Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana
Abstract
1. Introduction
2. Results
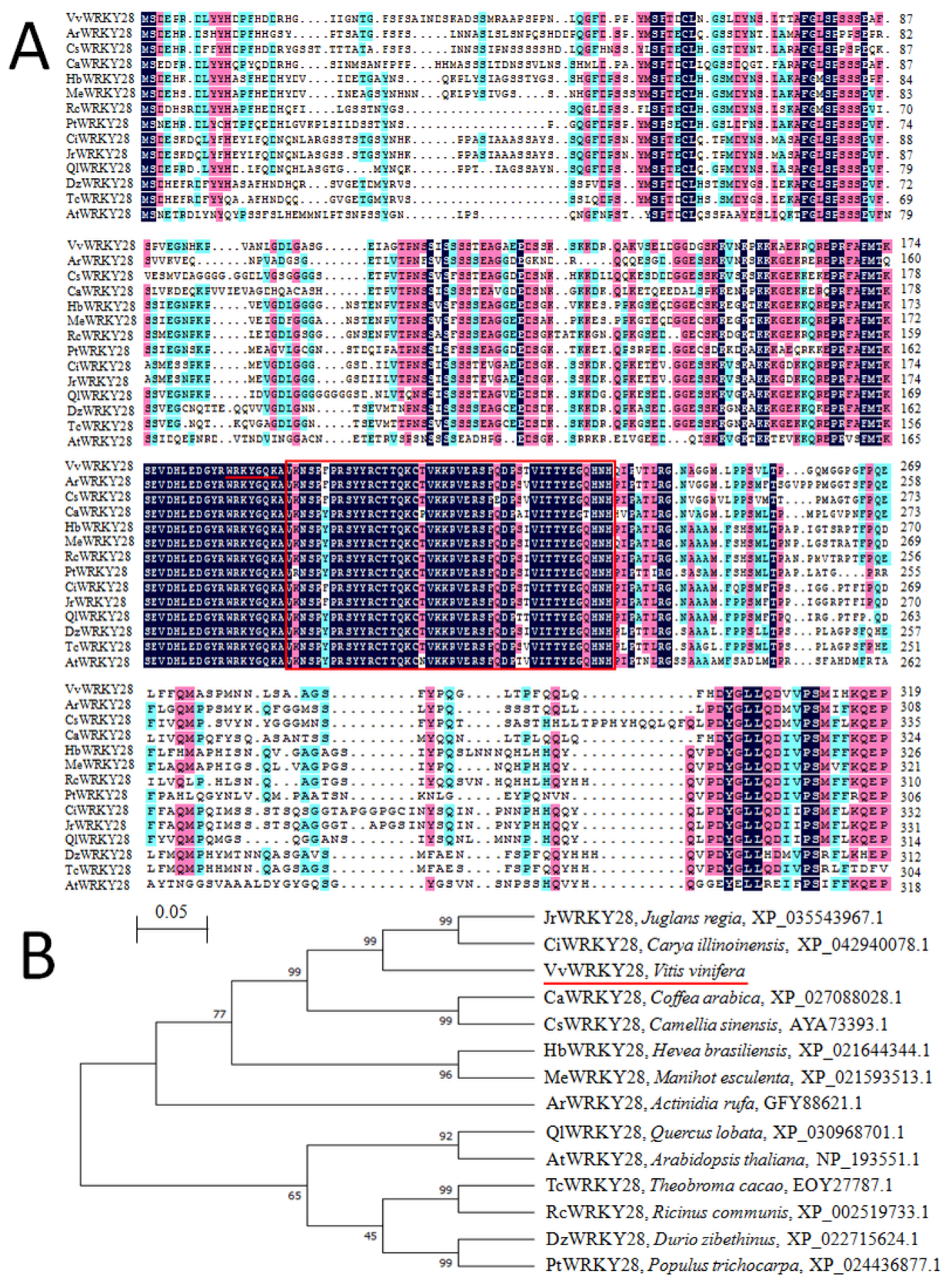
2.1. Cloning and Sequence Analysis of VvWRKY28
2.2. Phylogenetic Relationship of VvWRKY28 with Other WRKY Proteins
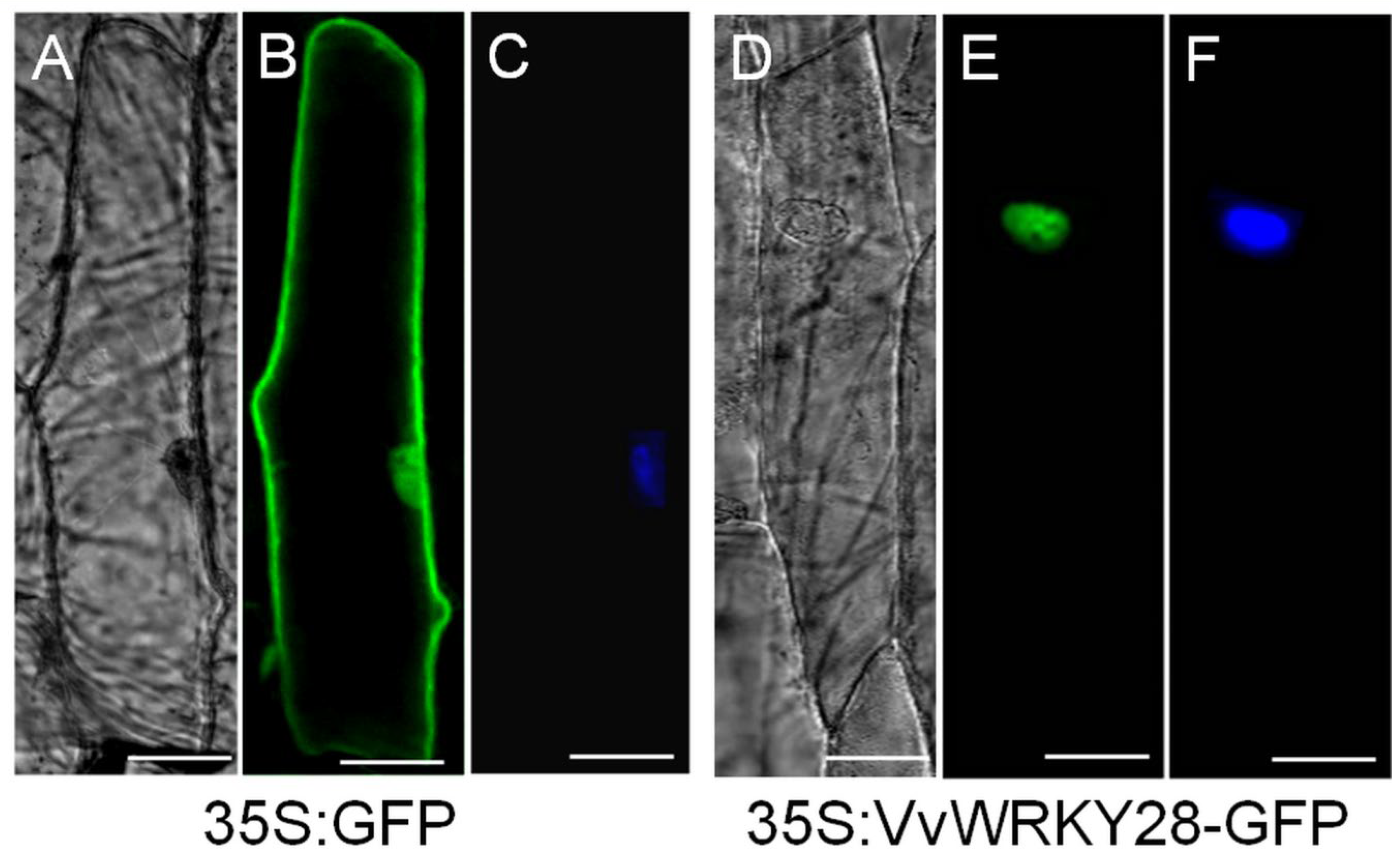
2.3. Subcellular Localization of VvWRKY28Protein
2.4. Expression Specificity Analysis of VvWRKY28
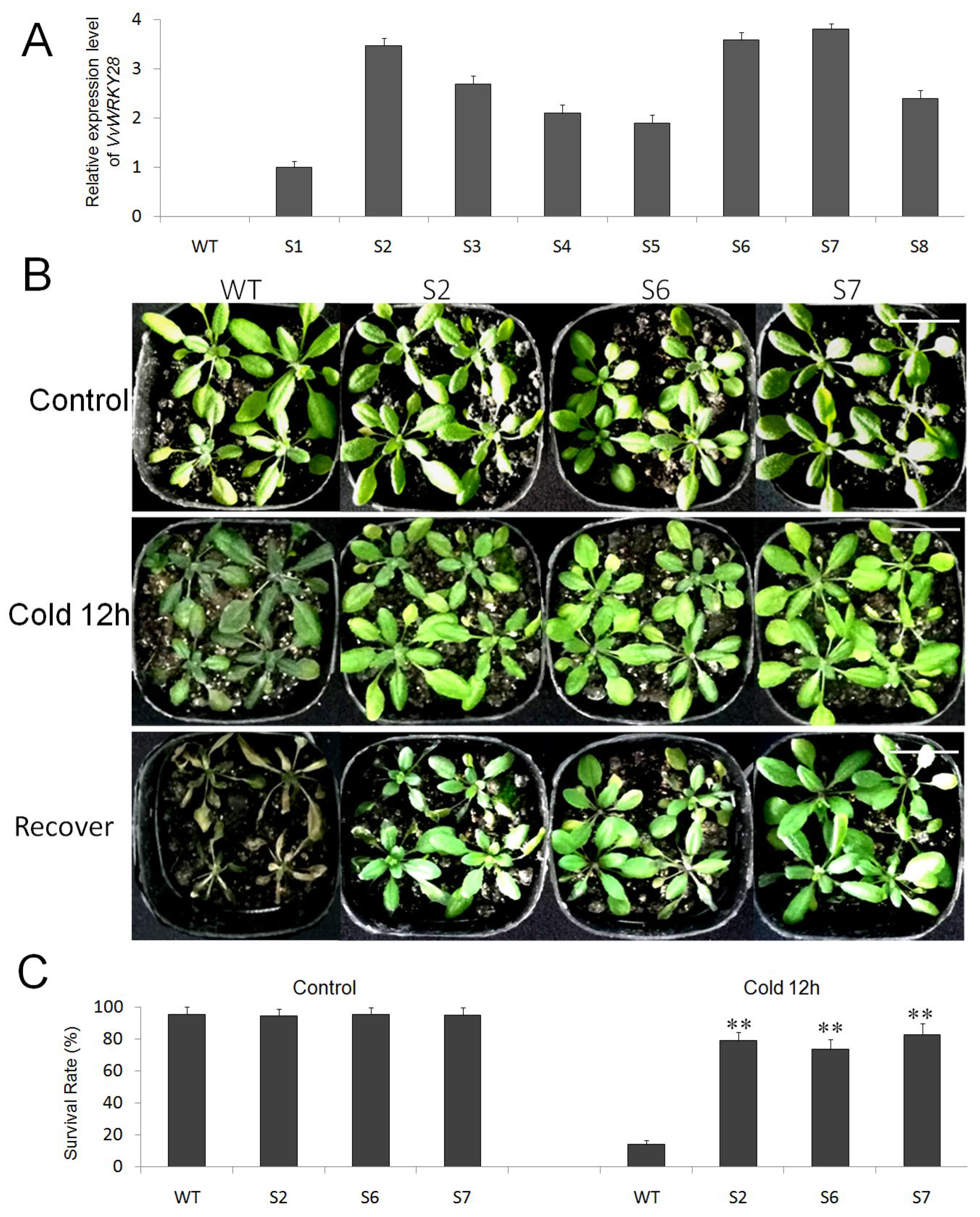
2.5. Analysis of the Ability of VvWRKY28 Transgenic A. thaliana to Cold Stress
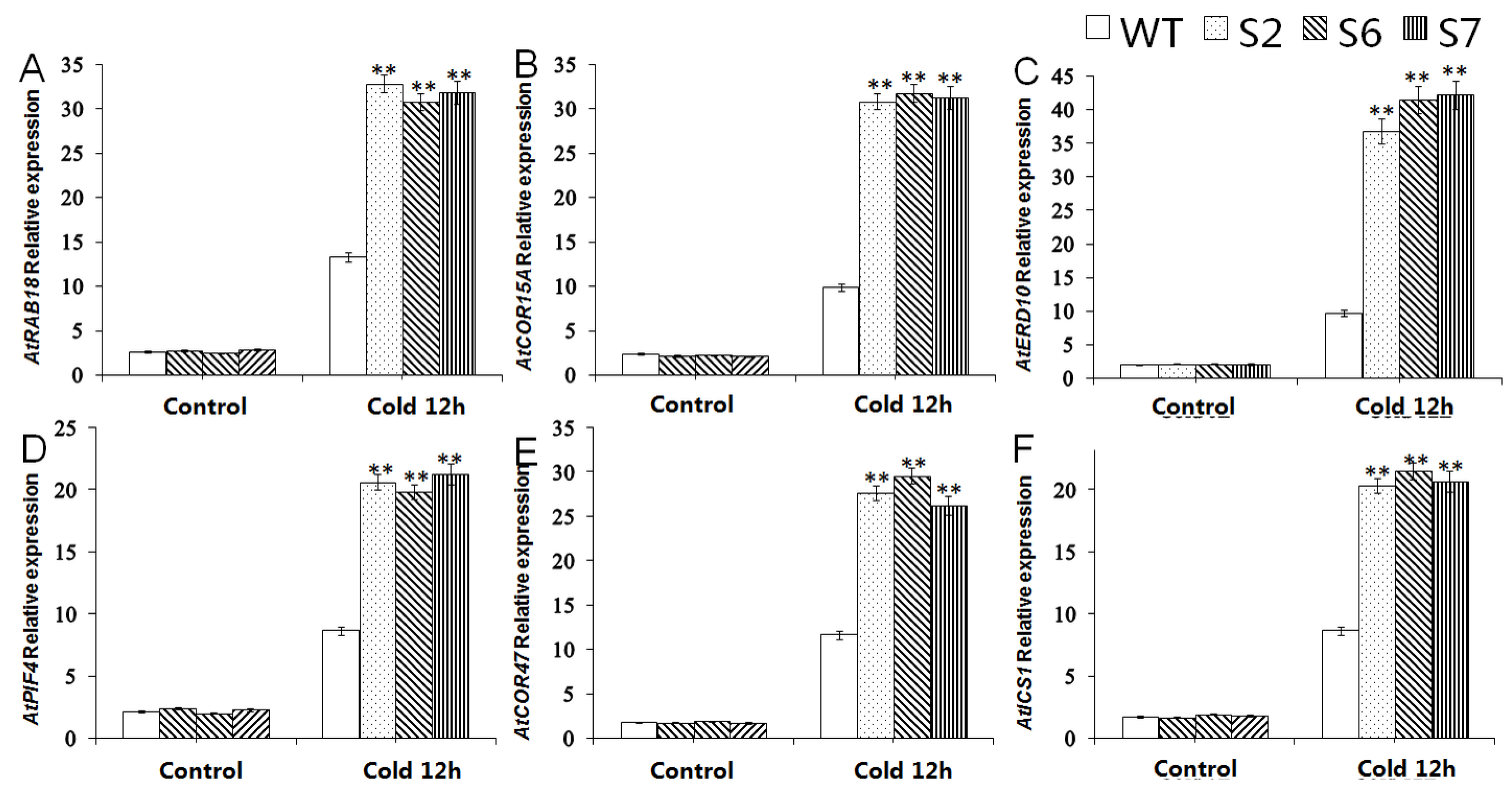
2.6. Expression Analysis of Cold-Resistant Downstream Genes in VvWRKY28-OE A. thaliana
2.7. Analysis of the Ability of VvWRKY28 Transgenic A. thaliana to Resist Salt Stress
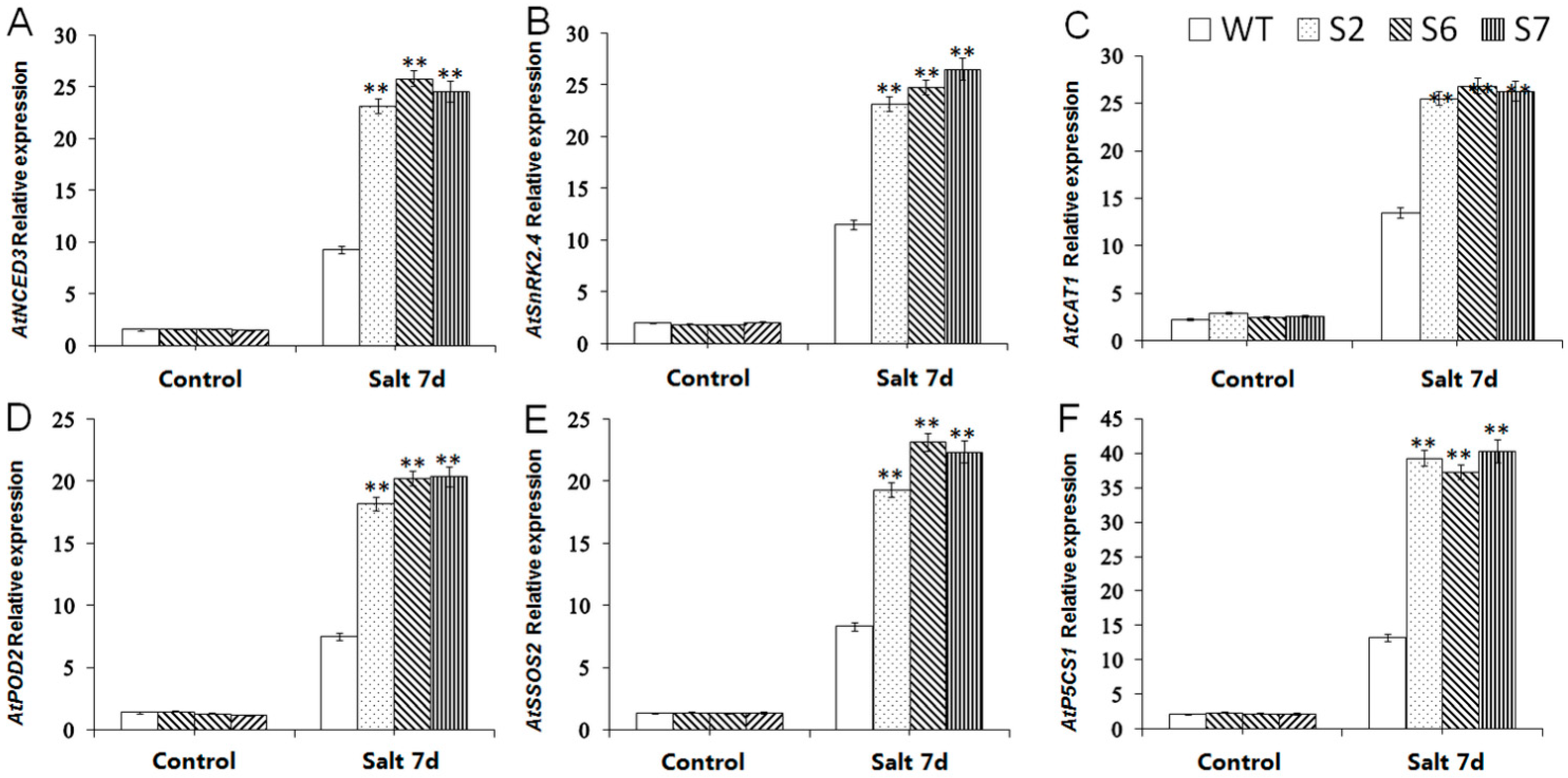
2.8. Expression Analysis of Salt-Resistant Downstream Genes in VvWRKY28-OE A. thaliana
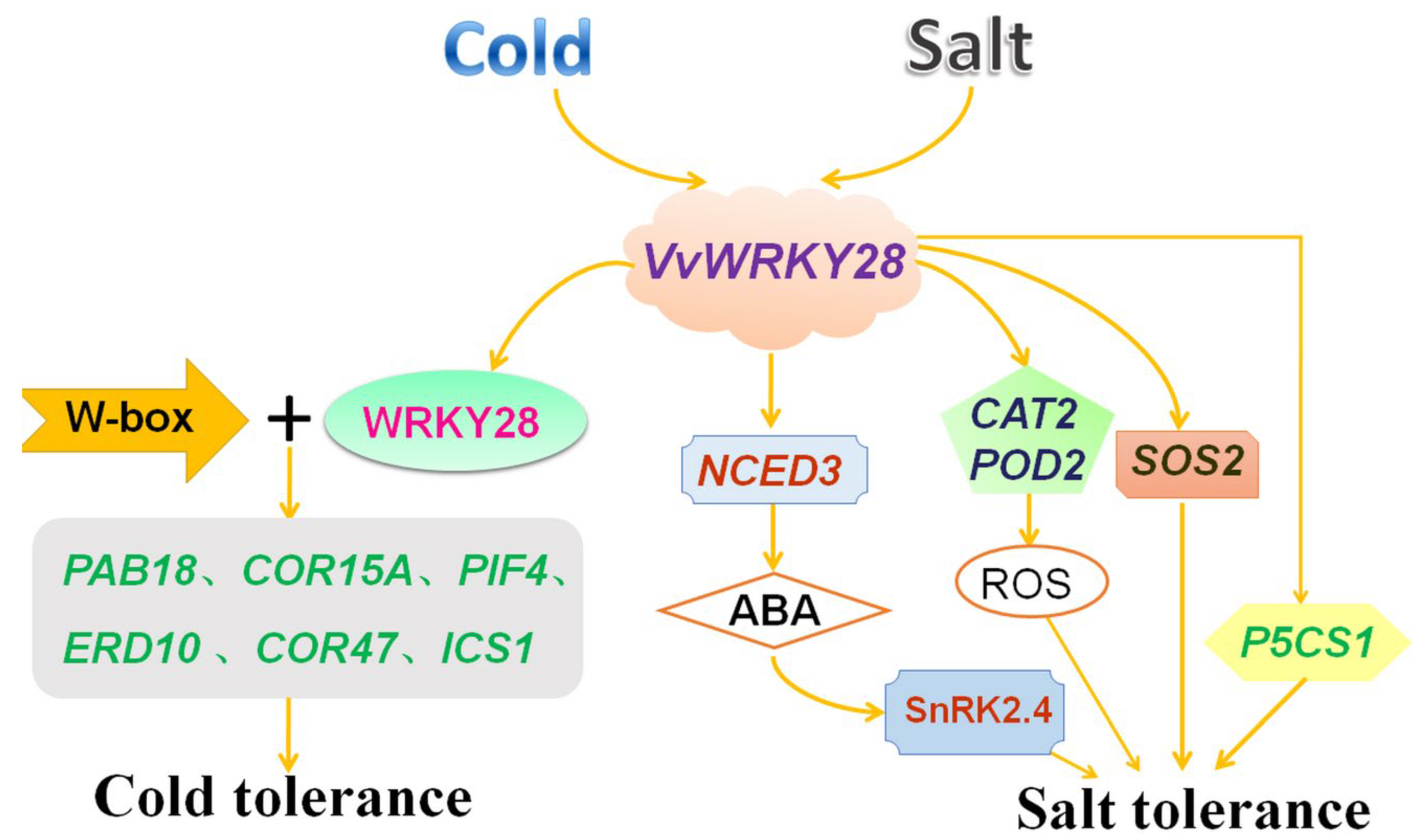
3. Discussion
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Cloning and qRT-PCR Expression Analysis of VvWRKY28
4.3. Bioinformatics Analysis of the VvWRKY28 Gene
4.4. Subcellular Localization Analysis of the VvWRKY28 Protein
4.5. Obtaining Transgenic A. thaliana
4.6. Determination of Relevant Physiological Indexes
4.7. Analysis of the Downstream Gene Expression of VvWRKY28
4.8. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ganapati, R.K.; Naveed, S.A.; Zafar, S.; Wang, W.S.; Xu, J. Saline-Alkali tolerancein Rice: Physiological Response, Molecular Mechanism, and QTL identification and application to breeding. Rice Sci. 2022, 29, 412–434. [Google Scholar] [CrossRef]
- Chen, K.; Tang, W.; Zhou, Y.; Chen, J.; Xu, Z.; Ma, R.; Dong, Y.; Ma, Y.; Chen, M. AP2/ERF transcription factor GmDREB1 confers drought tolerance in transgenic soybeanby interacting with GmERFs. Plant Physiol. Biochem. 2022, 170, 287–295. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Romero, I.; Alegria-Carrasco, E.; de Pradena, A.G.; Vazquez-Hernandez, M.; Escribano, M.I.; Merodio, C.; Sanchez-Ballesta, M.T. WRKY transcription factors in the response of table grapes (cv. Autumn royal) to high CO2 levels and low temperature. Postharvest Biol. Technol. 2019, 150, 42–51. [Google Scholar]
- Zhang, L.; Chen, L.; Yu, D. Transcription factor WRKY75 interacts with DELL Aproteinsto affect flowering. Plant Physiol. 2018, 176, 790–803. [Google Scholar] [CrossRef]
