Genome-Wide Analysis of SQUAMOSA-Promoter-Binding Protein-like Family in Flowering Pleioblastus pygmaeus
Abstract
1. Introduction
2. Results
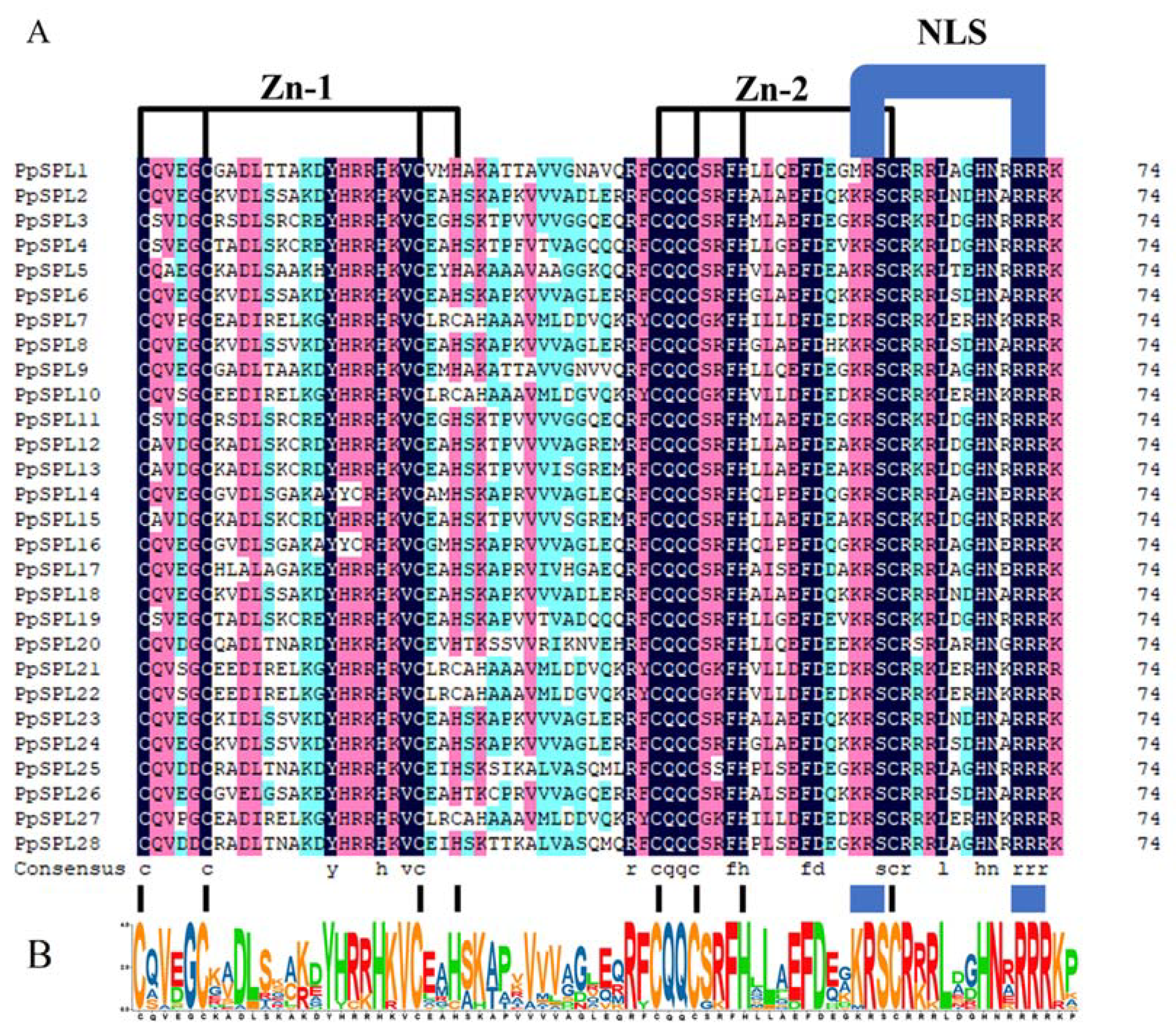
2.1. Identification and Sequence Analysis of SPL Genes from Pleioblastus pygmaeus
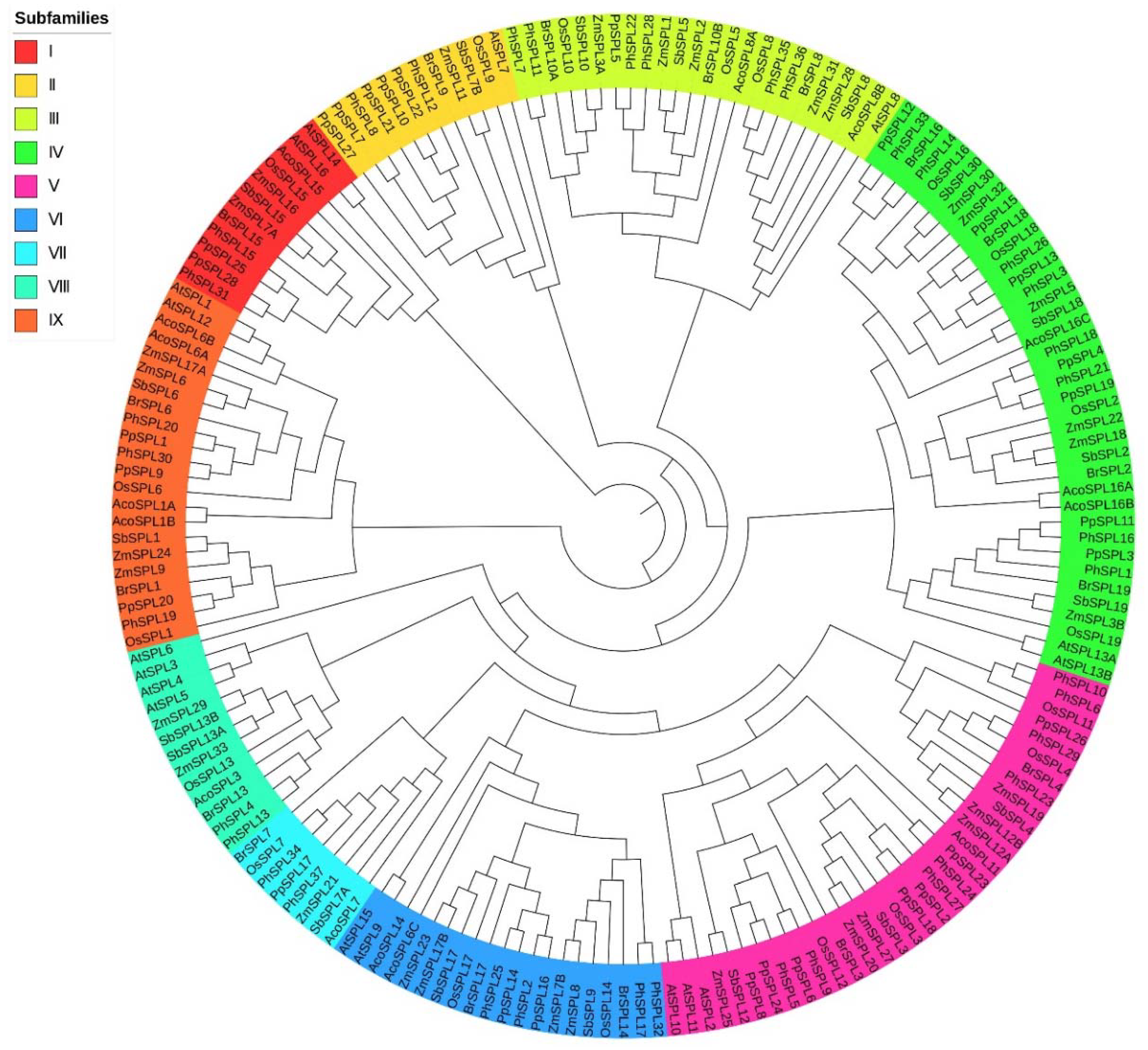
2.2. Phylogenetic Analysis of SPL Family Genes
2.3. Homologous Analysis of SPL Family Genes
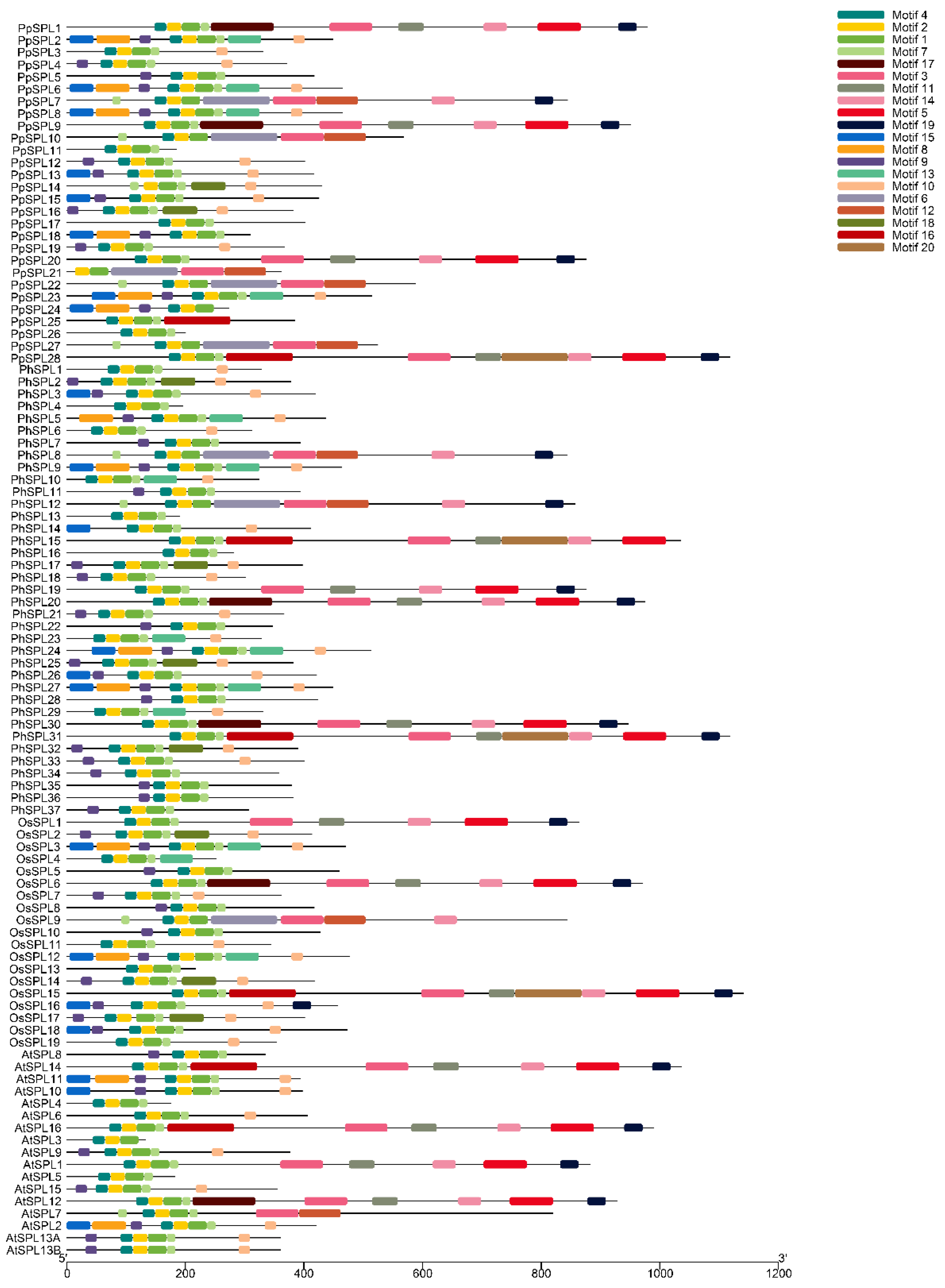
2.4. Motif Analysis of SPL Family Genes
2.5. miRNA Target Analysis
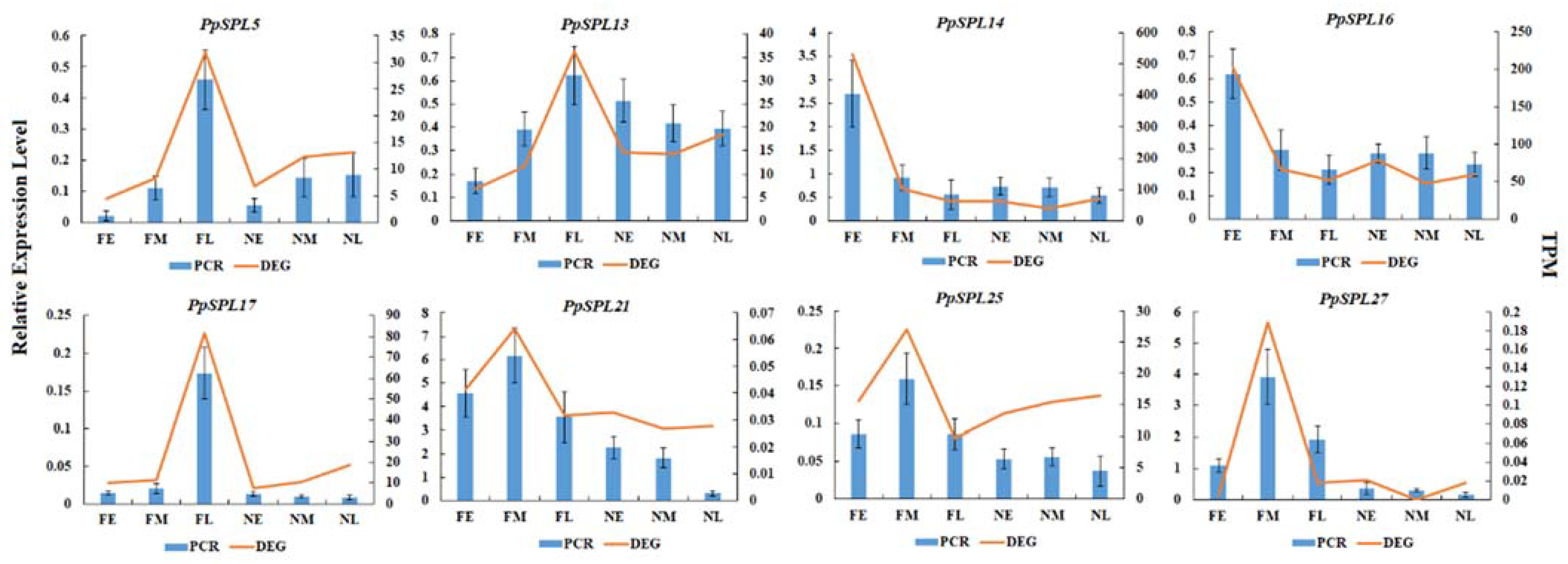
2.6. Expression Pattern Analysis of PpSPL Genes
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Library Preparation and Transcriptome Assembly
4.3. Identification and Sequence Analysis of SPL Family Genes from P. pygmaeu
4.4. Multiple Alignment and Phylogenetic Analysis of SPL Family Genes
4.5. Calculation of Equivalent (Ks) and Non-Equivalent (Ka) Substitutions
4.6. MiRNA-Targeting PpSPLs Prediction
4.7. Expression Pattern Analysis of PpSPLs by RNA-Seq
4.8. RT-qPCR Verification of PpSPLs
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liese, W.; Köhl, M. Bamboo: The Plant and its Uses; Springer: Heidelberg, Germany, 2015. [Google Scholar]
- Song, X.; Zhou, G.; Jiang, H.; Yu, S.; Fu, J.; Li, W.; Wang, W.; Ma, Z.; Peng, C. Carbon sequestration by Chinese bamboo forests and their ecological benefits: Assessment of potential, problems, and future challenges. Environ. Rev. 2011, 19, 418–428. [Google Scholar] [CrossRef]
- Zheng, X.; Lin, S.Y.; Fu, H.J.; Wan, Y.W.; Ding, Y.L. The bamboo flowering cycle sheds light on flowering diversity. Front. Plant Sci. 2020, 11, 381. [Google Scholar] [CrossRef] [PubMed]
- Biswas, P.; Chakraborty, S.; Dutta, S.; Pal, A.; Das, M. Bamboo flowering from the perspective of comparative genomics and transcriptomics. Front. Plant Sci. 2016, 7, 1900. [Google Scholar] [CrossRef] [PubMed]
