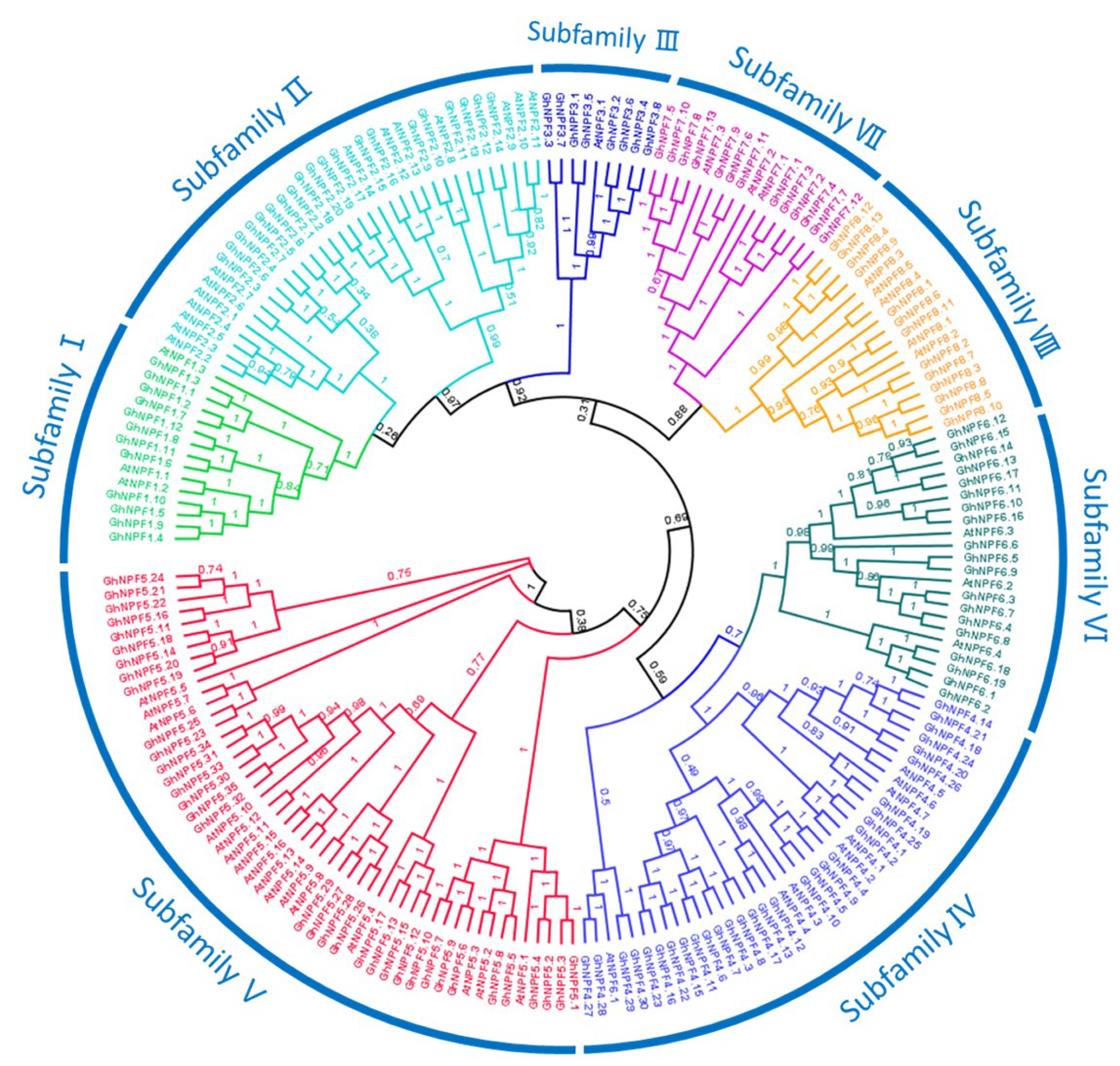
2.1. Identification and Phylogenetic Analysis of NPF Genes in the Three Gossypium Species
2.2. Gene Structures and Conserved Motifs Analysis
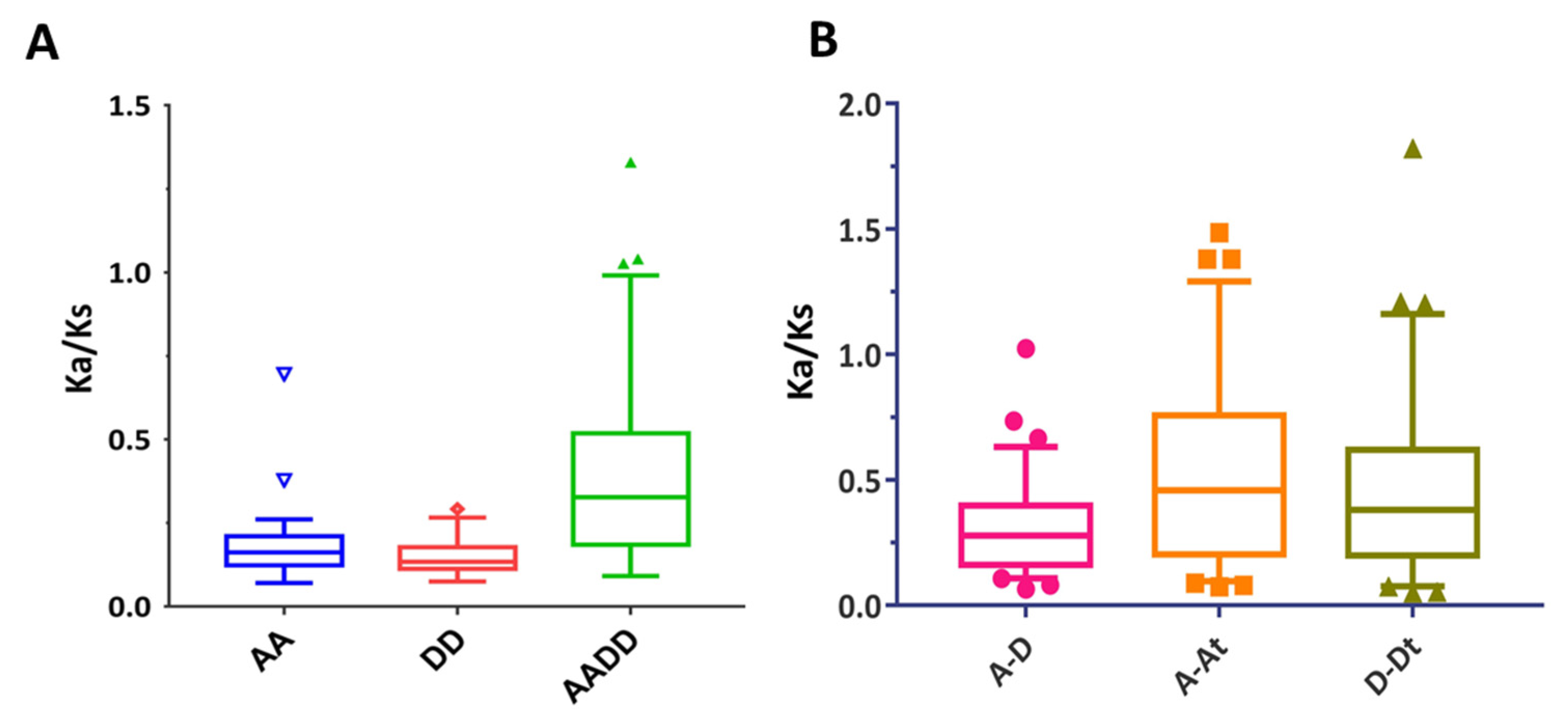
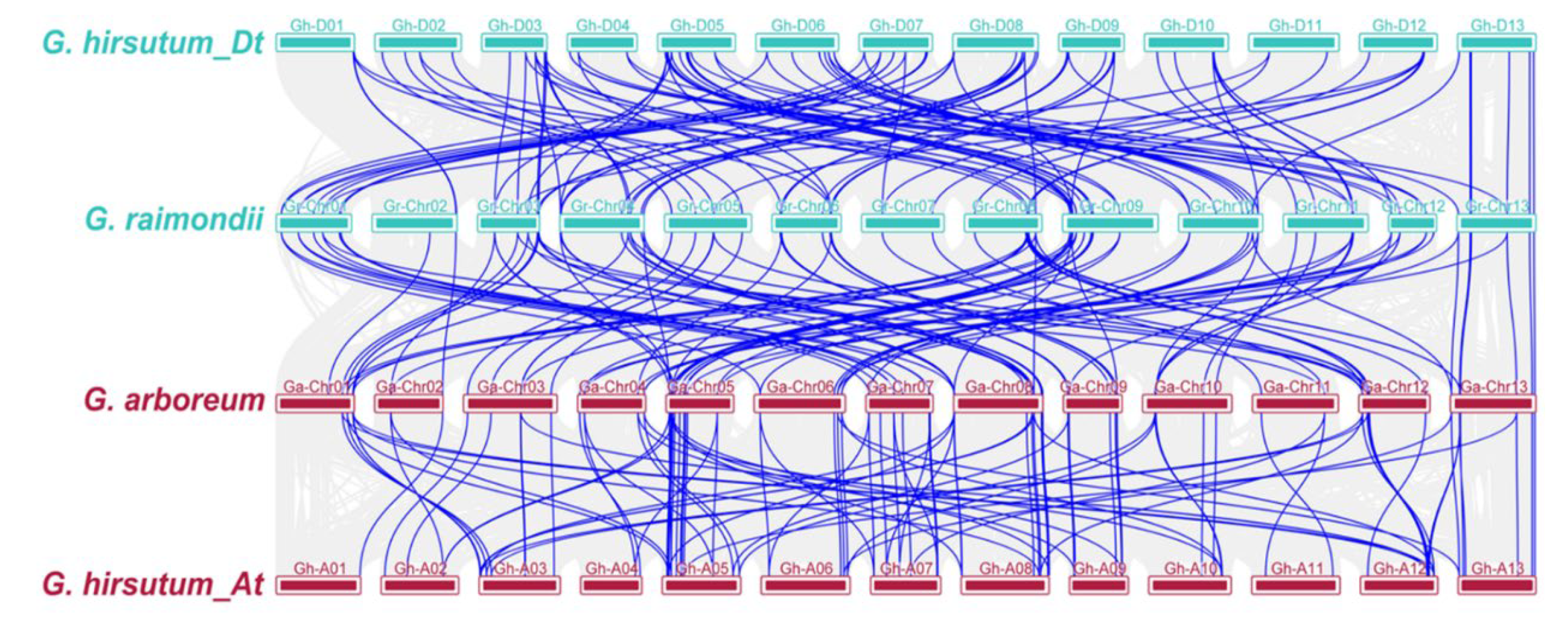
2.3. Chromosomal Position, Duplication, and Collinearity Analysis
2.4. Cis-Acting Elements Analysis of GhNPFs
2.5. Expression Patterns Analysis of the GhNPF Genes
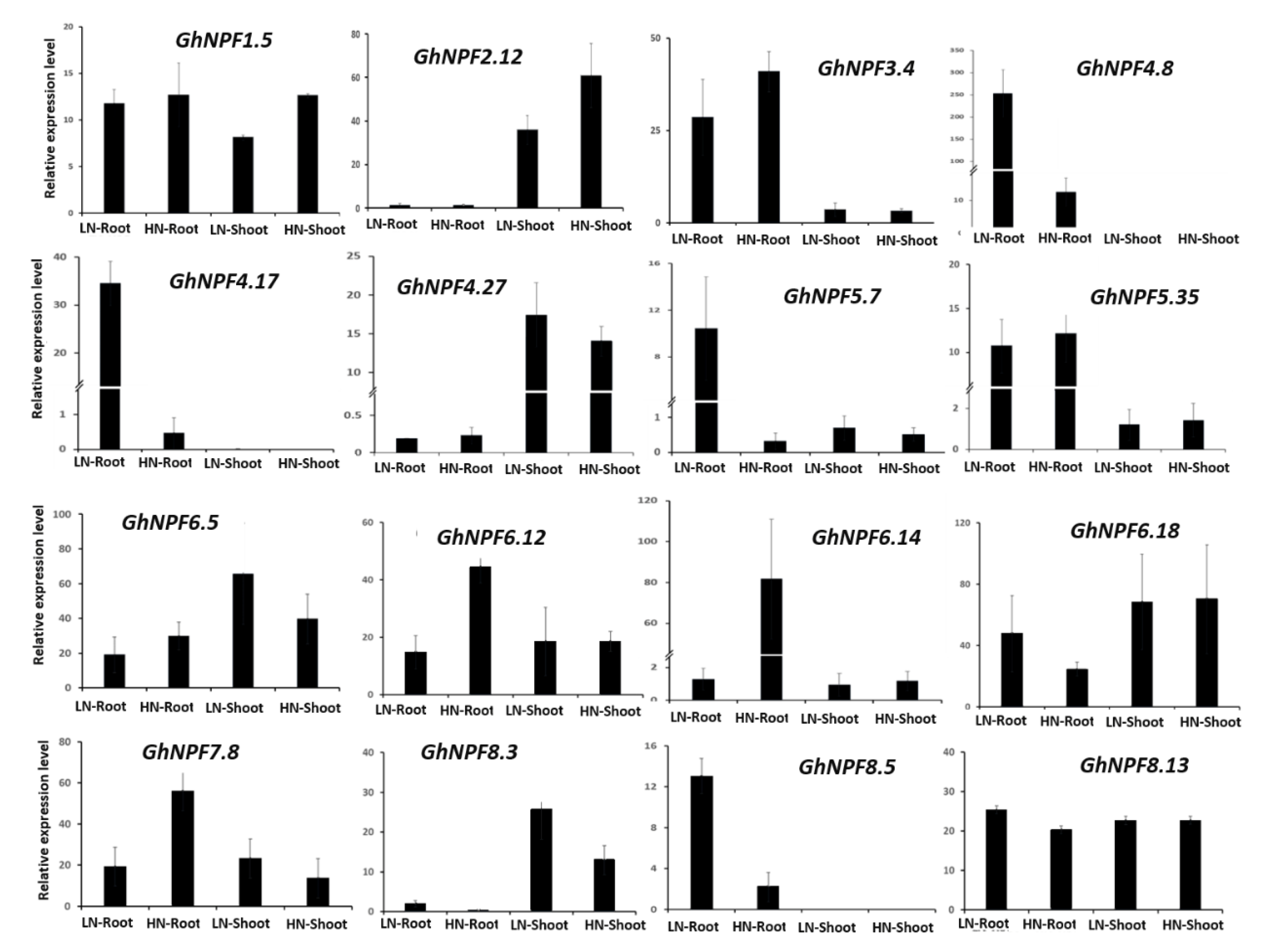
2.6. GhNPF Genes Expression Pattern under N Deficiency Conditions
2.7. Silencing of GhNPF6.14 in Cotton
3. Discussion
4. Materials and Methods
4.1. Identification of NPF Genes
4.2. Chromosomal Location and Sequences and Phylogenetic Analysis
4.3. Analysis of the Conserved Motifs, Gene Structure and Cis-Regulatory Elements
4.4. Evolutionary Analysis of NPF Genes
4.5. GhNPF-Expression Pattern Analysis
4.6. N deficiency Treatments and the qRT-PCR Analysis
4.7. VIGS Treatment and Measurement of N Concentration
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, Y.-Y.; Hsu, P.-K.; Tsay, Y.-F. Uptake, allocation and signaling of nitrate. Trends Plant Sci. 2012, 17, 458–467. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Fan, X.; Miller, A.J. Plant Nitrogen Assimilation and Use Efficiency. Annu. Rev. Plant Biol. 2012, 63, 153–182. [Google Scholar] [CrossRef] [PubMed]
- Tsay, Y.-F.; Chiu, C.-C.; Tsai, C.-B.; Ho, C.-H.; Hsu, P.-K. Nitrate transporters and peptide transporters. FEBS Lett. 2007, 581, 2290–2300. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-Y.; Cheng, Y.-H.; Chen, K.-E.; Tsay, Y.-F. Nitrate Transport, Signaling, and Use Efficiency. Annu. Rev. Plant Biol. 2018, 69, 85–122. [Google Scholar] [CrossRef] [PubMed]