- Zhang, W.; Zhao, S.; Gu, S.; Cao, X.; Zhang, Y.; Niu, J.; Liu, L.; Li, A.; Jia, W.; Qi, B.; et al. FvWRKY48 binds to the pectate lyase FvPLA promoter to control fruit softening in Fragaria vesca. Plant Physiol. 2022, 189, 1037–1049. [Google Scholar] [CrossRef] [PubMed]
- Niu, F.; Cui, X.; Zhao, P.; Sun, M.; Yang, B.; Deyholos, M.K.; Li, Y.; Zhao, X.; Jiang, Y. WRKY42 transcription factor positively regulates leaf senescence through modulating SA and ROS synthesisin Arabidopsis thaliana. Plant J. 2020, 104, 171–184. [Google Scholar] [CrossRef]
- Heeraha, S.; Katarib, M.; Penjorb, R.; Coruzzib, G.; Marshall-Colona. WRKY1 mediates transcriptional regulation of light and nitrogen signaling pathways. Plant Physiol. 2019, 181, 1371–1388. [Google Scholar]
- Zhou, M.; Lu, Y.; Bethke, G.; Harrison, B.T.; Hatsugai, N.; Katagiri, F.; Glazebrook, J. WRKY70 prevents axenic activation of plant immunity by direct repression of SARD1. New Phytol. 2018, 217, 700–712. [Google Scholar] [CrossRef]
- Han, D.; Han, J.; Yang, G.; Wang, S.; Xu, T.; Li, W. An ERF transcription factor gene from Malus baccata (L.) borkh, MbERF11, affects cold and salt stress tolerancein Arabidopsis. Forests 2020, 11, 514. [Google Scholar] [CrossRef]
- Wu, X.; Chen, Q.; Chen, L.; Tian, F.; Chen, X.; Han, C.; Mi, J.; Lin, X.; Wan, X.; Jiang, B.; et al. A WRKY transcription factor, PyWRKY75, enhanced cadmium accumulation and tolerance in poplar. Ecotoxicol. Environ. Saf. 2022, 239, 113630. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Jiang, Y.; Guo, Y.; Huang, J.; Zhou, M.; Tang, Y.; Sui, J.; Wang, J.; Qiao, L. An ovelsaltinducible WRKY transcription factor gene, AhWRKY75,confers salt tolerance in transgenic peanut. Plant Physiol. Biochem. 2021, 160, 175–183. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhou, Z.; Du, M.; Li, T.; Wu, X.; Yu, J.; Zhang, P.; Yang, G. Overexpression of a Malus xiaojinensis WRKY transcription factor gene (MxWRKY55) increased iron and high salinity stress tolerancein Arabidopsis thaliana. In Vitro Cell. Dev. Biol.-Plant 2020, 56, 600–609. [Google Scholar] [CrossRef]
