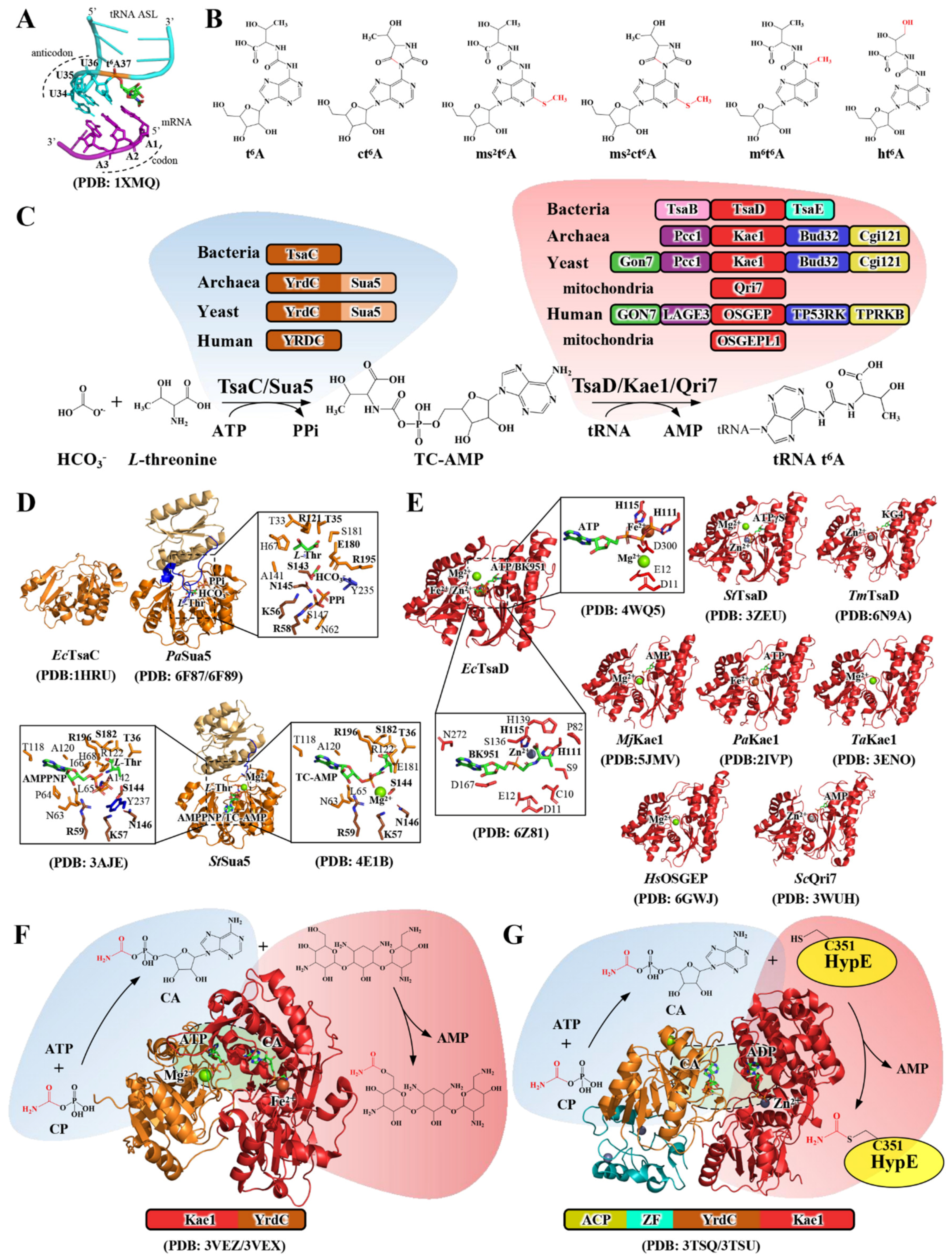
Conservation and Diversification of tRNA t6A-Modifying Enzymes across the Three Domains of Life
Abstract
1. Introduction
| Modified | Organisms | Enzymes | Functions and Deficit-Associated Phenotypes | Refs |
|---|---|---|---|---|
| t6A | Bacteria | E. coli TsaC and TsaDBE B. subtilisYwlCandYdiECB | tRNA t6A is required for recognizing the start codon (AUG) and prevents frameshift in translation; the depletion of tsaC and tsaD leads to cell death. | [17,60,66,67] |
| Archaea | P. abyssi/M. jannaschi/H. volcanii Sua5 and KEOPS | H. volcanii Pcc1 (KEOPS subunit) is not essential to cellular life; the deletion of pcc1 reduces the level of t6A modification and leads to decreased growth rate. | [39,71,77] | |
| Eukarya | S. cerevisiae Sua5 and KEOPS(CT) | Deletion of sua5 and KEOPS (kae1, bud32, cgi121 and pcc1) abolishes the tRNA t6A formation and leads to pleiotropic lethal phenotypes. | [41,65,68,70] | |
| S. cerevisiae Sua5 and Qri7(MT) | Qri7 is essential for yeast life. Deletion of qri7 leads to mitochondrial genome instability and abnormal morphology. | [18,30,73,78] | ||
| H. sapiens YRDC and KEOPS(CT) | Deletion or mutations of YRDC and KEOPS (OSGEP, TP53RK, TPRKB, LAGE3 and GON7) leads to depletion of tRNA t6A and is implicated in Galloway–Mowat syndrome. | [42,56,79,80] | ||
| H. sapiens YRDC and OSGEPL1(MT) | Deletion of YRDC and OSGEPL1 causes mitochondrial dysfunction (interferes with protein synthesis and respiratory activities) and is clinically associated with an onset of myoclonus epilepsy with ragged-red fibers (MERRF). | [29,74] | ||
| ct6A | Bacteria | E. coli TcdA/ B. subtilis YrvM | Bacterial ct6A-tRNALys (UUU) is involved in promoting decoding efficiency. | [46,48] |
| Eukarya | S. cerevisiae TCD1 and TCD2 | Yeast ct6A-tRNAs are required for yeast respiratory growth under nonfermenting conditions. | [46] | |
| A. thaliana enzymes N.D. | N.D. | [49] | ||
| ms2t6A | Bacteria | B. subtilis YqeV (MtaB) | B. subtilis ms2t6A-tRNALys (UUU) is required for the accurate translation of AAG codon. | [58] |
| Eukarya | T. brucei MtaB | Depletion of ms2t6A confers no significant growth. | [49] | |
| A. thaliana enzymes N.D. | N.D. | [49] | ||
| H. sapiens CDKAL1 | ms2t6A-tRNALys (UUU) is required for the accurate translation of AAA codon and its deficit is associated with type 2 diabetes. | [58] | ||
| ms2ct6A | Bacteria | B. subtilis YrvM and YqeV | Depletion of ms2ct6A confers no significant growth or temperature-sensitivity phenotypes. | [49] |
| Eukarya | T. brucei TcdA and MtaB | ms2ct6A-tRNA promotes decoding efficiency and contributes to cell growth in inhibitory conditions or under stress. | [49] | |
| A. thaliana enzymes N.D. | N.D. | [49] | ||
| m6t6A | Bacteria | E. coli TrmO | m6t6A enhances the decoding ability of tRNAThr and discriminates tRNAsThr for ACY codons from other isoacceptors. | [48] |
| Eukarya | R. norvegicus TrmO | m6t6A-tRNASer (GCU) is responsible for the AGY codons. | [48] | |
| H. sapiens TRMO | N.D. | [48] | ||
| ht6A | Eukarya | Echinoderm mitochondria t6A enzymes using the substrate of Hydroxy-L-threonine | M. nudus mt-tRNALys (UUU) is employed to decodes asparagine instead of lysine. | [28] |
2. The Biochemical and Structural Aspects of tRNA t6A Catalysis
2.1. Structure–Function Relationship of TsaC/Sua5 in TC-AMP Biosynthesis
2.2. Structure–Function Relationship of TsaD/Kae1/Qri7 in t6A Biosynthesis
2.3. Evolutionary Implications of a Functional Cooperation between YrdC-like Domain and Kae1-like Domain
3. The Diversification of tRNA t6A Biosynthetic Systems
3.1. The Dynamic TsaD–TsaB–TsaE Complex in Bacteria
3.2. The KEOPS Machinery in Archaea and Eukarya
4. Molecular Interactions between tRNAs and tRNA t6A-Modifying Enzymes
4.1. Sequence Motifs and Structural Determinants of t6A tRNA Substrate
4.2. Structural Models of t6A-Modifying Enzymes in Complex with tRNA
4.3. Binding of tRNAs to TsaC/Sua5
4.4. Interaction between tRNA and TsaD–TsaB–TsaE Complex
4.5. Interaction between tRNA and the KEOPS Complex
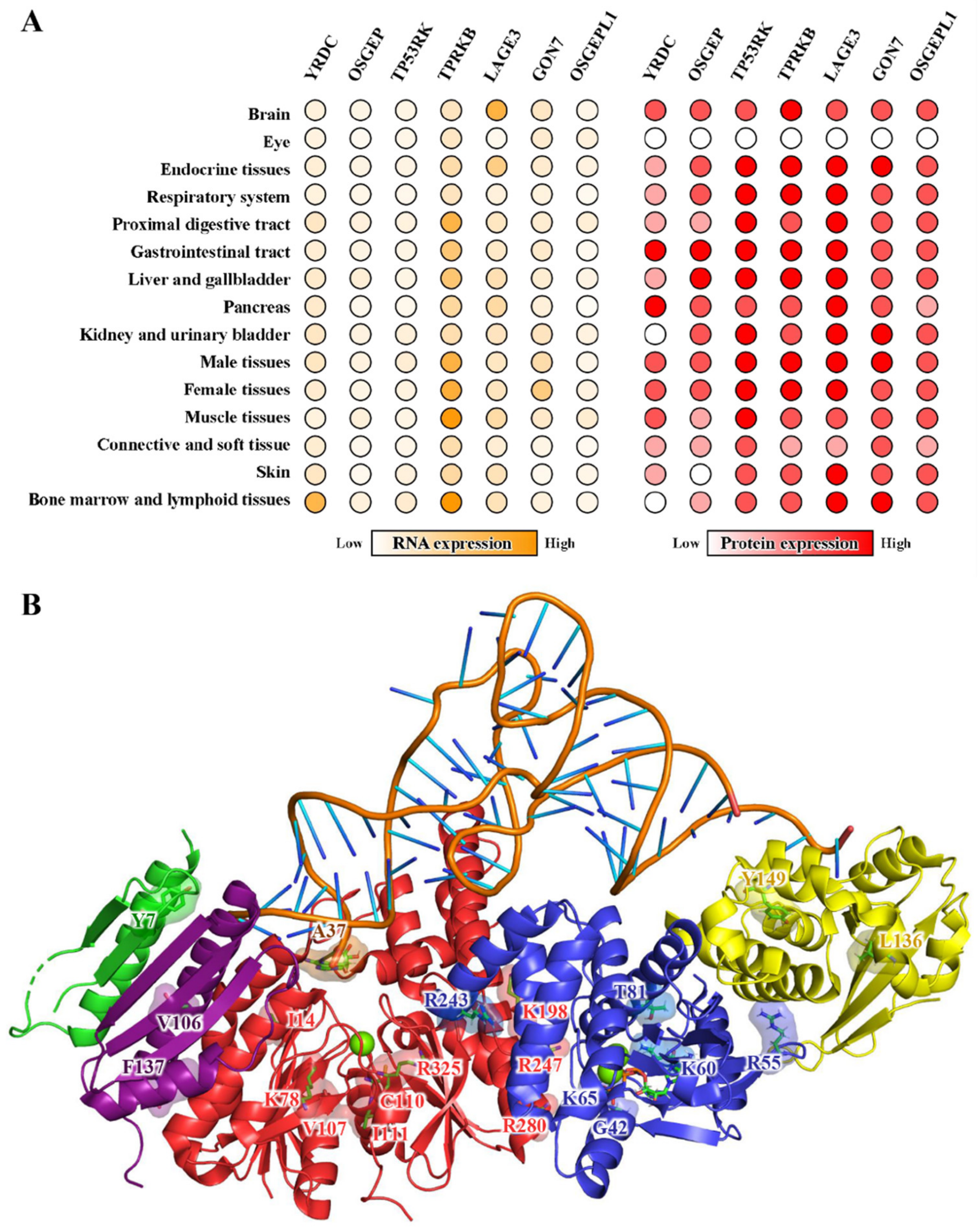
5. Diseases Implications
5.1. Mitochondrial Diseases Caused by tRNA t6A
5.2. KEOPS Mutations and Neurological Disorders
6. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Acknowledgments
Conflicts of Interest
References
- Crick, F. Central dogma of molecular biology. Nature 1970, 227, 561–563. [Google Scholar] [CrossRef]
- Moore, P.B.; Steitz, T.A. The roles of RNA in the synthesis of protein. Cold Spring Harb. Perspect. Biol. 2011, 3, a003780. [Google Scholar] [CrossRef]
- Jackman, J.E.; Alfonzo, J.D. Transfer RNA modifications: Nature’s combinatorial chemistry playground. Wiley Interdiscip. Rev. RNA 2013, 4, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Boccaletto, P.; Stefaniak, F.; Ray, A.; Cappannini, A.; Mukherjee, S.; Purta, E.; Kurkowska, M.; Shirvanizadeh, N.; Destefanis, E.; Groza, P.; et al. MODOMICS: A database of RNA modification pathways. 2021 update. Nucleic Acids Res. 2022, 50, D231–D235. [Google Scholar] [CrossRef] [PubMed]