- Sertse, D.; Disasa, T.; Bekele, K.; Alebachew, M.; Kebede, Y.; Eshete, N.; Eshetu, S. Mass flowering and death of bamboo: A potential threat to biodiversity and livelihoods in Ethiopia. J. Biodivers. Environ. Sci. 2011, 1, 16–25. [Google Scholar]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.C. General aspects of plant transcription factor families. In Plant Transcription Factors; Academic Press: Cambridge, MA, USA, 2016; pp. 35–56. [Google Scholar]
- Fryxell, K.J. The coevolution of gene family trees. Trends Genet. 1996, 12, 364–369. [Google Scholar] [CrossRef]
- Hahn, M.W.; De Bie, T.; Stajich, J.E.; Nguyen, C.; Cristianini, N. Estimating the tempo and mode of gene family evolution from comparative genomic data. Genome Res. 2005, 15, 1153–1160. [Google Scholar] [CrossRef] [PubMed]
- Annilo, T.; Chen, Z.-Q.; Shulenin, S.; Costantino, J.; Thomas, L.; Lou, H.; Stefanov, S.; Dean, M. Evolution of the vertebrate ABC gene family: Analysis of gene birth and death. Genomics 2006, 88, 1–11. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E.; et al. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Shikata, M.; Koyama, T.; Mitsuda, N.; Ohme-Takagi, M. Arabidopsis SBP-box genes SPL10, SPL11 and SPL2 control morphological change in association with shoot maturation in the reproductive phase. Plant Cell Physiol. 2009, 50, 2133–2145. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. MGG 1996, 250, 7–16. [Google Scholar] [PubMed]
- Preston, J.C.; Hileman, L.C. Functional evolution in the plant Squamosa-Promoter Binding Protein-Like (SPL) gene family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed]
- Riese, M.; Höhmann, S.; Saedler, H.; Münster, T.; Huijser, P. Comparative analysis of the SBP-box gene families in P. patens and seed plants. Gene 2007, 401, 28–37. [Google Scholar] [CrossRef]
- Zhang, W.; Bei, L.I.; Bin, Y.U. Genome-wide identification, phylogeny and expression analysis of the SBP-box gene family in maize (Zea mays). J. Integr. Agric. 2016, 15, 29–41. [Google Scholar] [CrossRef]