- Fan, C.; Yao, H.; Qiu, Z.; Ma, H.; Zeng, B. Genome-wide analysis of Eucalyptusgrandis WRKY genes family and their expression profiling in response to hormone and abiotic stress treatment. Gene 2018, 678, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Yu, H.; Yang, X.; Li, Q.; Ling, J.; Wang, H.; Gu, X.; Huang, S.; Jiang, W. CsWRKY46, a WRKY transcription factor from cucumber, confers cold resistance in transgenic-plant by regulating a set of cold-stress responsive genes in an ABA-dependent manner. Plant Physiol. Biochem. 2016, 108, 478–487. [Google Scholar] [CrossRef]
- Parvaiz, A.; Satyawati, S. Salt stress and phyto-biochemical responses of plants. Plant Soil Environ. 2008, 54, 89–99. [Google Scholar] [CrossRef]
- Yang, Z.; Chi, X.; Guo, F.; Jin, X.; Luo, H.; Hawar, A.; Chen, Y.; Feng, K.; Wang, B.; Qi, J.; et al. SbWRKY30 enhances the drought tolerance of plants and regulates a drought stress-responsive gene, SbRD19, insorghum. J. Plant Physiol. 2020, 246, 153142. [Google Scholar] [CrossRef]
- Fei, J.; Wang, Y.; Cheng, H.; Sun, F.; Sun, C. Comparative physiological and proteomicanalyses of mangrove plant Kandelia obovata under cold stress. Ecotoxicology 2021, 30, 1826–1840. [Google Scholar] [CrossRef]
- Cui, L.; Zheng, F.; Zhang, D.; Li, C.; Li, M.; Ye, J.; Zhang, Y.; Wang, T.; Bo, O.; Hong, Z.; et al. Tomato methionine sulfoxide reductase B2 functions in drought tolerance by promoting ROS scavenging and chlorophyll accumulation through interaction with Catalase2 and RBCS3B. Plant Sci. 2022, 318, 111206. [Google Scholar] [CrossRef]
- Han, D.; Hou, Y.; Wang, Y.; Ni, B.; Li, Z.; Yang, G. Overexpression of a Malus baccata WRKY transcription factor gene (MbWRKY5) increases drought and salt tolerance in transgenic tobacco. Can. J. Plant Sci. 2019, 99, 173–183. [Google Scholar] [CrossRef]
- Mhamdi, A.; Queval, G.; Chaouch, S.; Vanderauwera, S.; Breusegem, F.V.; Noctor, G. Catalase function in plants: a focus on Arabidopsis mutants as stress-mimic models. J. Exp. Bot. 2010, 61, 4197–4220. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.; Li, X.; Li, Y.; Yang, G.; Liu, W.; Shao, B.; Zhong, J.; Huang, P.; Han, D. Overexpression of a Malus baccata MYB transcription factor gene MbMYB4 increases cold and drought tolerancein Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 1794. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhang, Z.; Ding, H.; Wang, Y.; Liu, W.; Li, H.; Yang, G. Molecular cloning and function alanalysis of MbWRKY3 involved in improved drought tolerance in transformed tobacco. J. Plant Interact. 2018, 13, 329–337. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.X.J. WRKY transcription factors. Trends Plant Sci. 2010, 15, 247–258. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.; Rabara, R.C.; Rushton, P.J. Asystems biologype rspective on the role of WRKY transcription factors in drought responses in plants. Plant 2014, 239, 255–266. [Google Scholar] [CrossRef]
- Lin, H.X.; Yang, Y.Q.; Quan, R.D.; Mendoza, I.; Wu, Y.S.; Du, W.M.; Zhao, S.S.; Schumaker, K.S.; Pardo, J.M.; Guo, Y. Phosphorylation of SOS3-like calcium binding protein8 by SOS2 protein kinase stabilizes their protein complex and regulates salt tolerance in Arabidopsis. Plant Cell 2009, 21, 1607–1619. [Google Scholar] [CrossRef]
- Du, C.; Ma, B.; Wu, Z.; Li, N.; Zheng, L.; Wang, Y. Reaumuriatrigyna transcription factor RtWRKY23 enhances salt stress tolerance and delays flowering in plants. J. Plant Physiol. 2019, 239, 38–51. [Google Scholar] [CrossRef]
- Yan, Y.; Wei, C.; Zhang, W.; Cheng, H.; Liu, J. Cross-talk between calcium and reactive oxygen species signaling. Acta Pharmacol. Sin. 2006, 27, 821–826. [Google Scholar] [CrossRef]
- Zhu, J. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 53, 247–273. [Google Scholar] [CrossRef]
- Luo, P.; Li, Z.; Chen, W.; Xing, W.; Yang, J.; Cui, Y. Overexpression of RmICE1, a bHLH transcription factor from Rosa multiflora, enhances cold tolerance via modulating ROS levels and activating the expression of stress-responsive genes. Environ Exp. Bot. 2020, 178, 104160. [Google Scholar] [CrossRef]
- McLoughlin, F.; Galvan-Ampudia, C.S.; Julkowska, M.M.; Caarls, L.; Van Der Does, D.; Laurière, C.; Munnik, T.; Haring, M.A.; Testerink, C. The Snf1-related protein kinases SnRK2. 4 and SnRK2. 10 are involved in maintenance of root system architecture during salt stress. Plant J. 2012, 72, 436–449. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Li, S.; Ouyang, M.; Yang, L.; Sun, S.; Wang, Y.; Cai, X.; Wu, G.; Li, Y. Overexpression of watermelon ClWRKY20 in transgenic Arabidopsis improves salt and low-temperature tolerance. Sci. Hortic. 2022, 295, 110848. [Google Scholar] [CrossRef]