- Durant, P.C.; Bajji, A.C.; Sundaram, M.; Kumar, R.K.; Davis, D.R. Structural effects of hypermodified nucleosides in the Escherichia coli and human tRNALys anticodon loop: The effect of nucleosides s2U, mcm5U, mcm5s2U, mnm5s2U, t6A, and ms2t6A. Biochemistry 2005, 44, 8078–8089. [Google Scholar] [CrossRef]
- Motorin, Y.; Helm, M. tRNA stabilization by modified nucleotides. Biochemistry 2010, 49, 4934–4944. [Google Scholar] [CrossRef]
- Yarian, C.; Townsend, H.; Czestkowski, W.; Sochacka, E.; Malkiewicz, A.J.; Guenther, R.; Miskiewicz, A.; Agris, P.F. Accurate translation of the genetic code depends on tRNA modified nucleosides. J. Biol. Chem. 2002, 277, 16391–16395. [Google Scholar] [CrossRef]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef]
- Helm, M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006, 34, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Phizicky, E.M.; Hopper, A.K. tRNA biology charges to the front. Genes Dev. 2010, 24, 1832–1860. [Google Scholar] [CrossRef] [PubMed]
- Grosjean, H.; Sprinzl, M.; Steinberg, S. Posttranscriptionally modified nucleosides in transfer RNA: Their locations and frequencies. Biochimie 1995, 77, 139–141. [Google Scholar] [CrossRef]
- El Yacoubi, B.; Bailly, M.; de Crecy-Lagard, V. Biosynthesis and function of posttranscriptional modifications of transfer RNAs. Annu. Rev. Genet. 2012, 46, 69–95. [Google Scholar] [CrossRef]
- Suzuki, T. The expanding world of tRNA modifications and their disease relevance. Nat. Rev. Mol. Cell Biol. 2021, 22, 375–392. [Google Scholar] [CrossRef] [PubMed]
- Agris, P.F.; Vendeix, F.A.; Graham, W.D. tRNA’s wobble decoding of the genome: 40 years of modification. J. Mol. Biol. 2007, 366, 1–13. [Google Scholar] [CrossRef]
- Chheda, G.B. Isolation and characterization of a novel nucleoside, N-[(9-beta-D-ribofuranosyl-9H-purin-6-yl)carbamoyl]threonine, from human urine. Life Sci. 1969, 8, 979–987. [Google Scholar] [CrossRef]
- Schweizer, M.P.; Chheda, G.B.; Baczynskyj, L.; Hall, R.H. Aminoacyl nucleosides. VII. N-(Purin-6-ylcarbamoyl)threonine. A new component of transfer ribonucleic acid. Biochemistry 1969, 8, 3283–3289. [Google Scholar] [CrossRef]
- Thiaville, P.C.; El Yacoubi, B.; Kohrer, C.; Thiaville, J.J.; Deutsch, C.; Iwata-Reuyl, D.; Bacusmo, J.M.; Armengaud, J.; Bessho, Y.; Wetzel, C.; et al. Essentiality of threonylcarbamoyladenosine (t(6)A), a universal tRNA modification, in bacteria. Mol. Microbiol. 2015, 98, 1199–1221. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; El Yacoubi, B.; Perrochia, L.; Hecker, A.; Prigent, M.; Thiaville, J.J.; Forterre, P.; Namy, O.; Basta, T.; de Crecy-Lagard, V. Cross kingdom functional conservation of the core universally conserved threonylcarbamoyladenosine tRNA synthesis enzymes. Eukaryot. Cell 2014, 13, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Ohrt, J.M.; Chheda, G.B. Modified nucleosides and conformation of anticodon loops: Crystal structure of t6A and g6A. Biochemistry 1977, 16, 4999–5008. [Google Scholar] [CrossRef]
- Sundaram, M.; Durant, P.C.; Davis, D.R. Hypermodified nucleosides in the anticodon of tRNALys stabilize a canonical U-turn structure. Biochemistry 2000, 39, 12575–12584. [Google Scholar] [CrossRef]
- Han, L.; Phizicky, E.M. A rationale for tRNA modification circuits in the anticodon loop. Rna 2018, 24, 1277–1284. [Google Scholar] [CrossRef] [PubMed]
- Murphy, F.V.t.; Ramakrishnan, V.; Malkiewicz, A.; Agris, P.F. The role of modifications in codon discrimination by tRNA(Lys)UUU. Nat. Struct. Mol. Biol. 2004, 11, 1186–1191. [Google Scholar] [CrossRef] [PubMed]
- Weissenbach, J.; Grosjean, H. Effect of threonylcarbamoyl modification (t6A) in yeast tRNA Arg III on codon-anticodon and anticodon-anticodon interactions. A thermodynamic and kinetic evaluation. Eur. J. Biochem. 1981, 116, 207–213. [Google Scholar] [CrossRef] [PubMed]
- Konevega, A.L.; Soboleva, N.G.; Makhno, V.I.; Semenkov, Y.P.; Wintermeyer, W.; Rodnina, M.V.; Katunin, V.I. Purine bases at position 37 of tRNA stabilize codon-anticodon interaction in the ribosomal A site by stacking and Mg2+-dependent interactions. Rna 2004, 10, 90–101. [Google Scholar] [CrossRef]
- Lescrinier, E.; Nauwelaerts, K.; Zanier, K.; Poesen, K.; Sattler, M.; Herdewijn, P. The naturally occurring N6-threonyl adenine in anticodon loop of Schizosaccharomyces pombe tRNAi causes formation of a unique U-turn motif. Nucleic Acids Res. 2006, 34, 2878–2886. [Google Scholar] [CrossRef]
- Thiaville, P.C.; Legendre, R.; Rojas-Benitez, D.; Baudin-Baillieu, A.; Hatin, I.; Chalancon, G.; Glavic, A.; Namy, O.; de Crecy-Lagard, V. Global translational impacts of the loss of the tRNA modification t(6)A in yeast. Microb. Cell 2016, 3, 29–45. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.T.; Zhou, J.B.; Mao, X.L.; Zhou, L.; Chen, M.; Zhang, W.; Wang, E.D.; Zhou, X.L. Commonality and diversity in tRNA substrate recognition in t6A biogenesis by eukaryotic KEOPSs. Nucleic Acids Res. 2022, 50, 2223–2239. [Google Scholar] [CrossRef]
- Nagao, A.; Ohara, M.; Miyauchi, K.; Yokobori, S.I.; Yamagishi, A.; Watanabe, K.; Suzuki, T. Hydroxylation of a conserved tRNA modification establishes non-universal genetic code in echinoderm mitochondria. Nat. Struct. Mol. Biol. 2017, 24, 778–782. [Google Scholar] [CrossRef]
