Gene Expression in the Developing Seed of Wild and Domesticated Rice
Abstract
1. Introduction
2. Results
2.1. The Mapping Output of RNA-Seq Reads
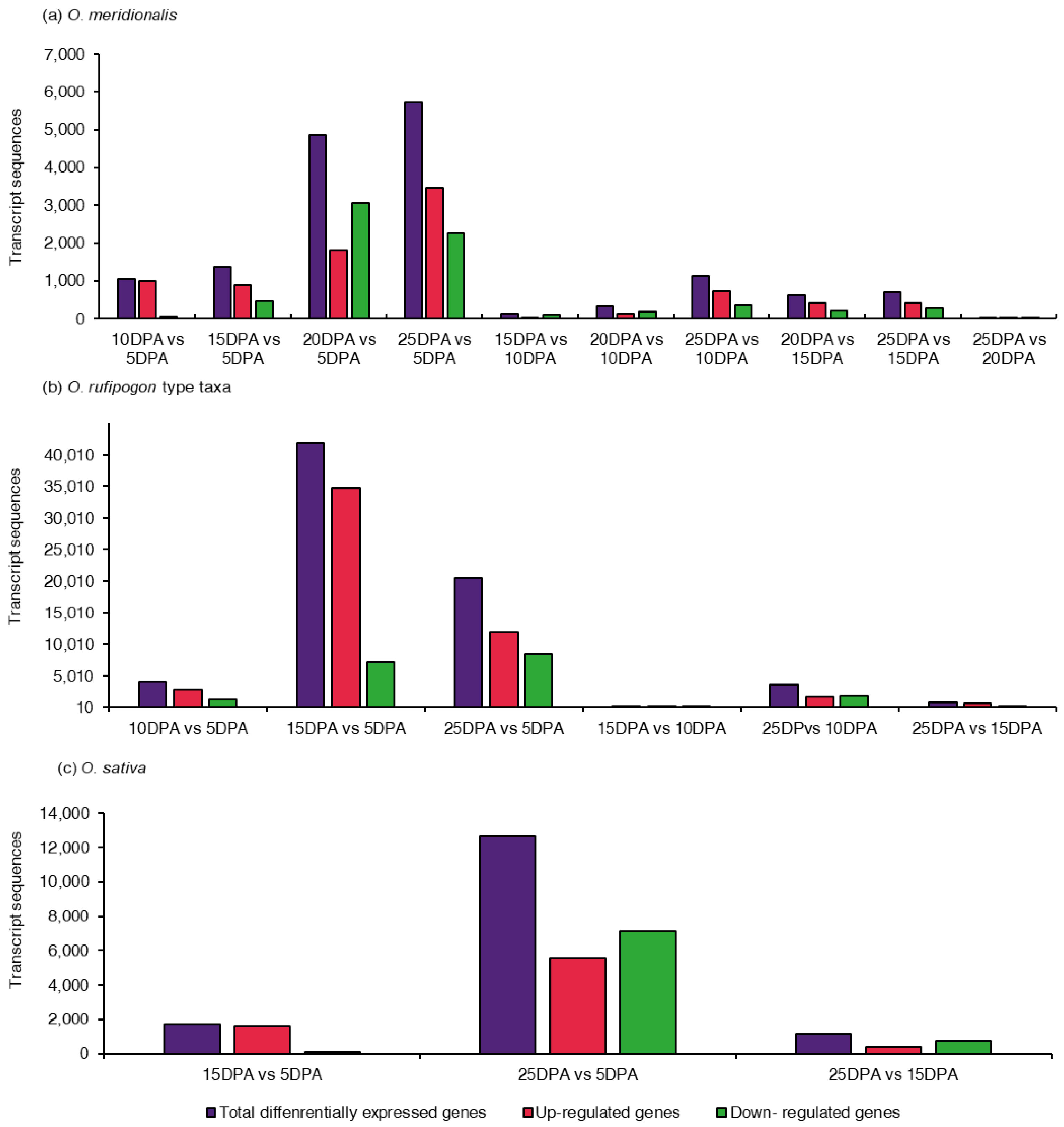
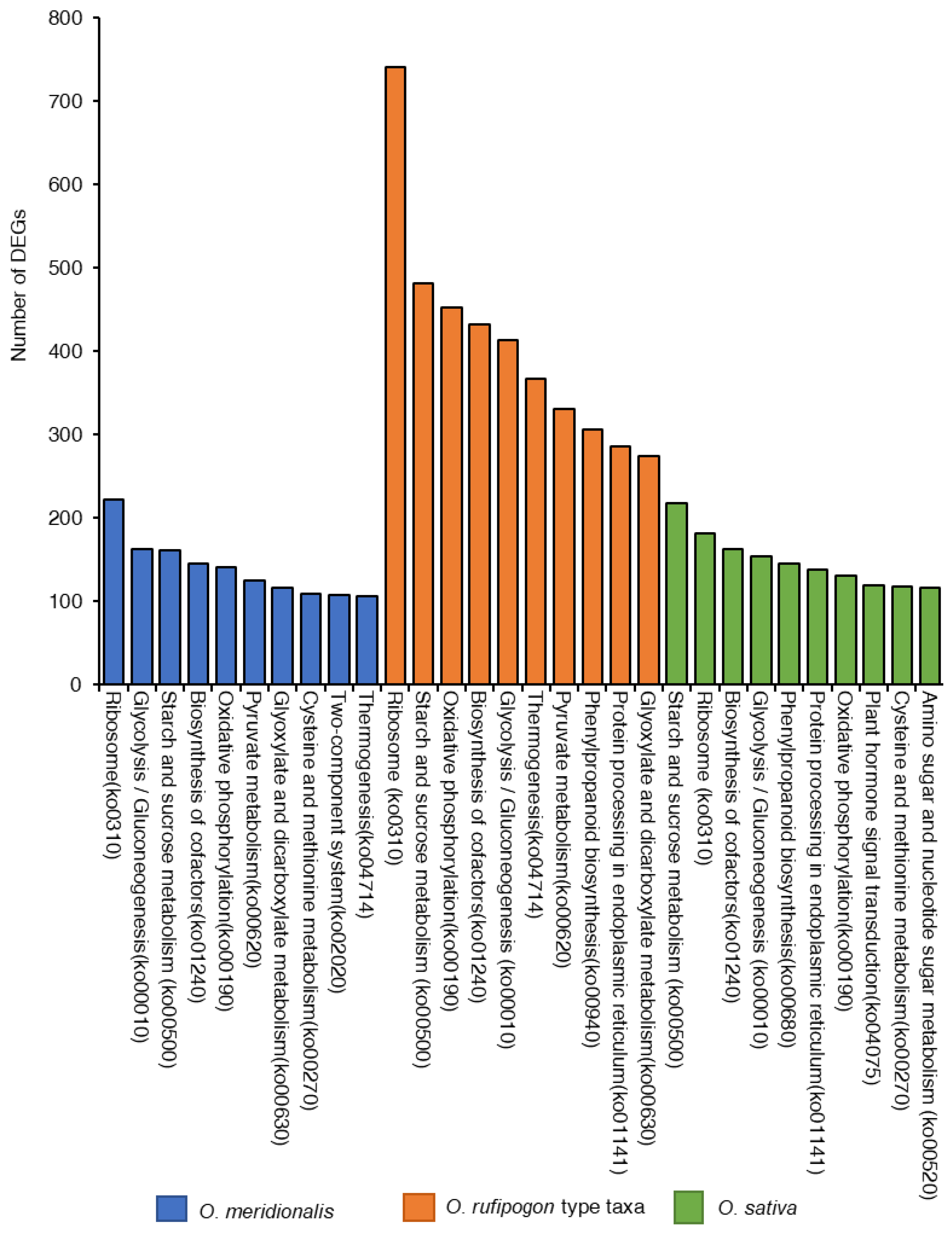
2.2. Analysis of Differentially Expressed Genes (DEGs)
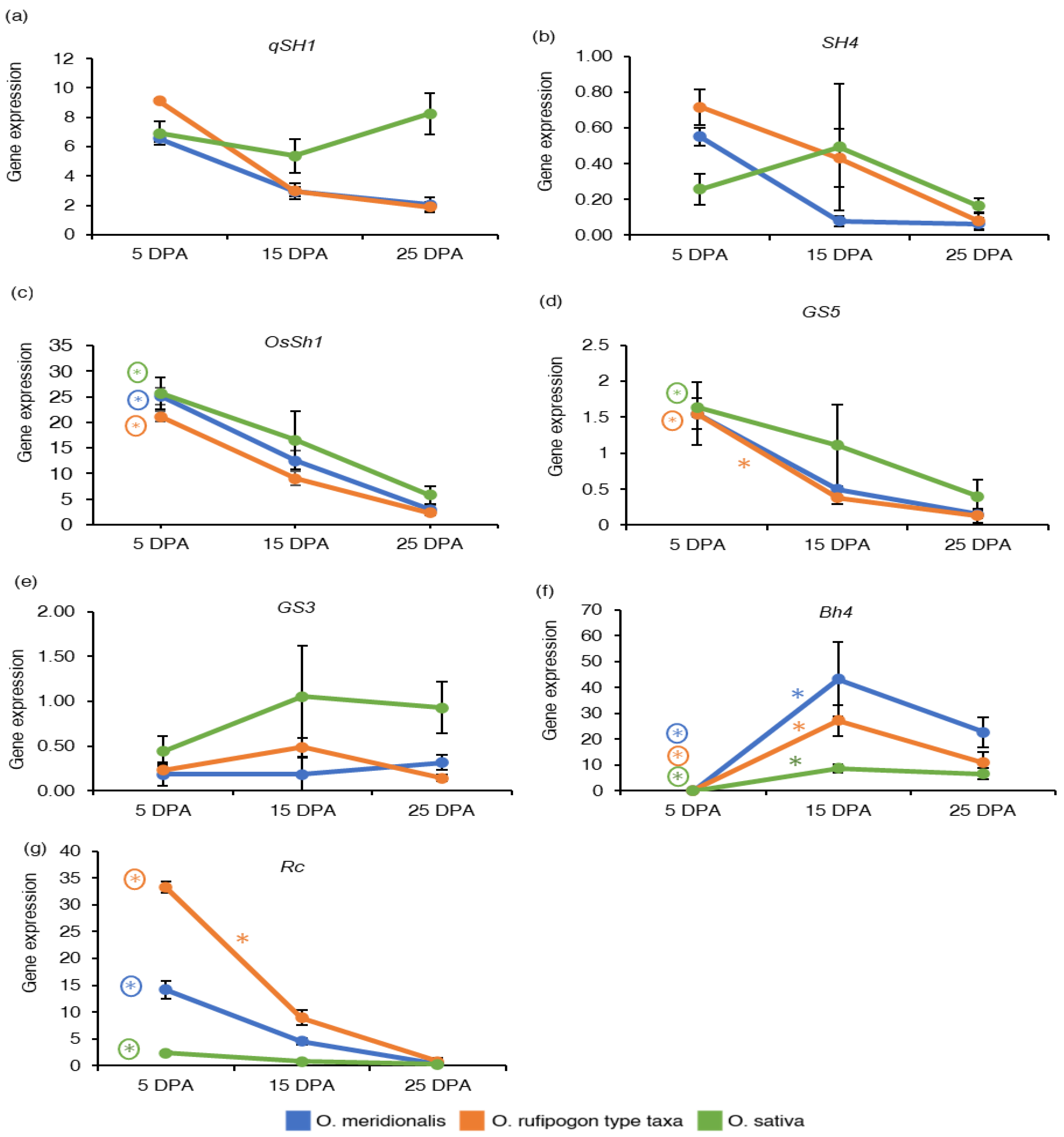
2.3. Expression Patterns of Genes Associated with Domestication Traits
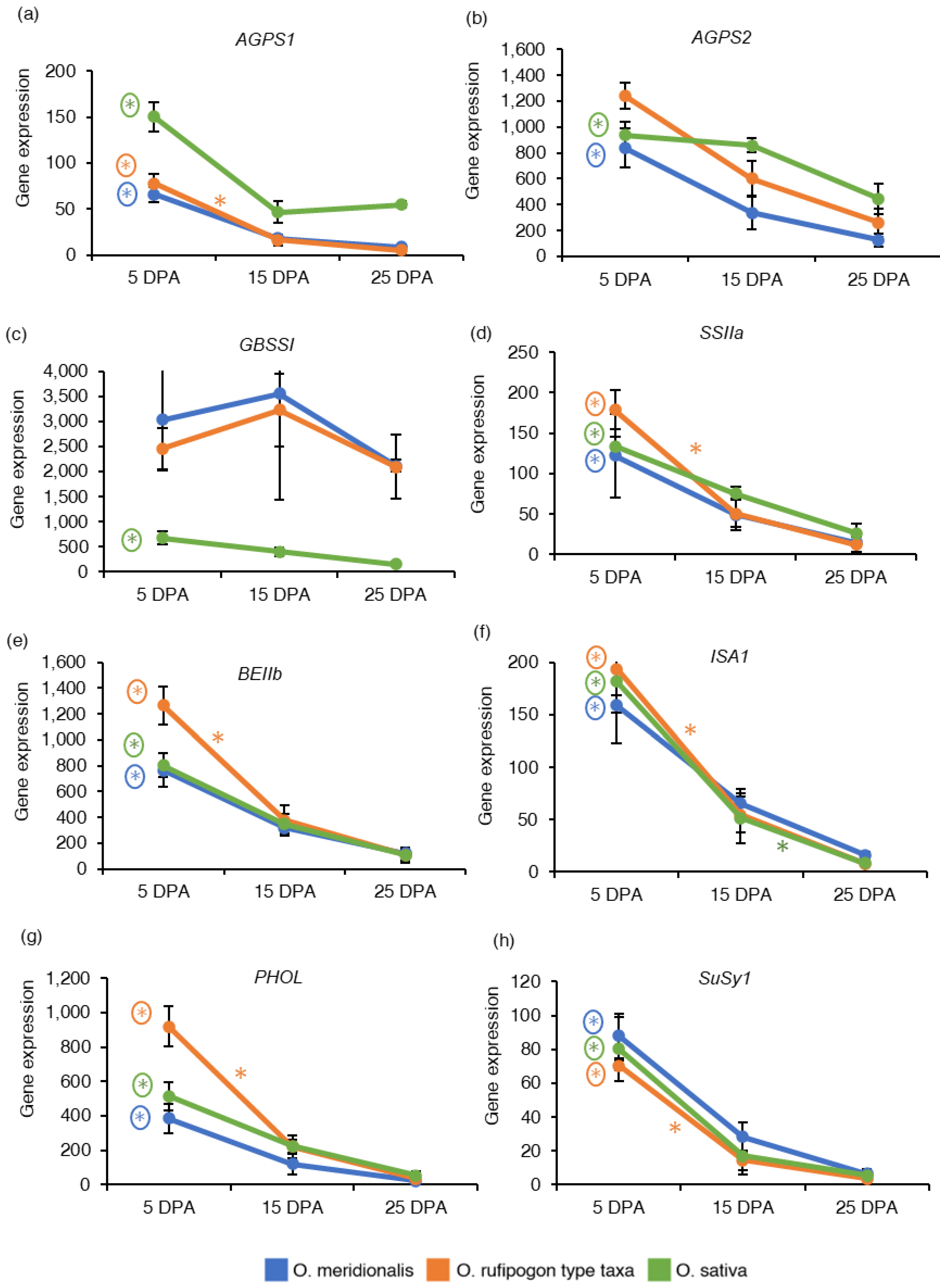
2.4. Expression Patterns of Genes Associated with Starch and Sucrose Metabolism
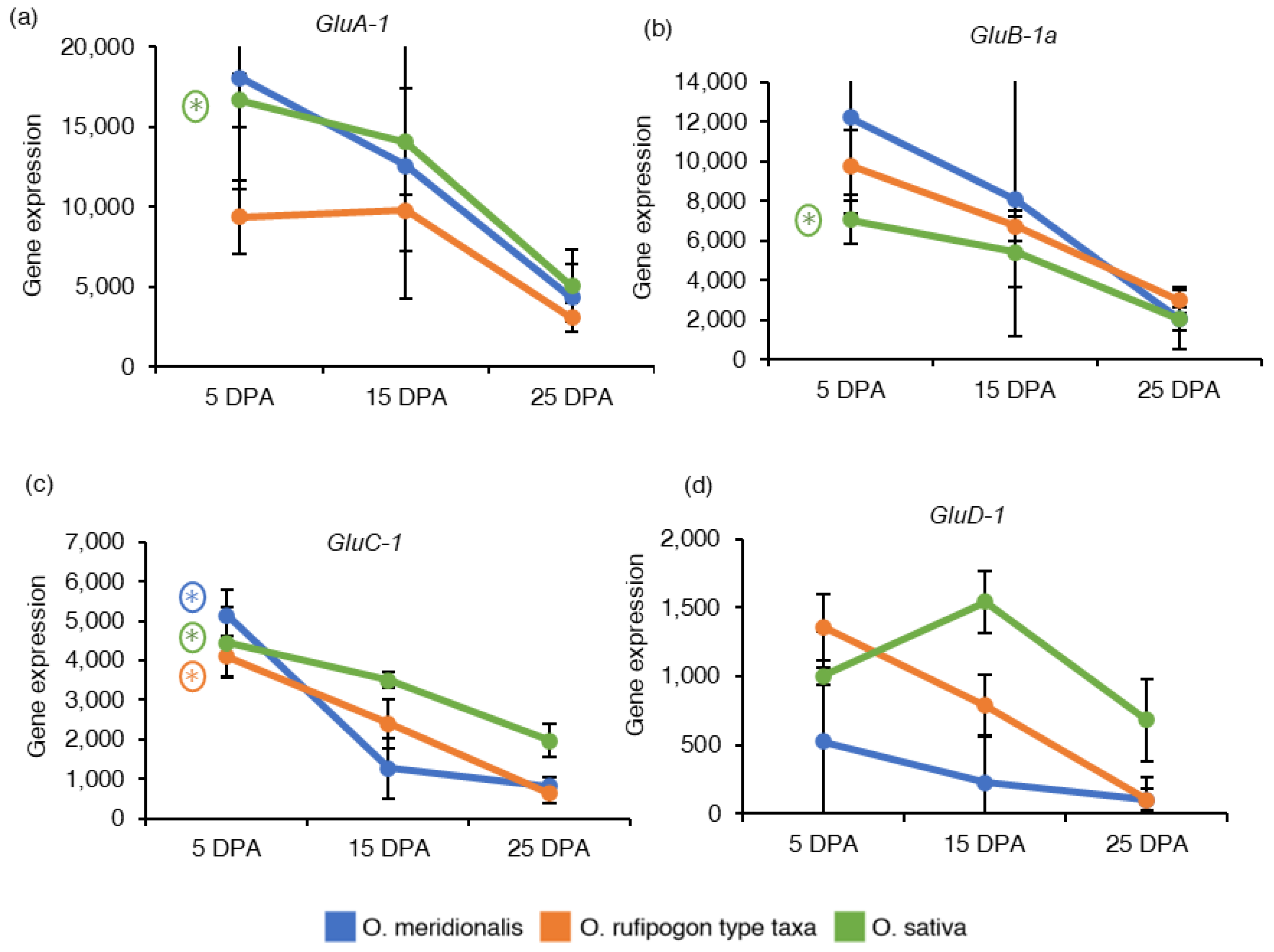
2.5. Expression Patterns of Genes Associated with Seed Storage Proteins
2.6. Discovery of Novel Transcripts