- Xie, K.; Wu, C.; Xiong, L. Genomic organization, differential expression, and interaction of SQUAMOSA promoter-binding-like transcription factors and microRNA156 in rice. Plant Physiol. 2006, 142, 280–293. [Google Scholar] [CrossRef]
- Ali, H.; Liu, Y.; Azam, S.M.; Priyadarshani, S.V.G.N.; Li, W.; Huang, X.; Hu, B.; Xiong, J.; Ali, U.; Qin, Y. Genomic survey, characterization, and expression profile analysis of the SBP genes in pineapple (Ananas comosus L.). Int. J. Genom. 2017, 2017, 1032846. [Google Scholar] [CrossRef]
- Tripathi, R.K.; Overbeek, W.; Singh, J. Global analysis of SBP gene family in Brachypodium distachyon reveals its association with spike development. Sci. Rep. 2020, 10, 1–15. [Google Scholar] [CrossRef]
- Chang, J.; Yan, F.; Qiao, L.; Zheng, J.; Zhang, F.; Liu, Q. Genome-wide identification and expression analysis of SBP-box gene family in Sorghum bicolor L. Yi Chuan=Hered. 2016, 38, 569–580. [Google Scholar]
- Pan, F.; Wang, Y.; Liu, H.; Wu, M.; Chu, W.; Chen, D.; Xiang, Y. Genome-wide identification and expression analysis of SBP-like transcription factor genes in Moso Bamboo (Phyllostachys edulis). BMC Genom. 2017, 18, 1–17. [Google Scholar] [CrossRef]
- Xing, S.; Salinas, M.; Höhmann, S.; Berndtgen, R.; Huijser, P. miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 2010, 22, 3935–3950. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Kohalmi, S.E.; Amyot, L.; Hannoufa, A. SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 2 controls floral organ development and plant fertility by activating ASYMMETRIC LEAVES 2 in Arabidopsis thaliana. Plant Mol. Biol. 2016, 92, 661–674. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wu, K.; Yuan, Q.; Liu, X.; Liu, Z.; Lin, X.; Zeng, R.; Zhu, H.; Dong, G.; Qian, Q.; et al. Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 2012, 44, 950–954. [Google Scholar] [CrossRef] [PubMed]
- Xing, S.; Salinas, M.; Garcia-Molina, A.; Höhmann, S.; Berndtgen, R.; Huijser, P. SPL8 and miR156-targeted SPL genes redundantly regulate Arabidopsis gynoecium differential atterning. Plant J. 2013, 75, 566–577. [Google Scholar] [CrossRef] [PubMed]