- Lin, H.; Miyauchi, K.; Harada, T.; Okita, R.; Takeshita, E.; Komaki, H.; Fujioka, K.; Yagasaki, H.; Goto, Y.I.; Yanaka, K.; et al. CO2-sensitive tRNA modification associated with human mitochondrial disease. Nat. Commun. 2018, 9, 1875. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zeng, Q.Y.; Zheng, W.Q.; Ji, Q.Q.; Zhou, X.L.; Wang, E.D. A natural non-Watson-Crick base pair in human mitochondrial tRNAThr causes structural and functional susceptibility to local mutations. Nucleic Acids Res. 2018, 46, 4662–4676. [Google Scholar] [CrossRef]
- Teplova, M.; Tereshko, V.; Sanishvili, R.; Joachimiak, A.; Bushueva, T.; Anderson, W.F.; Egli, M. The structure of the yrdC gene product from Escherichia coli reveals a new fold and suggests a role in RNA binding. Protein Sci. 2000, 9, 2557–2566. [Google Scholar] [CrossRef] [PubMed]
- Pichard-Kostuch, A.; Zhang, W.; Liger, D.; Daugeron, M.C.; Letoquart, J.; Li de la Sierra-Gallay, I.; Forterre, P.; Collinet, B.; van Tilbeurgh, H.; Basta, T. Structure-function analysis of Sua5 protein reveals novel functional motifs required for the biosynthesis of the universal t(6)A tRNA modification. RNA 2018, 24, 926–938. [Google Scholar] [CrossRef] [PubMed]
- Kuratani, M.; Kasai, T.; Akasaka, R.; Higashijima, K.; Terada, T.; Kigawa, T.; Shinkai, A.; Bessho, Y.; Yokoyama, S. Crystal structure of Sulfolobus tokodaii Sua5 complexed with L-threonine and AMPPNP. Proteins 2011, 79, 2065–2075. [Google Scholar] [CrossRef] [PubMed]
- Parthier, C.; Gorlich, S.; Jaenecke, F.; Breithaupt, C.; Brauer, U.; Fandrich, U.; Clausnitzer, D.; Wehmeier, U.F.; Bottcher, C.; Scheel, D.; et al. The O-carbamoyltransferase TobZ catalyzes an ancient enzymatic reaction. Angew. Chem. Int. Ed. 2012, 51, 4046–4052. [Google Scholar] [CrossRef]
- Zhang, W.; Collinet, B.; Perrochia, L.; Durand, D.; van Tilbeurgh, H. The ATP-mediated formation of the YgjD-YeaZ-YjeE complex is required for the biosynthesis of tRNA t6A in Escherichia coli. Nucleic Acids Res. 2015, 43, 1804–1817. [Google Scholar] [CrossRef]
- Kopina, B.J.; Missoury, S.; Collinet, B.; Fulton, M.G.; Cirio, C.; van Tilbeurgh, H.; Lauhon, C.T. Structure of a reaction intermediate mimic in t6A biosynthesis bound in the active site of the TsaBD heterodimer from Escherichia coli. Nucleic Acids Res. 2021, 49, 2141–2160. [Google Scholar] [CrossRef]
- Nichols, C.E.; Lamb, H.K.; Thompson, P.; El Omari, K.; Lockyer, M.; Charles, I.; Hawkins, A.R.; Stammers, D.K. Crystal structure of the dimer of two essential Salmonella typhimurium proteins, YgjD & YeaZ and calorimetric evidence for the formation of a ternary YgjD-YeaZ-YjeE complex. Protein Sci. 2013, 22, 628–640. [Google Scholar] [CrossRef]
- Luthra, A.; Paranagama, N.; Swinehart, W.; Bayooz, S.; Phan, P.; Quach, V.; Schiffer, J.M.; Stec, B.; Iwata-Reuyl, D.; Swairjo, M.A. Conformational communication mediates the reset step in t6A biosynthesis. Nucleic Acids Res. 2019, 47, 6551–6567. [Google Scholar] [CrossRef]
- Wan, L.C.; Pillon, M.C.; Thevakumaran, N.; Sun, Y.; Chakrabartty, A.; Guarne, A.; Kurinov, I.; Durocher, D.; Sicheri, F. Structural and functional characterization of KEOPS dimerization by Pcc1 and its role in t6A biosynthesis. Nucleic Acids Res. 2016, 44, 6971–6980. [Google Scholar] [CrossRef]
- Hecker, A.; Leulliot, N.; Gadelle, D.; Graille, M.; Justome, A.; Dorlet, P.; Brochier, C.; Quevillon-Cheruel, S.; Le Cam, E.; van Tilbeurgh, H.; et al. An archaeal orthologue of the universal protein Kae1 is an iron metalloprotein which exhibits atypical DNA-binding properties and apurinic-endonuclease activity in vitro. Nucleic Acids Res. 2007, 35, 6042–6051. [Google Scholar] [CrossRef]
- Mao, D.Y.; Neculai, D.; Downey, M.; Orlicky, S.; Haffani, Y.Z.; Ceccarelli, D.F.; Ho, J.S.; Szilard, R.K.; Zhang, W.; Ho, C.S.; et al. Atomic structure of the KEOPS complex: An ancient protein kinase-containing molecular machine. Mol. Cell 2008, 32, 259–275. [Google Scholar] [CrossRef] [PubMed]
- Arrondel, C.; Missoury, S.; Snoek, R.; Patat, J.; Menara, G.; Collinet, B.; Liger, D.; Durand, D.; Gribouval, O.; Boyer, O.; et al. Defects in t(6)A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat. Commun. 2019, 10, 3967. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, T.; Kobayashi, K.; Ishii, R.; Ishitani, R.; Nureki, O. Structure of Saccharomyces cerevisiae mitochondrial Qri7 in complex with AMP. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2014, 70, 1009–1014. [Google Scholar] [CrossRef]
- Petkun, S.; Shi, R.; Li, Y.; Asinas, A.; Munger, C.; Zhang, L.; Waclawek, M.; Soboh, B.; Sawers, R.G.; Cygler, M. Structure of hydrogenase maturation protein HypF with reaction intermediates shows two active sites. Structure 2011, 19, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Matuszewski, M.; Wojciechowski, J.; Miyauchi, K.; Gdaniec, Z.; Wolf, W.M.; Suzuki, T.; Sochacka, E. A hydantoin isoform of cyclic N6-threonylcarbamoyladenosine (ct6A) is present in tRNAs. Nucleic Acids Res. 2017, 45, 2137–2149. [Google Scholar] [CrossRef] [PubMed]