2.7. Functional Annotation of DEGs
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. RNA Extraction and Purity Determination
4.3. cDNA Library Preparation, RNA Sequencing, and Quality Check
4.4. Construction of a New Reference Genome and RNA-Seq Analysis
4.5. Gene Expression Analysis
4.6. Differentially Expressed Genes (DEGs) Analysis
4.7. Discovery of Novel Transcripts
4.8. Functional Annotation of the Rice Transcriptome
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feng, Q.; Zhang, Y.; Hao, P.; Wang, S.; Fu, G.; Huang, Y.; Li, Y.; Zhu, J.; Liu, Y.; Hu, X.; et al. Sequence and analysis of rice chromosome 4. Nature 2002, 420, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, P.; Kapoor, S.; Tyagi, A.K. Transcription factors regulating the progression of monocot and dicot seed development. Bioessays 2011, 33, 189–202. [Google Scholar] [CrossRef]
- Xue, L.J.; Zhang, J.J.; Xue, H.W. Genome-wide analysis of the complex transcriptional networks of rice developing seeds. PLoS ONE 2012, 7, e31081. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, R.B.; de Paiva, G.; Yadegari, R. Plant Embryogenesis: Zygote to Seed. Science 1994, 266, 605–614. [Google Scholar] [CrossRef] [PubMed]
- Kasem, S.; Waters, D.L.E.; Henry, R.J. Analysis of starch gene diversity in the wild relatives of Oryza sativa. Trop. Plant Biol. 2012, 5, 286–308. [Google Scholar] [CrossRef]