- Zha, Q.; Xi, X.; He, Y.; Yin, X.; Jiang, A. Interaction of VvbZIP60s and VvHSP83 in response to high-temperature stress in grapes. Gene 2022, 810, 146053. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Ding, H.; Chai, L.; Liu, W.; Zhang, Z.; Hou, Y.; Yang, G. Isolation and characterization of MbWRKY1, a WRKY transcription factor gene from Malus baccata (L.) borkh involved in drought tolerance. Can. J. Plant Sci. 2018, 98, 1023–1034. [Google Scholar] [CrossRef]
- Wan, Y.; Mao, M.; Wan, D.; Yang, Q.; Yang, F.; Li, G.; Wang, R. Identification of the WRKY gene family and functional analysis of two genes in Caragana intermedia. BMC Plant Biol. 2018, 18, 31. [Google Scholar] [CrossRef]
- Pandey, S.P.; Somssich, I.E. The role of WRKY transcription factors in plant immunity. Plant Physiol. 2009, 150, 1648–1655. [Google Scholar] [CrossRef]
- Han, D.; Hou, Y.; Ding, H.; Zhou, Z.; Li, H.; Yang, G. Isolation and preliminary functional analysis of MbWRKY4 gene involved in salt tolerance in transgenic tobacco. Int. J. Agric. Biol. 2018, 20, 2045–2052. [Google Scholar]
- He, G.; Xu, J.; Wang, Y.; Liu, J.; Li, P.; Chen, M.; Ma, Y.; Xu, Z. Drought-responsive WRKY transcription factor genes TaWRKY1 and TaWRKY33 from wheat confer drought and/or heat resistance in Arabidopsis. BMC Plant Biol. 2016, 16, 116. [Google Scholar] [CrossRef]
- Bankaji, I.; Sleimi, N.; Vives-Peris, V.; Gómez-Cadenas, A.; Pérez-Clemente, R.M. Identification and expression of the Cucurbita WRKY transcription factors in response to water deficit and salt stress. Sci. Hortic. 2019, 256, 108562. [Google Scholar] [CrossRef]
- Han, D.; Du, M.; Zhou, Z.; Wang, S.; Li, T.; Han, J.; Xu, T.; Yang, G. An NAC transcription factor gene from Malus baccata, MbNAC29, increases cold and high salinity tolerance in Arabidopsis. In Vitro Cell. Dev. Biol.-Plant 2020, 56, 588–599. [Google Scholar] [CrossRef]
- Ahmad, M.; Alabd, A.; Gao, Y.H.; Yu, W.J.; Jamil, W.; Wang, X.X.; Wei, J.; Ni, J.B.; Teng, Y.W.; Bai, S.L. Three stress-responsive NAC transcription factors, Pp-SNACs, differentially and synergistically regulate abiotic stress in pear. Sci. Hortic. 2022, 305, 111393. [Google Scholar] [CrossRef]
