Time-Resolved Proteomics of Germinating Spores of Bacillus cereus
Abstract
1. Introduction
2. Results
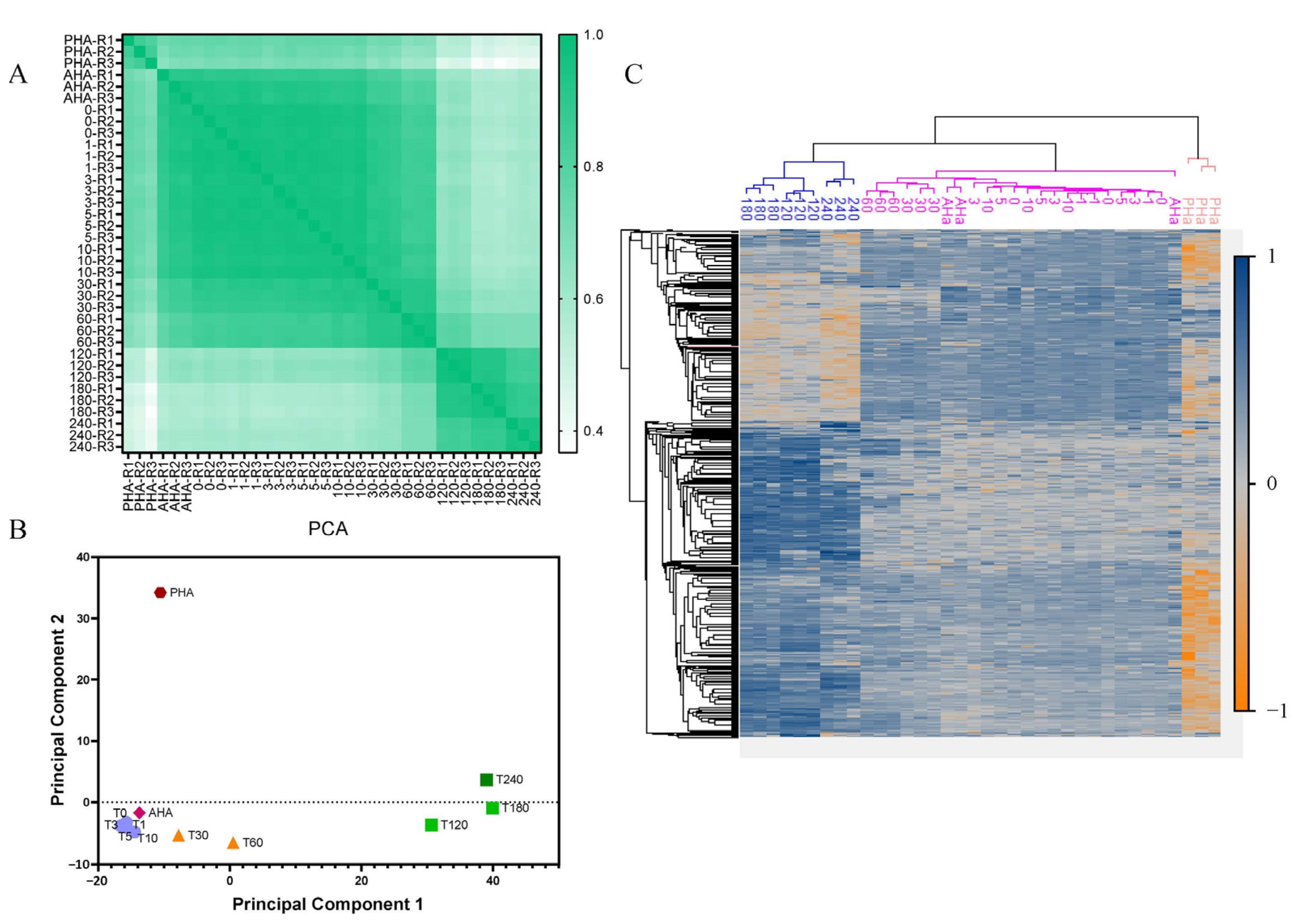
2.1. Morphological and Proteomic Studies on Spores during Germination and Outgrowth
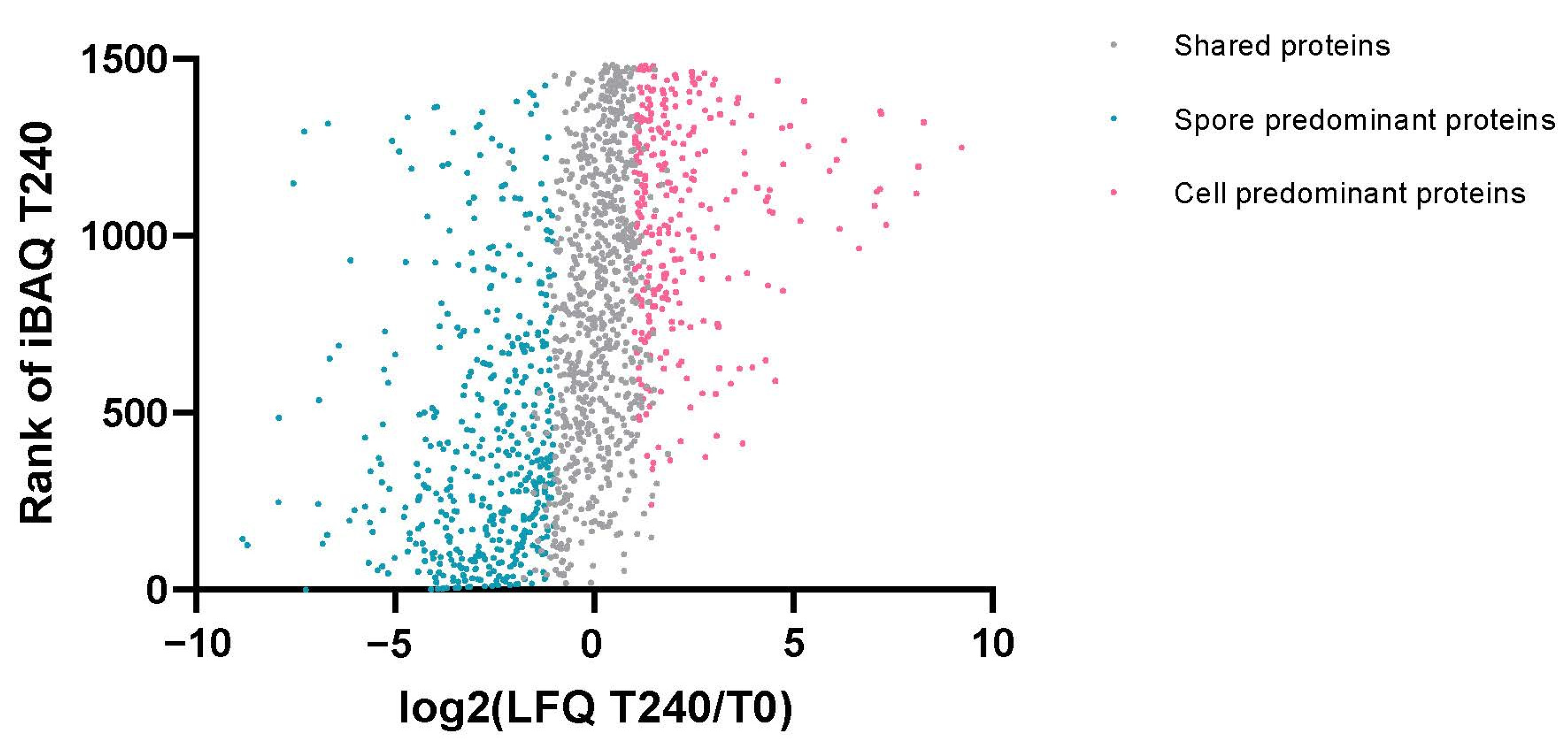
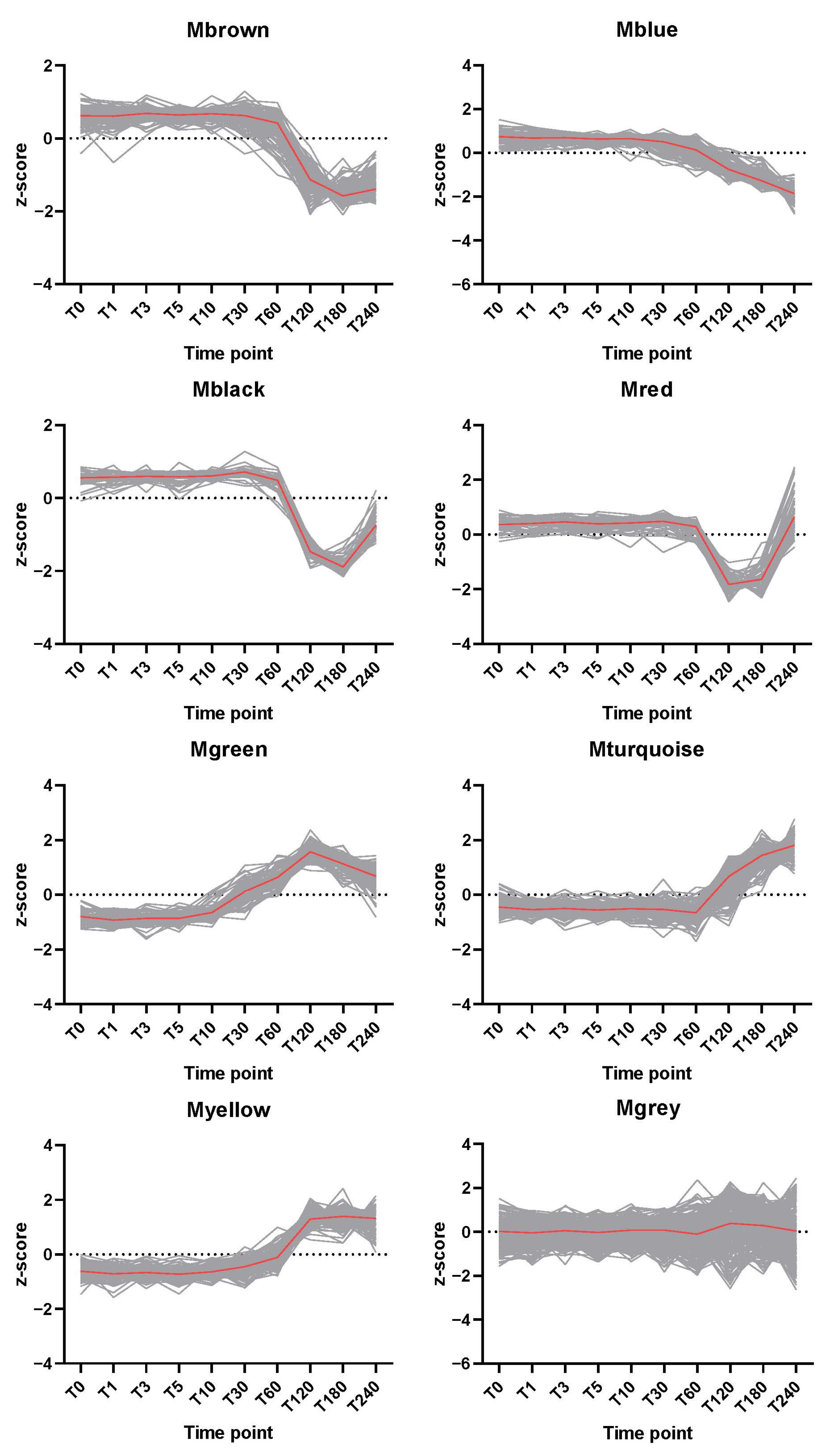
2.2. Dynamic Changes of Spore Specific Proteins
2.3. Co-Expression Network Construction
3. Discussion
4. Materials and Methods
4.1. Morphological and Proteomic Studies on Spores during Germination and Outgrowth
4.2. Sample Preparation and LC-MS/MS Analysis
4.3. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Christie, G.; Setlow, P. Bacillus spore germination: Knowns, unknowns and what we need to learn. Cell. Signal. 2020, 74, 109729. [Google Scholar] [CrossRef] [PubMed]
- Paredes-Sabja, D.; Setlow, P.; Sarker, M.R. Germination of spores of Bacillales and Clostridiales species: Mechanisms and proteins involved. Trends Microbiol. 2011, 19, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Swarge, B.; Abhyankar, W.; Jonker, M.; Hoefsloot, H.; Kramer, G.; Setlow, P.; Brul, S.; de Koning, L.J. Integrative Analysis of Proteome and Transcriptome Dynamics during Bacillus subtilis Spore Revival. mSphere 2020, 5, e00463-20. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Driks, A. Contrasting evolutionary patterns of spore coat proteins in two Bacillus species groups are linked to a difference in cellular structure. BMC Evol. Biol. 2013, 13, 261. [Google Scholar] [CrossRef] [PubMed]