- Miyauchi, K.; Kimura, S.; Suzuki, T. A cyclic form of N6-threonylcarbamoyladenosine as a widely distributed tRNA hypermodification. Nat. Chem. Biol. 2013, 9, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Arragain, S.; Handelman, S.K.; Forouhar, F.; Wei, F.Y.; Tomizawa, K.; Hunt, J.F.; Douki, T.; Fontecave, M.; Mulliez, E.; Atta, M. Identification of eukaryotic and prokaryotic methylthiotransferase for biosynthesis of 2-methylthio-N6-threonylcarbamoyladenosine in tRNA. J. Biol. Chem. 2010, 285, 28425–28433. [Google Scholar] [CrossRef] [PubMed]
- Kimura, S.; Miyauchi, K.; Ikeuchi, Y.; Thiaville, P.C.; Crecy-Lagard, V.; Suzuki, T. Discovery of the beta-barrel-type RNA methyltransferase responsible for N6-methylation of N6-threonylcarbamoyladenosine in tRNAs. Nucleic Acids Res. 2014, 42, 9350–9365. [Google Scholar] [CrossRef] [PubMed]
- Kang, B.I.; Miyauchi, K.; Matuszewski, M.; D’Almeida, G.S.; Rubio, M.A.T.; Alfonzo, J.D.; Inoue, K.; Sakaguchi, Y.; Suzuki, T.; Sochacka, E.; et al. Identification of 2-methylthio cyclic N6-threonylcarbamoyladenosine (ms2ct6A) as a novel RNA modification at position 37 of tRNAs. Nucleic Acids Res. 2017, 45, 2124–2136. [Google Scholar] [CrossRef] [PubMed]
- Rojas-Benitez, D.; Ibar, C.; Glavic, A. The Drosophila EKC/KEOPS complex: Roles in protein synthesis homeostasis and animal growth. Fly 2013, 7, 168–172. [Google Scholar] [CrossRef]
- Lin, C.J.; Smibert, P.; Zhao, X.; Hu, J.F.; Ramroop, J.; Kellner, S.M.; Benton, M.A.; Govind, S.; Dedon, P.C.; Sternglanz, R.; et al. An extensive allelic series of Drosophila kae1 mutants reveals diverse and tissue-specific requirements for t6A biogenesis. RNA 2015, 21, 2103–2118. [Google Scholar] [CrossRef] [PubMed]
- Chujo, T.; Tomizawa, K. Human transfer RNA modopathies: Diseases caused by aberrations in transfer RNA modifications. FEBS J. 2021, 288, 7096–7122. [Google Scholar] [CrossRef]
- Tahmasebi, S.; Khoutorsky, A.; Mathews, M.B.; Sonenberg, N. Translation deregulation in human disease. Nat. Rev. Mol. Cell Biol. 2018, 19, 791–807. [Google Scholar] [CrossRef] [PubMed]
- Pereira, M.; Francisco, S.; Varanda, A.S.; Santos, M.; Santos, M.A.S.; Soares, A.R. Impact of tRNA Modifications and tRNA-Modifying Enzymes on Proteostasis and Human Disease. Int. J. Mol. Sci. 2018, 19, 3738. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Nagao, A.; Suzuki, T. Human mitochondrial tRNAs: Biogenesis, function, structural aspects, and diseases. Annu. Rev. Genet. 2011, 45, 299–329. [Google Scholar] [CrossRef] [PubMed]
- Braun, D.A.; Rao, J.; Mollet, G.; Schapiro, D.; Daugeron, M.C.; Tan, W.; Gribouval, O.; Boyer, O.; Revy, P.; Jobst-Schwan, T.; et al. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat. Genet. 2017, 49, 1529–1538. [Google Scholar] [CrossRef] [PubMed]
- Steinthorsdottir, V.; Thorleifsson, G.; Reynisdottir, I.; Benediktsson, R.; Jonsdottir, T.; Walters, G.B.; Styrkarsdottir, U.; Gretarsdottir, S.; Emilsson, V.; Ghosh, S.; et al. A variant in CDKAL1 influences insulin response and risk of type 2 diabetes. Nat. Genet. 2007, 39, 770–775. [Google Scholar] [CrossRef]
- Wei, F.Y.; Suzuki, T.; Watanabe, S.; Kimura, S.; Kaitsuka, T.; Fujimura, A.; Matsui, H.; Atta, M.; Michiue, H.; Fontecave, M.; et al. Deficit of tRNA(Lys) modification by Cdkal1 causes the development of type 2 diabetes in mice. J. Clin. Investig. 2011, 121, 3598–3608. [Google Scholar] [CrossRef]
- El Yacoubi, B.; Lyons, B.; Cruz, Y.; Reddy, R.; Nordin, B.; Agnelli, F.; Williamson, J.R.; Schimmel, P.; Swairjo, M.A.; de Crecy-Lagard, V. The universal YrdC/Sua5 family is required for the formation of threonylcarbamoyladenosine in tRNA. Nucleic Acids Res. 2009, 37, 2894–2909. [Google Scholar] [CrossRef]
- El Yacoubi, B.; Hatin, I.; Deutsch, C.; Kahveci, T.; Rousset, J.P.; Iwata-Reuyl, D.; Murzin, A.G.; de Crecy-Lagard, V. A role for the universal Kae1/Qri7/YgjD (COG0533) family in tRNA modification. EMBO J. 2011, 30, 882–893. [Google Scholar] [CrossRef]
- Koonin, E.V. Comparative genomics, minimal gene-sets and the last universal common ancestor. Nat. Rev. Microbiol. 2003, 1, 127–136. [Google Scholar] [CrossRef] [PubMed]
- Arigoni, F.; Talabot, F.; Peitsch, M.; Edgerton, M.D.; Meldrum, E.; Allet, E.; Fish, R.; Jamotte, T.; Curchod, M.L.; Loferer, H. A genome-based approach for the identification of essential bacterial genes. Nat. Biotechnol. 1998, 16, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Butland, G.; Peregrin-Alvarez, J.M.; Li, J.; Yang, W.; Yang, X.; Canadien, V.; Starostine, A.; Richards, D.; Beattie, B.; Krogan, N.; et al. Interaction network containing conserved and essential protein complexes in Escherichia coli. Nature 2005, 433, 531–537. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Koonin, E.V. ‘Conserved hypothetical’ proteins: Prioritization of targets for experimental study. Nucleic Acids Res. 2004, 32, 5452–5463. [Google Scholar] [CrossRef]
- Perrochia, L.; Crozat, E.; Hecker, A.; Zhang, W.; Bareille, J.; Collinet, B.; van Tilbeurgh, H.; Forterre, P.; Basta, T. In vitro biosynthesis of a universal t6A tRNA modification in Archaea and Eukarya. Nucleic Acids Res. 2013, 41, 1953–1964. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, C.; El Yacoubi, B.; de Crecy-Lagard, V.; Iwata-Reuyl, D. Biosynthesis of threonylcarbamoyl adenosine (t6A), a universal tRNA nucleoside. J. Biol. Chem. 2012, 287, 13666–13673. [Google Scholar] [CrossRef] [PubMed]