- Di Meo, S.; Reed, T.T.; Venditti, P.; Victor, V.M. Role of ROS and RNS sources in physiological and pathological conditions. Oxid. Med. Cell. Longev. 2016, 2016, 1245049. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Montagu, M.V.; Inze, D. Superoxide Dismutase and Stress Tolerance. Annu. Rev. Plant Phys. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Bakshi, A.; Moin, M.; Madhav, M.S.; Datla, R.; Kirti, P.B. Target of Rapamycin (TOR) negatively regulates chlorophyll degradation and lipid peroxidation and controls responses under abiotic stress in Arabidopsis thaliana. Plant Stress 2021, 2, 100020. [Google Scholar] [CrossRef]
- Evans, J.R. Photosynthesis and Nitrogen Relationships in Leaves of C₃ Plants. Oecologia 1989, 78, 9–19. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Morales, A.; Berjón, A.; Agüera, J. Hyperspectral indices and model simulation for chlorophyll estimation in open-canopy tree crops. Remote Sens. Environ. 2004, 90, 463–476. [Google Scholar] [CrossRef]
- Feng, Z.; He, G.; Zheng, W.; Lu, P.; Chen, M.; Gong, Y.; Ma, Y.; Xu, Z. Foxtail millet NF-Y families: Genome-wide survey and evolution analyses identified two functional genes important in abiotic stresses. Front. Plant Sci. 2015, 6, 1142. [Google Scholar] [CrossRef]
- Sun, C.; Zhao, H.; Zheng, C. Effects of NaCl stress on osmotic and proline metabolism in Ginkgo biloba seeding. J. Plant Physiol. 2017, 53, 470–476. [Google Scholar]
- Li, X.; Liang, X.; Li, W.; Yao, A.; Liu, W.; Wang, Y.; Yang, G.; Han, D. Isolation and functional analysis of MbCBF2, a Malus baccata (L.) Borkh CBF transcription factor gene, with functions in tolerance to cold and salt stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 9827. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, W.; Liu, J.; Song, S.; Hou, X.; Jia, C.; Li, J.; Miao, H.; Wang, Z.; Tie, W.; et al. An aquaporin gene MaPIP2-7 is involved in tolerance to drought, cold and salt stresses in transgenic banana (Musa acuminata L.). Plant Physiol. Biochem. 2020, 147, 66–76. [Google Scholar] [CrossRef]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. 2017, 132, 1–25. [Google Scholar] [CrossRef]
- Yu, S.; Lan, X.; Zhou, J.; Gao, K.; Zhong, C.; Xie, J. Dioscorea composita WRKY3 positively regulates salt-stress tolerance in transgenic Arabidopsis thaliana. J. Plant Physiol. 2022, 269, 153592. [Google Scholar] [CrossRef] [PubMed]