- Chao, L.M.; Liu, Y.Q.; Chen, D.Y.; Xue, X.Y.; Mao, Y.B.; Chen, X.Y. Arabidopsis transcription factors SPL1 and SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef]
- Bäurle, I.; Dean, C. The timing of developmental transitions in plants. Cell 2006, 125, 655–664. [Google Scholar] [CrossRef]
- Wang, J.W.; Czech, B.; Weigel, D. miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 2009, 138, 738–749. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Wu, M.F.; Yang, L.; Wu, G.; Poethig, R.S.; Wagner, D. The microRNA-regulated SBP-Box transcription factor SPL3 is a direct upstream activator of LEAFY, FRUITFULL, and APETALA1. Dev. Cell 2009, 17, 268–278. [Google Scholar] [CrossRef]
- Jung, J.H.; Lee, H.J.; Ryu, J.Y.; Park, C.M. SPL3/4/5 integrate developmental aging and photoperiodic signals into the FT-FD module in Arabidopsis flowering. Mol. Plant 2016, 9, 1647–1659. [Google Scholar] [CrossRef]
- Wang, J.W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef]
- Gandikota, M.; Birkenbihl, R.P.; Höhmann, S.; Cardon, G.H.; Saedler, H.; Huijser, P. The miRNA156/157 recognition element in the 3′ UTR of the Arabidopsis SBP box gene SPL3 prevents early flowering by translational inhibition in seedlings. Plant J. 2007, 49, 683–693. [Google Scholar] [CrossRef]
- Gao, R.; Wang, Y.; Gruber, M.Y.; Hannoufa, A. miR156/SPL10 modulates lateral root development, branching and leaf morphology in Arabidopsis by silencing AGAMOUS-LIKE 79. Front. Plant Sci. 2018, 8, 2226. [Google Scholar] [CrossRef] [PubMed]
- Yao, W.J.; Li, C.Z.; Lin, S.Y.; Wang, J.P.; Fan, T.T.; Zhao, W.Q. The structures of floral organs and reproductive characteristics of ornamental bamboo species, Pleioblastus pygmaeus. Hortic. Plant J. 2022. [Google Scholar] [CrossRef]
- Yao, W.; Li, C.; Lin, S.; Ren, L.; Wan, Y.; Zhang, L.; Ding, Y. Morphological characteristics and transcriptome comparisons of the shoot buds from flowering and non-flowering Pleioblastus pygmaeus. Forests 2020, 11, 1229. [Google Scholar] [CrossRef]