- Jorgensen, M.E.; Xu, D.; Crocoll, C.; Ramirez, D.; Motawia, M.S.; Olsen, C.E.; Nour-Eldin, H.H.; Halkier, B.A. Origin and evolution of transporter substrate specificity within the NPF family. Elife 2017, 6, e19466. [Google Scholar] [CrossRef]
- Kanstrup, C.; Nour-Eldin, H.H. The emerging role of the nitrate and peptide transporter family: NPF in plant specialized metabolism. Curr. Opin. Plant Biol. 2022, 68, 102243. [Google Scholar] [CrossRef]
- Leran, S.; Varala, K.; Boyer, J.-C.; Chiurazzi, M.; Crawford, N.; Daniel-Vedele, F.; David, L.; Dickstein, R.; Fernandez, E.; Forde, B.; et al. A unified nomenclature of Nitrate Transporter 1/Peptide Transporter family members in plants. Trends Plant Sci. 2014, 19, 5–9. [Google Scholar] [CrossRef]
- Ho, C.-H.; Lin, S.-H.; Hu, H.-C.; Tsay, Y.-F. CHL1 Functions as a Nitrate Sensor in Plants. Cell 2009, 138, 1184–1194. [Google Scholar] [CrossRef]
- Li, B.; Byrt, C.; Qiu, J.; Baumann, U.; Hrmova, M.; Evrard, A.; Johnson, A.A.T.; Birnbaum, K.D.; Mayo, G.M.; Jha, D.; et al. Identification of a Stelar-Localized Transport Protein That Facilitates Root-to-Shoot Transfer of Chloride in Arabidopsis. Plant Physiol. 2016, 170, 1014–1029. [Google Scholar] [CrossRef]
- Lin, S.-H.; Kuo, H.-F.; Canivenc, G.; Lin, C.-S.; Lepetit, M.; Hsu, P.-K.; Tillard, P.; Lin, H.-L.; Wang, Y.-Y.; Tsai, C.-B.; et al. Mutation of the Arabidopsis NRT1.5 Nitrate Transporter Causes Defective Root-to-Shoot Nitrate Transport. Plant Cell 2008, 20, 2514–2528. [Google Scholar] [CrossRef]
- Li, J.-Y.; Fu, Y.-L.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.-Z.; Zhang, Y.; Li, H.-M.; Huang, J.; et al. The Arabidopsis Nitrate Transporter NRT1.8 Functions in Nitrate Removal from the Xylem Sap and Mediates Cadmium Tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef]