- Liang, X.; Li, Y.; Yao, A.; Liu, W.; Yang, T.; Zhao, M.; Zhang, B.; Han, D. Overexpression of MxbHLH18 increased iron and high salinity stress tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 8007. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Fang, L.; Karungo, S.; Zhang, L.; Gao, Y.; Li, S.; Xin, H. Overexpression of VaPAT1, a GRAS transcription factor from Vitisamurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2016, 35, 655–666. [Google Scholar] [CrossRef]
- Yang, Y.; Guang, Y.; Wang, F.; Chen, Y.; Yang, W.; Xiao, X.; Luo, S.; Zhou, Y. Characterization of Phytochrome-Interacting factor genes in pepper and functional analysis of CaPIF8 in cold and salt stress. Front. Plant Sci. 2021, 12, 746517. [Google Scholar] [CrossRef] [PubMed]
- Han, D.; Zhang, Z.; Ni, B.; Ding, H.; Liu, W.; Li, W.; Chai, L.; Yang, G. Isolation and functional analysis of MxNAS3 involved in enhanced iron stress tolerance and abnormal flower in transgenic Arabidopsis. J. Plant Interact. 2018, 13, 433–441. [Google Scholar] [CrossRef]
- Han, D.; Shi, Y.; Wang, B.; Liu, W.; Yu, Z.; Lv, B.; Yang, G. Isolation and preliminary functional analysis of MxCS2: a gene encoding a citrate synthase in Malus xiaojinensis. Plant Mol. Biol. Rep. 2015, 33, 133–142. [Google Scholar] [CrossRef]
- Han, D.; Han, J.; Xu, T.; Li, T.; Yao, C.; Wang, Y.; Luo, D.; Yang, G. Isolation and preliminary functional characterization of MxWRKY64, a new WRKY transcription factor gene from Malus xiaojinensis Cheng et Jiang. In Vitro Cell. Dev. Biol.-Plant 2021, 57, 202–213. [Google Scholar] [CrossRef]
- Yao, C.; Li, W.; Liang, X.; Ren, C.; Liu, W.; Yang, G.; Zhao, M.; Yang, T.; Li, X.; Han, D. Molecular cloning and characterization of MbMYB108, a Malus baccata MYB transcription factor gene, with functions in tolerance to cold and drought stress in transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 4846. [Google Scholar] [CrossRef]
- Lu, S.; Zhang, Y.; Zhu, K.; Yang, W.; Ye, J.; Chai, L.; Xu, Q.; Deng, X. The citrus transcription factor CsMADS6 modulates carotenoid metabolism by directly regulating carotenogenic genes. Plant Physiol. 2018, 176, 2657–2676. [Google Scholar] [CrossRef]