- Feng, X.; Wang, Y.; Zhang, N.; Gao, S.; Wu, J.; Liu, R.; Huang, Y.; Zhang, J.; Qi, Y. Comparative phylogenetic analysis of CBL reveals the gene family evolution and functional divergence in Saccharum spontaneum. BMC Plant Biol. 2021, 21, 1–14. [Google Scholar] [CrossRef]
- Li, B.; Zhao, Y.; Wang, S.; Zhang, X.; Wang, Y.; Shen, Y.; Yuan, Z. Genome-wide identification, gene cloning, subcellular location and expression analysis of SPL gene family in P. granatum L. BMC Plant Biol. 2021, 21, 1–19. [Google Scholar] [CrossRef]
- Yu, N.; Yang, J.C.; Yin, G.T.; Li, R.S.; Zou, W.T. Genome-wide characterization of the SPL gene family involved in the age development of Jatropha curcas. BMC Genom. 2020, 21, 1–14. [Google Scholar] [CrossRef]
- Padmanabhan, M.S.; Ma, S.; Burch-Smith, T.M.; Czymmek, K.; Huijser, P.; Dinesh-Kumar, S.P. Novel positive regulatory role for the SPL6 transcription factor in the NTIR-NB-LRR receptor-mediated plant innate immunity. PLoS Pathog. 2013, 9, e1003235. [Google Scholar] [CrossRef]
- Martin, R.C.; Asahina, M.; Liu, P.P.; Kristof, J.R.; Coppersmith, J.L.; Pluskota, W.E.; Bassel, G.W.; Goloviznina, N.A.; Nguyen, T.T.; Martínez-Andújar, C.; et al. The regulation of post-germinative transition from the cotyledon-to vegetative-leaf stages by microRNA-targeted SQUAMOSA PROMOTER-BINDING PROTEIN LIKE13 in Arabidopsis. Seed Sci. Res. 2010, 20, 89–96. [Google Scholar] [CrossRef]
- Yamasaki, H.; Hayashi, M.; Fukazawa, M.; Kobayashi, Y.; Shikanai, T. SQUAMOSA promoter binding protein-like7 is a central regulator for copper homeostasis in Arabidopsis. Plant Cell 2009, 21, 347–361. [Google Scholar] [CrossRef]
- Stone, J.M.; Liang, X.; Nekl, E.R.; Stiers, J.J. Arabidopsis AtSPL14, a plant-specific SBP-domain transcription factor, participates in plant development and sensitivity to fumonisin B1. Plant J. 2005, 41, 744–754. [Google Scholar] [CrossRef]