- Zhang, G.-B.; Yi, H.-Y.; Gong, J.-M. The Arabidopsis Ethylene/Jasmonic Acid-NRT Signaling Module Coordinates Nitrate Reallocation and the Trade-Off between Growth and Environmental Adaptation. Plant Cell 2014, 26, 3984–3998. [Google Scholar] [CrossRef]
- Guan, Y.; Liu, D.-F.; Qiu, J.; Liu, Z.-J.; He, Y.-N.; Fang, Z.-J.; Huang, X.-H.; Gong, J.-M. The nitrate transporter OsNPF7.9 mediates nitrate allocation and the divergent nitrate use efficiency between indica and japonica rice. Plant Physiol. 2022, 189, 215–229. [Google Scholar] [CrossRef]
- Iqbal, A.; Qiang, D.; Alamzeb, M.; Xiangru, W.; Huiping, G.; Hengheng, Z.; Nianchang, P.; Xiling, Z.; Meizhen, S. Untangling the molecular mechanisms and functions of nitrate to improve nitrogen use efficiency. J. Sci. Food Agric. 2020, 100, 904–914. [Google Scholar] [CrossRef]
- Watts, D.B.; Runion, G.B.; Balkcom, K.S. Nitrogen fertilizer sources and tillage effects on cotton growth, yield, and fiber quality in a coastal plain soil. Field Crops Res. 2017, 201, 184–191. [Google Scholar] [CrossRef]
- Luo, Z.; Hu, Q.; Tang, W.; Wang, X.; Lu, H.; Zhang, Z.; Liu, T.; Kong, X. Effects of N fertilizer rate and planting density on short-season cotton yield, N agronomic efficiency and soil N using N-15 tracing technique. Eur. J. Agron. 2022, 138, 126546. [Google Scholar] [CrossRef]
- Chen, W.; Hou, Z.; Wu, L.; Liang, Y.; Wei, C. Effects of salinity and nitrogen on cotton growth in arid environment. Plant Soil 2010, 326, 61–73. [Google Scholar] [CrossRef]