- Soni, A.; Oey, I.; Silcock, P.; Bremer, P.J. Impact of temperature, nutrients, pH and cold storage on the germination, growth and resistance of Bacillus cereus spores in egg white. Food Res. Int. 2018, 106, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Jagtap, P.; Michailidis, G.; Zielke, R.; Walker, A.K.; Patel, N.; Strahler, J.R.; Driks, A.; Andrews, P.C.; Maddock, J.R. Early events of Bacillus anthracis germination identified by time-course quantitative proteomics. Proteomics 2006, 6, 5199–5211. [Google Scholar] [CrossRef] [PubMed]
- Arrieta-Ortiz, M.L.; Hafemeister, C.; Bate, A.R.; Chu, T.; Greenfield, A.; Shuster, B.; Barry, S.N.; Gallitto, M.; Liu, B.; Kacmarczyk, T.; et al. An experimentally supported model of the Bacillus subtilis global transcriptional regulatory network. Mol. Syst. Biol. 2015, 11, 839. [Google Scholar] [CrossRef]
- Bagyan, I.; Setlow, P.; Fau-Setlow, B.; Setlow, P. New small, acid-soluble proteins unique to spores of Bacillus subtilis: Identification of the coding genes and regulation and function of two of these genes. J. Bacteriol. 1998, 180, 6704–6712. [Google Scholar] [CrossRef]
- Pei, G.; Chen, L.; Zhang, W. WGCNA application to proteomic and metabolomic data analysis. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2017; Volume 585, pp. 135–158. [Google Scholar]
- Li, H.; Li, J.; Dong, Y.; Hao, H.; Ling, Z.; Bai, H.; Wang, H.; Cui, H.; Shi, L. Time-series transcriptome provides insights into the gene regulation network involved in the volatile terpenoid metabolism during the flower development of lavender. BMC Plant Biol. 2019, 19, 313. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef]
- Fernández, A.; Ocio, M.; Fernández, P.; Martínez, A. Effect of heat activation and inactivation conditions on germination and thermal resistance parameters of Bacillus cereus spores. Int. J. Food Microbiol. 2001, 63, 257–264. [Google Scholar] [CrossRef]