- Guo, A.Y.; Zhu, Q.H.; Gu, X.; Ge, S.; Yang, J.; Luo, J. Genome-wide identification and evolutionary analysis of the plant specific SBP-box transcription factor family. Gene 2008, 418, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Unte, U.S.; Sorensen, A.-M.; Pesaresi, P.; Gandikota, M.; Leister, D.; Saedler, H.; Huijser, P. SPL8, an SBP-box gene that affects pollen sac development in Arabidopsis. Plant Cell 2003, 15, 1009–1019. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, S.; Grande, A.V.; Bujdoso, N.; Saedler, H.; Huijser, P. The microRNA regulated SBP-box genes SPL9 and SPL15 control shoot maturation in Arabidopsis. Plant Mol. Biol. 2008, 67, 183–195. [Google Scholar] [CrossRef] [PubMed]
- Morea, E.G.O.; da Silva, E.M.; Valente, G.T.; Barrera Rojas, C.H.; Vincentz, M.; Nogueira, F.T.S. Functional and evolutionary analyses of the miR156 and miR529 families in land plants. BMC Plant Biol. 2016, 16, 1–13. [Google Scholar]
- Chen, Y.; Song, W.; Xie, X.; Wang, Z.; Guan, P.; Peng, H.; Jiao, Y.; Ni, Z.; Sun, Q.; Guo, W. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the triticeae tribe as a pilot practice in the plant pangenomic era. Mol. Plant 2020, 13, 1694–1708. [Google Scholar] [CrossRef]
- Zhong, H.; Kong, W.; Gong, Z.; Fang, X.; Deng, X.; Liu, C.; Li, Y. Evolutionary Analyses Reveal Diverged Patterns of SQUAMOSA Promoter Binding Protein-Like (SPL) Gene Family in Oryza Genus. Front. Plant Sci. 2019, 10, 565. [Google Scholar] [CrossRef]
- Yuan, H.; Qin, P.; Hu, L.; Zhan, S.; Wang, S.; Gao, P.; Li, J.; Jin, M.; Xu, Z.; Gao, Q.; et al. OsSPL18 controls grain weight and grain number in rice. J. Genet. Genom. 2019, 46, 41–51. [Google Scholar] [CrossRef]
- Yang, Z.; Wang, X.; Gu, S.; Hu, Z.; Xu, H.; Xu, C. Comparative study of SBP-box gene family in Arabidopsis and rice. Gene 2008, 407, 1–11. [Google Scholar] [CrossRef]
- Shao, F.; Lu, Q.; Wilson, I.W.; Qiu, D. Genome-wide identification and characterization of the SPL gene family in Ziziphus jujuba. Gene 2017, 627, 315–321. [Google Scholar] [CrossRef]