- Yang, X.; Xia, X.; Zeng, Y.; Nong, B.; Zhang, Z.; Wu, Y.; Tian, Q.; Zeng, W.; Gao, J.; Zhou, W.; et al. Genome-wide identification of the peptide transporter family in rice and analysis of the PTR expression modulation in two near-isogenic lines with different nitrogen use efficiency. Bmc Plant Biol. 2020, 20, 193. [Google Scholar] [CrossRef]
- Bai, H.; Euring, D.; Volmer, K.; Janz, D.; Polle, A. The Nitrate Transporter (NRT) Gene Family in Poplar. PLoS ONE 2013, 8, e72126. [Google Scholar] [CrossRef]
- Buchner, P.; Hawkesford, M.J. Complex phylogeny and gene expression patterns of members of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER family (NPF) in wheat. J. Exp. Bot. 2014, 65, 5697–5710. [Google Scholar] [CrossRef]
- Wang, H.; Wan, Y.; Buchner, P.; King, R.; Ma, H.; Hawkesford, M.J. Phylogeny and gene expression of the complete Nitrate Transporter 1/Peptide Transporter FamilY in Triticum aestivum. J. Exp. Bot. 2020, 71, 4531–4546. [Google Scholar] [CrossRef]
- Liu, X.; Gao, Y.; Li, K.; Yin, Y.; Liu, J.; Zhu, Y. Complex Phylogeny and Expression Patterns of the Nitrate Transporter 1/Peptide TransporteR Family Genes in Tomato. Russ. J. Plant Physiol. 2022, 69, 47. [Google Scholar] [CrossRef]
- Pellizzaro, A.; Alibert, B.; Planchet, E.; Limami, A.M.; Morere-Le Paven, M.-C. Nitrate transporters: An overview in legumes. Planta 2017, 246, 585–595. [Google Scholar] [CrossRef]
- Wang, Q.; Liu, C.; Dong, Q.; Huang, D.; Li, C.; Li, P.; Ma, F. Genome-Wide Identification and Analysis of Apple Nitrate Transporter 1/Peptide Transporter Family (NPF) Genes Reveals MdNPF6.5 Confers High Capacity for Nitrogen Uptake under Low-Nitrogen Conditions. Int. J. Mol. Sci. 2018, 19, 2761. [Google Scholar] [CrossRef]