- Lauhon, C.T. Mechanism of N6-threonylcarbamoyladenonsine (t(6)A) biosynthesis: Isolation and characterization of the intermediate threonylcarbamoyl-AMP. Biochemistry 2012, 51, 8950–8963. [Google Scholar] [CrossRef] [PubMed]
- Downey, M.; Houlsworth, R.; Maringele, L.; Rollie, A.; Brehme, M.; Galicia, S.; Guillard, S.; Partington, M.; Zubko, M.K.; Krogan, N.J.; et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell 2006, 124, 1155–1168. [Google Scholar] [CrossRef]
- Kisseleva-Romanova, E.; Lopreiato, R.; Baudin-Baillieu, A.; Rousselle, J.C.; Ilan, L.; Hofmann, K.; Namane, A.; Mann, C.; Libri, D. Yeast homolog of a cancer-testis antigen defines a new transcription complex. EMBO J. 2006, 25, 3576–3585. [Google Scholar] [CrossRef]
- Srinivasan, M.; Mehta, P.; Yu, Y.; Prugar, E.; Koonin, E.V.; Karzai, A.W.; Sternglanz, R. The highly conserved KEOPS/EKC complex is essential for a universal tRNA modification, t6A. EMBO J. 2011, 30, 873–881. [Google Scholar] [CrossRef]
- Perrochia, L.; Guetta, D.; Hecker, A.; Forterre, P.; Basta, T. Functional assignment of KEOPS/EKC complex subunits in the biosynthesis of the universal t6A tRNA modification. Nucleic Acids Res. 2013, 41, 9484–9499. [Google Scholar] [CrossRef]
- Beenstock, J.; Ona, S.M.; Porat, J.; Orlicky, S.; Wan, L.C.K.; Ceccarelli, D.F.; Maisonneuve, P.; Szilard, R.K.; Yin, Z.; Setiaputra, D.; et al. A substrate binding model for the KEOPS tRNA modifying complex. Nat. Commun. 2020, 11, 6233. [Google Scholar] [CrossRef]
- Wan, L.C.; Mao, D.Y.; Neculai, D.; Strecker, J.; Chiovitti, D.; Kurinov, I.; Poda, G.; Thevakumaran, N.; Yuan, F.; Szilard, R.K.; et al. Reconstitution and characterization of eukaryotic N6-threonylcarbamoylation of tRNA using a minimal enzyme system. Nucleic Acids Res. 2013, 41, 6332–6346. [Google Scholar] [CrossRef]
- Zhou, J.B.; Wang, Y.; Zeng, Q.Y.; Meng, S.X.; Wang, E.D.; Zhou, X.L. Molecular basis for t6A modification in human mitochondria. Nucleic Acids Res. 2020, 48, 3181–3194. [Google Scholar] [CrossRef]
- Harris, K.A.; Bobay, B.G.; Sarachan, K.L.; Sims, A.F.; Bilbille, Y.; Deutsch, C.; Iwata-Reuyl, D.; Agris, P.F. NMR-based Structural Analysis of Threonylcarbamoyl-AMP Synthase and Its Substrate Interactions. J. Biol. Chem. 2015, 290, 20032–20043. [Google Scholar] [CrossRef]
- Agari, Y.; Sato, S.; Wakamatsu, T.; Bessho, Y.; Ebihara, A.; Yokoyama, S.; Kuramitsu, S.; Shinkai, A. X-ray crystal structure of a hypothetical Sua5 protein from Sulfolobus tokodaii strain 7. Proteins 2008, 70, 1108–1111. [Google Scholar] [CrossRef] [PubMed]
- Naor, A.; Thiaville, P.C.; Altman-Price, N.; Cohen-Or, I.; Allers, T.; de Crecy-Lagard, V.; Gophna, U. A genetic investigation of the KEOPS complex in halophilic Archaea. PLoS ONE 2012, 7, e43013. [Google Scholar] [CrossRef]
- Oberto, J.; Breuil, N.; Hecker, A.; Farina, F.; Brochier-Armanet, C.; Culetto, E.; Forterre, P. Qri7/OSGEPL, the mitochondrial version of the universal Kae1/YgjD protein, is essential for mitochondrial genome maintenance. Nucleic Acids Res. 2009, 37, 5343–5352. [Google Scholar] [CrossRef] [PubMed]
- Wan, L.C.; Maisonneuve, P.; Szilard, R.K.; Lambert, J.P.; Ng, T.F.; Manczyk, N.; Huang, H.; Laister, R.; Caudy, A.A.; Gingras, A.C.; et al. Proteomic analysis of the human KEOPS complex identifies C14ORF142 as a core subunit homologous to yeast Gon7. Nucleic Acids Res. 2017, 45, 805–817. [Google Scholar] [CrossRef] [PubMed]
- Treimer, E.; Kalayci, T.; Schumann, S.; Suer, I.; Greco, S.; Schanze, D.; Schmeisser, M.J.; Kuhl, S.J.; Zenker, M. Functional characterization of a novel TP53RK mutation identified in a family with Galloway-Mowat syndrome. Hum. Mutat. 2022. [Google Scholar] [CrossRef] [PubMed]
- Thiaville, P.C.; Iwata-Reuyl, D.; de Crecy-Lagard, V. Diversity of the biosynthesis pathway for threonylcarbamoyladenosine (t(6)A), a universal modification of tRNA. RNA Biol. 2014, 11, 1529–1539. [Google Scholar] [CrossRef]
- Lin, C.A.; Ellis, S.R.; True, H.L. The Sua5 protein is essential for normal translational regulation in yeast. Mol. Cell. Biol. 2010, 30, 354–363. [Google Scholar] [CrossRef]
- Harris, K.A.; Jones, V.; Bilbille, Y.; Swairjo, M.A.; Agris, P.F. YrdC exhibits properties expected of a subunit for a tRNA threonylcarbamoyl transferase. Rna 2011, 17, 1678–1687. [Google Scholar] [CrossRef] [PubMed]
- Allali-Hassani, A.; Campbell, T.L.; Ho, A.; Schertzer, J.W.; Brown, E.D. Probing the active site of YjeE: A vital Escherichia coli protein of unknown function. Biochem. J. 2004, 384, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Luthra, A.; Swinehart, W.; Bayooz, S.; Phan, P.; Stec, B.; Iwata-Reuyl, D.; Swairjo, M.A. Structure and mechanism of a bacterial t6A biosynthesis system. Nucleic Acids Res. 2018, 46, 1395–1411. [Google Scholar] [CrossRef] [PubMed]
- Galperin, M.Y.; Koonin, E.V. From complete genome sequence to ‘complete’ understanding? Trends Biotechnol. 2010, 28, 398–406. [Google Scholar] [CrossRef]
- Missoury, S.; Plancqueel, S.; Li de la Sierra-Gallay, I.; Zhang, W.; Liger, D.; Durand, D.; Dammak, R.; Collinet, B.; van Tilbeurgh, H. The structure of the TsaB/TsaD/TsaE complex reveals an unexpected mechanism for the bacterial t6A tRNA-modification. Nucleic Acids Res. 2018, 46, 5850–5860. [Google Scholar] [CrossRef]