- Jiang, M.; He, Y.; Chen, X.; Zhang, X.; Guo, Y.; Yang, S.; Huang, J.; Traw, M.B. CRISPR-based assessment of genomic structure in the conserved SQUAMOSA promoter-binding-like gene clusters in rice. Plant J. 2020, 104, 1301–1314. [Google Scholar] [CrossRef]
- Yan, Y.; Wei, M.; Li, Y.; Tao, H.; Wu, H.; Chen, Z.; Li, C.; Xu, J.-H. MiR529a controls plant height, tiller number, panicle architecture and grain size by regulating SPL target genes in rice (Oryza sativa L.). Plant Sci. 2021, 302, 110728. [Google Scholar] [CrossRef] [PubMed]
- Dai, Z.; Wang, J.; Yang, X.; Lu, H.; Miao, X.; Shi, Z. Modulation of plant architecture by the miR156f-OsSPL7-OsGH3. 8 pathway in rice. J. Exp. Bot. 2018, 69, 5117–5130. [Google Scholar] [CrossRef] [PubMed]
- Jiao, Y.; Wang, Y.; Xue, D.; Wang, J.; Yan, M.; Liu, G.; Dong, G.; Zeng, D.; Lu, Z.; Zhu, X.; et al. Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 2010, 42, 541–544. [Google Scholar] [CrossRef] [PubMed]
- Miura, K.; Ikeda, M.; Matsubara, A.; Song, X.-J.; Ito, M.; Asano, K.; Matsuoka, M.; Kitano, H.; Ashikari, M. OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 2010, 42, 545–549. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Huang, L.; Chen, G.; Liu, H.; Zhang, Y.; Zhang, R.; Zhang, S.; Liu, J.; Hu, Q.; Hu, F.; et al. The elite alleles of OsSPL4 regulate grain size and increase grain yield in rice. Rice 2021, 14, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Chen, W.; Yang, W.; Li, X.; Zhang, C.; Zhang, X.; Zheng, L.; Zhu, X.; Yin, J.; Qin, P.; et al. OsSPL9 regulates grain number and grain yield in rice. Front. Plant Sci. 2021, 12, 682018. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Waheed, S.; Zeng, L. The critical role of miRNAs in regulation of flowering time and flower development. Genes 2020, 11, 319. [Google Scholar] [CrossRef]
- Yu, N.; Niu, Q.W.; Ng, K.H.; Chua, N.H. The role of miR156/SPLs modules in Arabidopsis lateral root development. Plant J. 2015, 83, 673–685. [Google Scholar] [CrossRef]