- Wang, J.; Li, Y.; Zhu, F.; Ming, R.; Chen, L.-Q. Genome-Wide Analysis of Nitrate Transporter (NRT/NPF) Family in Sugarcane Saccharum spontaneum L. Trop. Plant Biol. 2019, 12, 133–149. [Google Scholar] [CrossRef]
- Zhang, H.; Li, S.; Shi, M.; Wang, S.; Shi, L.; Xu, F.; Ding, G. Genome-Wide Systematic Characterization of theNPFFamily Genes and Their Transcriptional Responses to Multiple Nutrient Stresses in Allotetraploid Rapeseed. Int. J. Mol. Sci. 2020, 21, 5947. [Google Scholar] [CrossRef]
- Wang, X.; Cai, X.; Xu, C.; Wang, Q. Identification and characterization of the NPF, NRT2 and NRT3 in spinach. Plant Physiol. Biochem. 2021, 158, 297–307. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wei, K.; Ruan, L.; Bai, P.; Wu, L.; Wang, L.; Cheng, H. Systematic Investigation and Expression Profiles of the Nitrate Transporter 1/Peptide Transporter Family (NPF) in Tea Plant (Camellia sinensis). Int. J. Mol. Sci. 2022, 23, 6663. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.; Liang, Y.; Huang, Y.; Xi, J.; Huang, X.; Yang, X.; Yi, K. Phylogeny and Expression Atlas of the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY in Agave. Plants 2022, 11, 1434. [Google Scholar] [CrossRef] [PubMed]
- Xia, X.; Fan, X.; Wei, J.; Feng, H.; Qu, H.; Xie, D.; Miller, A.J.; Xu, G. Rice nitrate transporter OsNPF2.4 functions in low-affinity acquisition and long-distance transport. J. Exp. Bot. 2015, 66, 317–331. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Hu, Y.; Jiang, W.; Fang, L.; Guan, X.; Chen, J.; Zhang, J.; Saski, C.A.; Scheffler, B.E.; Stelly, D.M.; et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 2015, 33, 531–537. [Google Scholar] [CrossRef]