- Bork, P.; Sander, C.; Valencia, A. An ATPase domain common to prokaryotic cell cycle proteins, sugar kinases, actin, and hsp70 heat shock proteins. Proc. Natl. Acad. Sci. USA 1992, 89, 7290–7294. [Google Scholar] [CrossRef]
- Buss, K.A.; Cooper, D.R.; Ingram-Smith, C.; Ferry, J.G.; Sanders, D.A.; Hasson, M.S. Urkinase: Structure of acetate kinase, a member of the ASKHA superfamily of phosphotransferases. J. Bacteriol. 2001, 183, 680–686. [Google Scholar] [CrossRef]
- Hecker, A.; Graille, M.; Madec, E.; Gadelle, D.; Le Cam, E.; van Tilbergh, H.; Forterre, P. The universal Kae1 protein and the associated Bud32 kinase (PRPK), a mysterious protein couple probably essential for genome maintenance in Archaea and Eukarya. Biochem. Soc. Trans. 2009, 37, 29–35. [Google Scholar] [CrossRef]
- Harding, M.M.; Nowicki, M.W.; Walkinshaw, M.D. Metals in protein structures: A review of their principal features. Crystallogr. Rev. 2010, 16, 247–302. [Google Scholar] [CrossRef]
- Shomura, Y.; Higuchi, Y. Structural basis for the reaction mechanism of S-carbamoylation of HypE by HypF in the maturation of [NiFe]-hydrogenases. J. Biol. Chem. 2012, 287, 28409–28419. [Google Scholar] [CrossRef] [PubMed]
- Handford, J.I.; Ize, B.; Buchanan, G.; Butland, G.P.; Greenblatt, J.; Emili, A.; Palmer, T. Conserved network of proteins essential for bacterial viability. J. Bacteriol. 2009, 191, 4732–4749. [Google Scholar] [CrossRef]
- Msadek, T. Grasping at shadows: Revealing the elusive nature of essential genes. J. Bacteriol. 2009, 191, 4701–4704. [Google Scholar] [CrossRef] [PubMed]
- Aydin, I.; Saijo-Hamano, Y.; Namba, K.; Thomas, C.; Roujeinikova, A. Structural analysis of the essential resuscitation promoting factor YeaZ suggests a mechanism of nucleotide regulation through dimer reorganization. PLoS ONE 2011, 6, e23245. [Google Scholar] [CrossRef] [PubMed]
- Nichols, C.E.; Johnson, C.; Lockyer, M.; Charles, I.G.; Lamb, H.K.; Hawkins, A.R.; Stammers, D.K. Structural characterization of Salmonella typhimurium YeaZ, an M22 O-sialoglycoprotein endopeptidase homolog. Proteins 2006, 64, 111–123. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; McMullan, D.; Jaroszewski, L.; Krishna, S.S.; Elsliger, M.A.; Yeh, A.P.; Abdubek, P.; Astakhova, T.; Axelrod, H.L.; Carlton, D.; et al. Structure of an essential bacterial protein YeaZ (TM0874) from Thermotoga maritima at 2.5 A resolution. Acta Crystallogr. Sect. F Struct. Biol. Commun. 2010, 66, 1230–1236. [Google Scholar] [CrossRef]
- Vecchietti, D.; Ferrara, S.; Rusmini, R.; Macchi, R.; Milani, M.; Bertoni, G. Crystal structure of YeaZ from Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 2016, 470, 460–465. [Google Scholar] [CrossRef]
- Hecker, A.; Lopreiato, R.; Graille, M.; Collinet, B.; Forterre, P.; Libri, D.; van Tilbeurgh, H. Structure of the archaeal Kae1/Bud32 fusion protein MJ1130: A model for the eukaryotic EKC/KEOPS subcomplex. EMBO J. 2008, 27, 2340–2351. [Google Scholar] [CrossRef]
- Li, J.; Ma, X.; Banerjee, S.; Chen, H.; Ma, W.; Bode, A.M.; Dong, Z. Crystal structure of the human PRPK-TPRKB complex. Commun. Biol. 2021, 4, 167. [Google Scholar] [CrossRef]
- Zhang, W.; Collinet, B.; Graille, M.; Daugeron, M.C.; Lazar, N.; Libri, D.; Durand, D.; van Tilbeurgh, H. Crystal structures of the Gon7/Pcc1 and Bud32/Cgi121 complexes provide a model for the complete yeast KEOPS complex. Nucleic Acids Res. 2015, 43, 3358–3372. [Google Scholar] [CrossRef]
- Teplyakov, A.; Obmolova, G.; Tordova, M.; Thanki, N.; Bonander, N.; Eisenstein, E.; Howard, A.J.; Gilliland, G.L. Crystal structure of the YjeE protein from Haemophilus influenzae: A putative Atpase involved in cell wall synthesis. Proteins 2002, 48, 220–226. [Google Scholar] [CrossRef]
- Brown, E.D. Conserved P-loop GTPases of unknown function in bacteria: An emerging and vital ensemble in bacterial physiology. Biochem. Cell Biol. 2005, 83, 738–746. [Google Scholar] [CrossRef]
- Leipe, D.D.; Wolf, Y.I.; Koonin, E.V.; Aravind, L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002, 317, 41–72. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Tang, H.B.; Liu, N.N.; Tong, X.J.; Dang, W.; Duan, Y.M.; Fu, X.H.; Zhang, Y.; Peng, J.; Meng, F.L.; et al. Telomerase-null survivor screening identifies novel telomere recombination regulators. PLoS Genet. 2013, 9, e1003208. [Google Scholar] [CrossRef] [PubMed]
- He, M.H.; Liu, J.C.; Lu, Y.S.; Wu, Z.J.; Liu, Y.Y.; Wu, Z.; Peng, J.; Zhou, J.Q. KEOPS complex promotes homologous recombination via DNA resection. Nucleic Acids Res. 2019, 47, 5684–5697. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; He, M.H.; Duan, Y.M.; Liu, Y.T.; Zhou, J.Q. Inhibition of telomere recombination by inactivation of KEOPS subunit Cgi121 promotes cell longevity. PLoS Genet. 2015, 11, e1005071. [Google Scholar] [CrossRef]
- Daugeron, M.C.; Lenstra, T.L.; Frizzarin, M.; El Yacoubi, B.; Liu, X.; Baudin-Baillieu, A.; Lijnzaad, P.; Decourty, L.; Saveanu, C.; Jacquier, A.; et al. Gcn4 misregulation reveals a direct role for the evolutionary conserved EKC/KEOPS in the t6A modification of tRNAs. Nucleic Acids Res. 2011, 39, 6148–6160. [Google Scholar] [CrossRef]