- Zheng, C.; Ye, M.; Sang, M.; Wu, R. A regulatory network for miR156-SPL module in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 6166. [Google Scholar] [CrossRef]
- Li, C.; Lu, S. Molecular characterization of the SPL gene family in Populus trichocarpa. BMC Plant Biol. 2014, 14, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, A.; Abe, M. Regulation of reproductive development by non-coding RNA in Arabidopsis: To flower or not to flower. J. Plant Res. 2012, 6, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Blanc, G.; Wolfe, K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 2004, 16, 1679–1691. [Google Scholar] [CrossRef] [PubMed]
- Guo, Q.; Li, L.; Zhao, K.; Yao, W.; Cheng, Z.; Zhou, B.; Jiang, T. Genome-wide analysis of poplar SQUAMOSA-promoter-binding protein (SBP) family under salt stress. Forests 2021, 12, 413. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yao, W.; Li, C.; Fu, H.; Yang, M.; Wu, H.; Ding, Y.; Li, L.; Lin, S. Genome-Wide Analysis of SQUAMOSA-Promoter-Binding Protein-like Family in Flowering Pleioblastus pygmaeus. Int. J. Mol. Sci. 2022, 23, 14035. https://doi.org/10.3390/ijms232214035
Yao W, Li C, Fu H, Yang M, Wu H, Ding Y, Li L, Lin S. Genome-Wide Analysis of SQUAMOSA-Promoter-Binding Protein-like Family in Flowering Pleioblastus pygmaeus. International Journal of Molecular Sciences. 2022; 23(22):14035. https://doi.org/10.3390/ijms232214035
Chicago/Turabian StyleYao, Wenjing, Chuanzhe Li, Huajun Fu, Meng Yang, Hongyu Wu, Yulong Ding, Long Li, and Shuyan Lin. 2022. "Genome-Wide Analysis of SQUAMOSA-Promoter-Binding Protein-like Family in Flowering Pleioblastus pygmaeus" International Journal of Molecular Sciences 23, no. 22: 14035. https://doi.org/10.3390/ijms232214035
APA StyleYao, W., Li, C., Fu, H., Yang, M., Wu, H., Ding, Y., Li, L., & Lin, S. (2022). Genome-Wide Analysis of SQUAMOSA-Promoter-Binding Protein-like Family in Flowering Pleioblastus pygmaeus. International Journal of Molecular Sciences, 23(22), 14035. https://doi.org/10.3390/ijms232214035