- Soni, A.; Oey, I.; Silcock, P.; Permina, E.; Bremer, P.J. Differential gene expression for investigation of the effect of germinants and heat activation to induce ger-mination in Bacillus cereus spores. Food Res. Int. 2019, 119, 462–468. [Google Scholar] [CrossRef] [PubMed]
- Hornstra, L.M.; de Vries, Y.P.; Wells-Bennik, M.H.J.; de Vos, W.M.; Abee, T. Characterization of Germination Receptors of Bacillus cereus ATCC. Appl. Environ. Microbiol. 2006, 72, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Setlow, P.; Li, Y. Characterization of single heat-activated Bacillus spores using laser tweezers Raman spectroscopy. Opt. Express 2009, 17, 16480. [Google Scholar] [CrossRef]
- Hoch, J.A. A Life in Bacillus subtilis Signal Transduction. Annu. Rev. Microbiol. 2017, 71, 1–19. [Google Scholar] [CrossRef]
- Miethke, M.; Schmidt, S.; Marahiel, M.A. The Major Facilitator Superfamily-Type Transporter YmfE and the Multidrug-Efflux Activator Mta Mediate Bacillibactin Secretion in Bacillus subtilis. J. Bacteriol. 2008, 190, 5143–5152. [Google Scholar] [CrossRef]
- Driks, A.; Eichenberger, P. The spore coat. In The Bacterial Spore: From Molecules to Systems; Wiley Online Library: Hoboken, NJ, USA, 2016; pp. 179–200. [Google Scholar]
- Isticato, R.; Sirec, T.; Giglio, R.; Baccigalupi, L.; Rusciano, G.; Pesce, G.; Zito, G.; Sasso, A.; De Felice, M.; Ricca, E. Correction: Flexibility of the Programme of Spore Coat Formation in Bacillus subtilis: Bypass of CotE Requirement by Over-Production of CotH. PLoS ONE 2013, 8, e74949. [Google Scholar] [CrossRef]