- Liu, Y.Y.; He, M.H.; Liu, J.C.; Lu, Y.S.; Peng, J.; Zhou, J.Q. Yeast KEOPS complex regulates telomere length independently of its t(6)A modification function. J. Genet. Genom. 2018, 45, 247–257. [Google Scholar] [CrossRef]
- Rojas-Benitez, D.; Eggers, C.; Glavic, A. Modulation of the Proteostasis Machinery to Overcome Stress Caused by Diminished Levels of t6A-Modified tRNAs in Drosophila. Biomolecules 2017, 7, 25. [Google Scholar] [CrossRef]
- Hyun, H.S.; Kim, S.H.; Park, E.; Cho, M.H.; Kang, H.G.; Lee, H.S.; Miyake, N.; Matsumoto, N.; Tsukaguchi, H.; Cheong, H.I. A familial case of Galloway-Mowat syndrome due to a novel TP53RK mutation: A case report. BMC Med. Genet. 2018, 19, 131. [Google Scholar] [CrossRef]
- Ferreira-Cerca, S.; Sagar, V.; Schafer, T.; Diop, M.; Wesseling, A.M.; Lu, H.; Chai, E.; Hurt, E.; LaRonde-LeBlanc, N. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat. Struct. Mol. Biol. 2012, 19, 1316–1323. [Google Scholar] [CrossRef]
- Lee, K.T.; Hong, J.; Lee, D.G.; Lee, M.; Cha, S.; Lim, Y.G.; Jung, K.W.; Hwangbo, A.; Lee, Y.; Yu, S.J.; et al. Fungal kinases and transcription factors regulating brain infection in Cryptococcus neoformans. Nat. Commun. 2020, 11, 1521. [Google Scholar] [CrossRef]
- Uhlen, M.; Fagerberg, L.; Hallstrom, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, A.; Kampf, C.; Sjostedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef]
- Karlsson, M.; Zhang, C.; Mear, L.; Zhong, W.; Digre, A.; Katona, B.; Sjostedt, E.; Butler, L.; Odeberg, J.; Dusart, P.; et al. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021, 7, eabh2169. [Google Scholar] [CrossRef]
- Beenstock, J.; Sicheri, F. The structural and functional workings of KEOPS. Nucleic Acids Res. 2021, 49, 10818–10834. [Google Scholar] [CrossRef]
- Morin, A.; Auxilien, S.; Senger, B.; Tewari, R.; Grosjean, H. Structural requirements for enzymatic formation of threonylcarbamoyladenosine (t6A) in tRNA: An in vivo study with Xenopus laevis oocytes. RNA 1998, 4, 24–37. [Google Scholar]
- Karst, J.C.; Foucher, A.E.; Campbell, T.L.; Di Guilmi, A.M.; Stroebel, D.; Mangat, C.S.; Brown, E.D.; Jault, J.M. The ATPase activity of an ‘essential’ Bacillus subtilis enzyme, YdiB, is required for its cellular function and is modulated by oligomerization. Microbiology 2009, 155, 944–956. [Google Scholar] [CrossRef] [PubMed]
- Arroyo, M.N.; Green, J.A.; Cnop, M.; Igoillo-Esteve, M. tRNA Biology in the Pathogenesis of Diabetes: Role of Genetic and Environmental Factors. Int. J. Mol. Sci. 2021, 22, 496. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Suzuki, T. A complete landscape of post-transcriptional modifications in mammalian mitochondrial tRNAs. Nucleic Acids Res. 2014, 42, 7346–7357. [Google Scholar] [CrossRef] [PubMed]
- Lipska-Zietkiewicz, B.S.; Ozaltin, F.; Holtta, T.; Bockenhauer, D.; Berody, S.; Levtchenko, E.; Vivarelli, M.; Webb, H.; Haffner, D.; Schaefer, F.; et al. Genetic aspects of congenital nephrotic syndrome: A consensus statement from the ERKNet-ESPN inherited glomerulopathy working group. Eur. J. Hum. Genet. 2020, 28, 1368–1378. [Google Scholar] [CrossRef] [PubMed]
- Edvardson, S.; Prunetti, L.; Arraf, A.; Haas, D.; Bacusmo, J.M.; Hu, J.F.; Ta-Shma, A.; Dedon, P.C.; de Crecy-Lagard, V.; Elpeleg, O. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur. J. Hum. Genet. 2017, 25, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Carney, E.F. Nephrotic syndrome: Novel monogenic causes of Galloway-Mowat syndrome. Nat. Rev. Nephrol. 2017, 13, 661. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Hu, L.; Yang, L.; Wu, B.; Cao, Y.; Zhang, R.; Xu, X.; Ma, H.; Zhou, W.; Cheng, G.; et al. Galloway-Mowat Syndrome Type 3 Caused by OSGEP Gene Variants: A Case Report and Literature Review. Front. Pediatr. 2022, 10, 899991. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Yashiro, Y.; Kikuchi, I.; Ishigami, Y.; Saito, H.; Matsuzawa, I.; Okada, S.; Mito, M.; Iwasaki, S.; Ma, D.; et al. Complete chemical structures of human mitochondrial tRNAs. Nat. Commun. 2020, 11, 4269. [Google Scholar] [CrossRef]
- Abel, M.E.; Zhang, X.; Asah, S.M.; Wolfinger, A.; McCullumsmith, R.E.; O’Donovan, S.M. KEOPS complex expression in the frontal cortex in major depression and schizophrenia. World J. Biol. Psychiatry 2021, 22, 446–455. [Google Scholar] [CrossRef]
- Treimer, E.; Niedermayer, K.; Schumann, S.; Zenker, M.; Schmeisser, M.J.; Kuhl, S.J. Galloway-Mowat syndrome: New insights from bioinformatics and expression during Xenopus embryogenesis. Gene Expr. Patterns 2021, 42, 119215. [Google Scholar] [CrossRef]
- Braun, D.A.; Shril, S.; Sinha, A.; Schneider, R.; Tan, W.; Ashraf, S.; Hermle, T.; Jobst-Schwan, T.; Widmeier, E.; Majmundar, A.J.; et al. Mutations in WDR4 as a new cause of Galloway-Mowat syndrome. Am. J. Med. Genet. A 2018, 176, 2460–2465. [Google Scholar] [CrossRef]
- Alexandrov, A.; Martzen, M.R.; Phizicky, E.M. Two proteins that form a complex are required for 7-methylguanosine modification of yeast tRNA. RNA 2002, 8, 1253–1266. [Google Scholar] [CrossRef]

| Enzymes | Ligands | KD (μM) | Methods | Refs |
|---|---|---|---|---|
| EcTsaC | ATP | 1.04 ± 0.67 | FQ | [83] |