- Corratge-Faillie, C.; Lacombe, B. Substrate (un)specificity of Arabidopsis NRT1/PTR FAMILY (NPF) proteins. J. Exp. Bot. 2017, 68, 3107–3113. [Google Scholar] [CrossRef]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef]
- Paterson, A.H.; Wendel, J.F.; Gundlach, H.; Guo, H.; Jenkins, J.; Jin, D.; Llewellyn, D.; Showmaker, K.C.; Shu, S.; Udall, J.; et al. Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 2012, 492, 423–427. [Google Scholar] [CrossRef]
- Lynch, M.; Conery, J.S. The evolutionary fate and consequences of duplicate genes. Science 2000, 290, 1151–1155. [Google Scholar] [CrossRef]
- Roulin, A.; Auer, P.L.; Libault, M.; Schlueter, J.; Farmer, A.; May, G.; Stacey, G.; Doerge, R.W.; Jackson, S.A. The fate of duplicated genes in a polyploid plant genome. Plant J. 2013, 73, 143–153. [Google Scholar] [CrossRef]
- Imagawa, M. Negative regulation of gene expression in eukaryotes. Neurochem. Int. 1996, 29, 565–572. [Google Scholar] [CrossRef]
- Wang, J.; Wan, R.; Nie, H.; Xue, S.; Fang, Z. OsNPF5.16, a nitrate transporter gene with natural variation, is essential for rice growth and yield. Crop J. 2022, 10, 397–406. [Google Scholar] [CrossRef]
- Banedjschafie, S.; Bastani, S.; Widmoser, P.; Mengel, K. Improvement of water use and N fertilizer efficiency by subsoil irrigation of winter wheat. Eur. J. Agron. 2008, 28, 1–7. [Google Scholar] [CrossRef]
- Guo, F.O.; Young, J.; Crawford, N.M. The nitrate transporter AtNRT1.1 (CHL1) functions in stomatal opening and contributes to drought susceptibility in arabidopsis. Plant Cell 2003, 15, 107–117. [Google Scholar] [CrossRef]