- Henriques, A.O.; Moran, J.C.P. Structure, Assembly, and Function of the Spore Surface Layers. Annu. Rev. Microbiol. 2007, 61, 555–588. [Google Scholar] [CrossRef]
- Saggese, A.; Scamardella, V.; Sirec, T.; Cangiano, G.; Isticato, R.; Pane, F.; Amoresano, A.; Ricca, E.; Baccigalupi, L. Antagonistic Role of CotG and CotH on Spore Germination and Coat Formation in Bacillus subtilis. PLoS ONE 2014, 9, e104900. [Google Scholar]
- Ramaniuk, O.; Černý, M.; Krásný, L.; Vohradský, J. Kinetic modelling and meta-analysis of the B. subtilis SigA regulatory network during spore germination and outgrowth. Biochim. et Biophys. Acta (BBA)-Gene Regul. Mech. 2017, 1860, 894–904. [Google Scholar] [CrossRef]
- Eijlander, R.T.; Holsappel, S.; De Jong, A.; Ghosh, A.; Christie, G.; Kuipers, O.P. SpoVT: From Fine-Tuning Regulator in Bacillus subtilis to Essential Sporulation Protein in Bacillus cereus. Front. Microbiol. 2016, 7, 1607. [Google Scholar] [CrossRef] [PubMed]
- Van Dam, S.; Võsa, U.; Van Der Graaf, A.; Franke, L.; De Magalhães, J.P. Gene co-expression analysis for functional classification and gene–disease predictions. Brief. Bioinform. 2018, 19, 575–592. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.F.; Strange, R.E. Biochemical changes occurring during sporulation in Bacillus species. Biochem. J. 1956, 63, 661–668. [Google Scholar] [CrossRef] [PubMed]
- Swarge, B.N.; Roseboom, W.; Zheng, L.; Abhyankar, W.; Brul, S.; De Koster, C.G.; De Koning, L.J. “One-Pot” Sample Processing Method for Proteome-Wide Analysis of Microbial Cells and Spores. Proteom.-Clin. Appl. 2018, 12, e1700169. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized ppb-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Whiteside, M.D.; Winsor, G.L.; Laird, M.R.; Brinkman, F.S.L. OrtholugeDB: A bacterial and archaeal orthology resource for improved comparative genomic analysis. Nucleic Acids Res. 2012, 41, D366–D376. [Google Scholar] [CrossRef]
- Altenhoff, A.M.; Train, C.-M.; Gilbert, K.J.; Mediratta, I.; De Farias, T.M.; Moi, D.; Nevers, Y.; Radoykova, H.-S.; Rossier, V.; Vesztrocy, A.W.; et al. OMA orthology in 2021: Website overhaul, conserved isoforms, ancestral gene order and more. Nucleic Acids Res. 2020, 49, D373–D379. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Belinda, P.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, X.; Swarge, B.N.; Roseboom, W.; Setlow, P.; Brul, S.; Kramer, G. Time-Resolved Proteomics of Germinating Spores of Bacillus cereus. Int. J. Mol. Sci. 2022, 23, 13614. https://doi.org/10.3390/ijms232113614
Gao X, Swarge BN, Roseboom W, Setlow P, Brul S, Kramer G. Time-Resolved Proteomics of Germinating Spores of Bacillus cereus. International Journal of Molecular Sciences. 2022; 23(21):13614. https://doi.org/10.3390/ijms232113614
Chicago/Turabian StyleGao, Xiaowei, Bhagyashree N. Swarge, Winfried Roseboom, Peter Setlow, Stanley Brul, and Gertjan Kramer. 2022. "Time-Resolved Proteomics of Germinating Spores of Bacillus cereus" International Journal of Molecular Sciences 23, no. 21: 13614. https://doi.org/10.3390/ijms232113614
APA StyleGao, X., Swarge, B. N., Roseboom, W., Setlow, P., Brul, S., & Kramer, G. (2022). Time-Resolved Proteomics of Germinating Spores of Bacillus cereus. International Journal of Molecular Sciences, 23(21), 13614. https://doi.org/10.3390/ijms232113614