| L-Threonine | 0.04 ± 0.10 | |||
| StSua5 | ATP | 61.3 ± 3.0 | ITC | [33] |
| ADP | 101.0 ± 3.3 | |||
| AMP | 1420 ± 170 | |||
| L-Threonine | 9.3 ± 0.3 | |||
| HsYRDC | ATP | 170 ± 220 * | Kinetic analysis | [29] |
| L-Threonine | 190 ± 60 * | |||
| HCO3− | 13000 ± 3800 * | |||
| EcTsaD | AMPCPP | 1.62 ± 0.088 | ITC | [35] |
| EcTsaDB | AMPCPP | 0.70 ± 0.011 | ITC | [35] |
| BK951 | 3.4 ± 0.6 | MST | [36] | |
| EcTsaE | MANT-ADP | 8 ± 0.9 | FRET | [84] |
| ADP | 6.54 ± 0.053 | ITC | [35] | |
| ATPγS | 20.41 ± 1.32 | ITC | [35] | |
| TmTsaDBE | AMPPCP | 6.1 ± 1.0 | ITC | [85] |
| TmTsaE | AMPPCP | 9.0 ± 1.3 | ITC | [85] |
| HiTsaE | MANT-ADP | 18 ± 0.4 | FRET | [84] |
| Organisms | Enzymes [Refs] | t6A-Modified tRNAs |
|---|---|---|
| Bacteria | BsTsaC and BsTsaBDE [79] | BstRNALys (UUU) and BstRNAThr (GGU) |
| ScSua5/PaSua5/EcTsaC and EcTsaBDE [17,18,27,29,66,72] | Hsmt-tRNAAsn (GUU), Hsmt-tRNAThr (UGU), Hsmt-tRNAIle (GAU), Hsmt-tRNASer (AGY)(GCU), Hsmt-tRNALys (UUU) EctRNAiMet (CAU), EctRNAIle (GAU), EctRNALys (UUU), EctRNAAsn (GUU), MjtRNALys (UUU) and EctRNAThr (GGU) | |
| TmTsaC and TmTsaBDE [38,85] | EctRNAThr (CGU) and EctRNALys (UUU) | |
| Archaea | ScSua5/PaSua5/EcTsaC and PaKEOPS [65] | EctRNAAsn (GUU), EctRNAIle (GAU) EctRNAfMet (CAU), EctRNALys (UUU), PatRNALys (UUU) and PatRNAIle (GAU) |
| MjSua5 and MjKEOPS [72] | MjtRNAAsn (GUU), MjtRNAMet (CAU), SctRNAIle (AAU), MjtRNALys (UUU) and MjtRNAThr (GGU) | |
| Eukarya | ScSua5/PaSua5/EcTsaC/BsTsaC and ScKEOPS [18,27,65,74] | HstRNAAsn (GUU), HstRNASer (GCU), HstRNALys (CUU/UUU), HstRNAiMet (CAU), HstRNAArg (CCU/UCU), HstRNAThr (AGU/CGU/UGU), SctRNAiMet (CAU), Scmt-tRNAArg (UCU), HstRNAIle (AAU/GAU/UAU), EctRNALys (UUU), EctRNAAsn (GUU), EctRNAThr (AGU/CGU/UGU) and Hsmt-tRNAThr (UGU) |
| HsYRDC and CeKEOPS [27] | HstRNAiMet (CAU) and HstRNAThr (UGU) | |
| HsYRDC and HsKEOPS [27,72,79] | HstRNAIle (UAU), HstRNAiMet (CAU), HstRNAThr (UGU), HstRNAArg (UCU), HstRNALys (UUU) and Scmt-tRNAArg (UCU) | |
| ScSua5/EcTsaC and ScQri7 [18,72,74] | EctRNALys (UUU), MjtRNALys (UUU), SctRNAIle (AAU), Hsmt-tRNAThr (UGU) and Scmt-tRNAArg (UCU) | |
| HsYRDC and HsOSGEPL1 [29,74] | Hsmt-tRNASer (AGY)(GCU), Hsmt-tRNALys (UUU), Hsmt-tRNAIle (GAU), Hsmt-tRNAAsn (GUU) and Hsmt-tRNAThr (UGU) |
| Proteins | tRNAs | KD (μM) | Methods | Refs |
|---|---|---|---|---|
| EcTsaC | Modified EctRNAThr (CGU) | 0.62 ± 0.13 | FQ | [59] |
| EctRNAThr (CGU) | 0.11 ± 0.05 | [59] | ||
| EctRNAs * | 0.68 ± 0.15 | [31] | ||
| EcASLLys (UUU) | 0.27 ± 0.20 | [83] | ||
| EcTsaDB | EctRNALys (UUU) | 0.087 ± 0.011 | FQ | [36] |
| TmTsaB2D2 | EctRNAThr (CGU) | 1.3 ± 0.07 | EMSA | [85] |
| TmTsaB2D2E | EctRNAThr (CGU) | 0.8 ± 0.02 | ||
| PaKEOPS | EctRNALys (UUU) | 0.1~0.5 | EMSA | [65] |
| PaPcc1–Kae1 | 0.2~0.4 | |||
| PaKae1–Bud32 | ~2.0 | |||
| PaCgi121 | No binding | |||
| MjKEOPS | MjtRNALys (UUU) | 0.263 ± 0.064 | FP | [72] |
| MjKBC | 0.153 ± 0.068 | |||
| MjBC | 0.229 ± 0.027 | |||
| MjCgi121 | 0.730 ± 0.060 | |||
| HsKEOPS | MjtRNALys (UUU) | 4.30 ± 0.64 | FP | [72] |
| HsBC | 0.45 ± 0.05 | |||
| HsTPRKB | 28.3 ± 10 | |||
| ScQri7–Qri7 | Hsmt-tRNAThr (CGU) | 0.0269 ± 0.0012 | BLI | [30] |
| HsOSGEPL1 | Hsmt-tRNAThr (UGU) | 6.7 ± 0.1 | BLI | [74] |
| Proteins | Mutations | Effect | Refs |
|---|---|---|---|
| OSGEP | Ile14Phe Cys110Arg Ile111Thr | May affect catalytic activity of OSGEP | [56,124] |
| Lys78Glu Val107Met | May interfere with protein folding | [56] | |
| Lys198Arg Arg247Gln Arg280His | May interfere with the interaction with TP53RK | [42,124] | |
| Arg325Gln | May interfere with the interaction with the tRNA substrate | [56] | |
| TP53RK | Gly42Asp | May affect the ATPase activity of TP53RK | [80] |
| Arg55Gly Lys60Ser Thr81Arg | May interfere with the interaction with TPRKB | [56,80,100] | |
| Lys65Met | May interfere with the interaction with OSGEP | [100,111] | |
| Arg243Leu | May affect the t6A-catalytic activity of OSGEP | [100] | |
| TPRKB | Leu136Pro Tyr149Cys | May affect protein structural integrity | [100] |
| LAGE3 | Val106Phe | May affect protein structural integrity | [56] |
| Phe137Ser | May interfere with the interaction with OSGEP | [42] | |
| GON7 | Try7* | Loss of GON7, affects KEOPS stability | [42] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, C.; Jin, M.; Zhang, W. Conservation and Diversification of tRNA t6A-Modifying Enzymes across the Three Domains of Life. Int. J. Mol. Sci. 2022, 23, 13600. https://doi.org/10.3390/ijms232113600
Su C, Jin M, Zhang W. Conservation and Diversification of tRNA t6A-Modifying Enzymes across the Three Domains of Life. International Journal of Molecular Sciences. 2022; 23(21):13600. https://doi.org/10.3390/ijms232113600
Chicago/Turabian StyleSu, Chenchen, Mengqi Jin, and Wenhua Zhang. 2022. "Conservation and Diversification of tRNA t6A-Modifying Enzymes across the Three Domains of Life" International Journal of Molecular Sciences 23, no. 21: 13600. https://doi.org/10.3390/ijms232113600
APA StyleSu, C., Jin, M., & Zhang, W. (2022). Conservation and Diversification of tRNA t6A-Modifying Enzymes across the Three Domains of Life. International Journal of Molecular Sciences, 23(21), 13600. https://doi.org/10.3390/ijms232113600