- Zhang, Z.; Ma, W.; Ren, Z.; Wang, X.; Zhao, J.; Pei, X.; Liu, Y.; He, K.; Zhang, F.; Huo, W.; et al. Characterization and expression analysis of wall-associated kinase (WAK) and WAK-like family in cotton. Int. J. Biol. Macromol. 2021, 187, 867–879. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.-L. Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 2013, 73, 941–951. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Cui, F.; Han, X.; He, Y.; Zhao, L.; Zhang, N.; Zhang, H.; Zhu, H.; Liu, Z.; Ma, B.; et al. Comparative genomic and transcriptomic analyses uncover the molecular basis of high nitrogen-use efficiency in the wheat cultivar Kenong 9204. Mol. Plant 2022, 15, 1440–1456. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Rehman, A.; Peng, Z.; Li, H.; Qin, G.; Jia, Y.; Pan, Z.; He, S.; Qayyum, A.; Du, X. Genome wide analysis of IQD gene family in diploid and tetraploid species of cotton (Gossypium spp.). Int. J. Biol. Macromol. 2021, 184, 1035–1061. [Google Scholar] [CrossRef]
- Zhu, S.; Wang, X.; Chen, W.; Yao, J.; Li, Y.; Fang, S.; Lv, Y.; Li, X.; Pan, J.; Liu, C.; et al. Cotton DMP gene family: Characterization, evolution, and expression profiles during development and stress. Int. J. Biol. Macromol. 2021, 183, 1257–1269. [Google Scholar] [CrossRef]
- Iqbal, A.; Dong, Q.; Wang, Z.; Wang, X.; Gui, H.; Zhang, H.; Pang, N.; Zhang, X.; Song, M. Growth and nitrogen metabolism are associated with nitrogen-use efficiency in cotton genotypes. Plant Physiol. Biochem. 2020, 149, 61–74. [Google Scholar] [CrossRef]
- Iqbal, A.; Dong, Q.; Wang, X.; Gui, H.P.; Zhang, H.; Pang, N.; Zhang, X.; Song, M. Nitrogen preference and genetic variation of cotton genotypes for nitrogen use efficiency. J. Sci. Food Agric. 2020, 100, 2761–2773. [Google Scholar] [CrossRef]