- Ou, L.; Wei, G.; Zhang, Z.; Dai, X.; Zou, X. Effects of low temperature and low irradiance on the physiological characteristics and related gene expression of different pepper species. Photosynthetica 2015, 53, 85–94. [Google Scholar] [CrossRef]
- Wang, Y.; Tian, S.; Xu, Y.; Qin, G.; Yao, H. Changes in the activities of pro-and anti-oxidant enzymes in peach fruit inoculated with Cryptococcus laurentii or Penicillium expansum at 0 or 20 °C. Postharvest Biol. Technol. 2004, 34, 21–28. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, Z.; Qu, M.; Cheng, F. Research on physicochemical properties, microscopic characterization and detection of different freezing-damaged corn seeds. Food Chem. 2022, 14, 100338. [Google Scholar] [CrossRef] [PubMed]
- Kavian, S.; Safarzadeh, S.; Yasrebi, J. Zinc improves growth and antioxidant enzyme activity in Aloe vera plant under salt stress. S. Afr. J. Bot. 2022, 147, 1221–1229. [Google Scholar] [CrossRef]
- Wang, Y.; Ye, Z.; Li, J.; Zhang, Y.; Guo, Y.; Cheng, J. Effects of dielectric barrier discharge cold plasma on the activity, structure and conformation of horseradish peroxidase (HRP) and on the activity of litchi peroxidase (POD). LWT-Food Sci. Technol. 2021, 141, 111078. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, W.; Liang, X.; Cai, W.; Wang, H.; Liu, X.; Cheng, L.; Song, P.; Luo, G.; Han, D. Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 13418. https://doi.org/10.3390/ijms232113418
Liu W, Liang X, Cai W, Wang H, Liu X, Cheng L, Song P, Luo G, Han D. Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(21):13418. https://doi.org/10.3390/ijms232113418
Chicago/Turabian StyleLiu, Wei, Xiaoqi Liang, Weijia Cai, Hao Wang, Xu Liu, Longfei Cheng, Penghui Song, Guijie Luo, and Deguo Han. 2022. "Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 21: 13418. https://doi.org/10.3390/ijms232113418
APA StyleLiu, W., Liang, X., Cai, W., Wang, H., Liu, X., Cheng, L., Song, P., Luo, G., & Han, D. (2022). Isolation and Functional Analysis of VvWRKY28, a Vitis vinifera WRKY Transcription Factor Gene, with Functions in Tolerance to Cold and Salt Stress in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences, 23(21), 13418. https://doi.org/10.3390/ijms232113418