- Iqbal, A.; Dong, Q.; Wang, X.; Gui, H.; Zhang, H.; Zhang, X.; Song, M. Transcriptome Analysis Reveals Differences in Key Genes and Pathways Regulating Carbon and Nitrogen Metabolism in Cotton Genotypes under N Starvation and Resupply. Int. J. Mol. Sci. 2020, 21, 1500. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Iqbal, A.; Gui, H.; Wang, X.; Zhang, H.; Zhang, X.; Song, M. Genome-wide expression analysis reveals involvement of asparagine synthetase family in cotton development and nitrogen metabolism. BMC Plant Biol. 2022, 22, 122. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, Q.; Wang, G.; Iqbal, A.; Muhammad, N.; Wang, X.; Gui, H.; Zhang, H.; Kayoumu, M.; Li, X.; Zhang, X.; et al. Identification and Expression Analysis of the NPF Genes in Cotton. Int. J. Mol. Sci. 2022, 23, 14262. https://doi.org/10.3390/ijms232214262
Dong Q, Wang G, Iqbal A, Muhammad N, Wang X, Gui H, Zhang H, Kayoumu M, Li X, Zhang X, et al. Identification and Expression Analysis of the NPF Genes in Cotton. International Journal of Molecular Sciences. 2022; 23(22):14262. https://doi.org/10.3390/ijms232214262
Chicago/Turabian StyleDong, Qiang, Guoxin Wang, Asif Iqbal, Noor Muhammad, Xiangru Wang, Huiping Gui, Hengheng Zhang, Mirezhatijiang Kayoumu, Xiaotong Li, Xiling Zhang, and et al. 2022. "Identification and Expression Analysis of the NPF Genes in Cotton" International Journal of Molecular Sciences 23, no. 22: 14262. https://doi.org/10.3390/ijms232214262
APA StyleDong, Q., Wang, G., Iqbal, A., Muhammad, N., Wang, X., Gui, H., Zhang, H., Kayoumu, M., Li, X., Zhang, X., & Song, M. (2022). Identification and Expression Analysis of the NPF Genes in Cotton. International Journal of Molecular Sciences, 23(22), 14262. https://doi.org/10.3390/ijms232214262