- Kasem, S.; Waters, D.L.E.; Ward, R.M.; Rice, N.F.; Henry, R.J. Wild Oryza grain physico-chemical properties. Trop. Plant Biol. 2014, 7, 13–18. [Google Scholar] [CrossRef]
- Kurata, N.; Miyoshi, K.; Nonomura, K.; Yamazaki, Y.; Ito, Y. Rice mutants and genes related to organ development, morphogenesis and physiological traits. Plant Cell Physiol. 2005, 46, 48–62. [Google Scholar] [CrossRef]
- Xu, H.; Gao, Y.; Wang, J. Transcriptomic analysis of rice (Oryza sativa) developing embryos using the RNA-Seq technique. PLoS ONE 2012, 7, e30646. [Google Scholar] [CrossRef]
- Conesa, A.; Madrigal, P.; Tarazona, S.; Gomez-Cabrero, D.; Cervera, A.; McPherson, A.E.; Szcześniak, M.W.; Gaffney, D.J.; Elo, L.L.; Zhang, X.; et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016, 17, 13. [Google Scholar] [CrossRef]
- Alwine, J.C.; Kemp, D.J.; Stark, G.R. Method for detection of specific RNAs in agarose gels by transfer to diazobenzyloxymethyl-paper and hybridization with DNA probes. Proc. Natl. Acad. Sci. USA 1977, 74, 5350. [Google Scholar] [CrossRef]
- Becker-André, M.; Hahlbrock, K. Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcript titration assay (PATTY). Nucleic Acids Res. 1989, 17, 9437–9446. [Google Scholar] [CrossRef] [PubMed]
- Adams, M.D.; Kelley, J.M.; Gocayne, J.D.; Dubnick, M.; Polymeropoulos, M.H.; Xiao, H.; Merril, C.R.; Wu, A.; Olde, B.; Moreno, R.F.; et al. Complementary DNA sequencing: Expressed sequence tags and human genome project. Science 1991, 252, 1651–1656. [Google Scholar] [CrossRef] [PubMed]
- Velculescu, V.E.; Zhang, L.; Vogelstein, B.; Kinzler, K.W. Serial analysis of gene expression. Science 1995, 270, 484–487. [Google Scholar] [CrossRef] [PubMed]
- Shiraki, T.; Kondo, S.; Katayama, S.; Waki, K.; Kasukawa, T.; Kawaji, H.; Kodzius, R.; Watahiki, A.; Nakamura, M.; Arakawa, T. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. USA 2003, 100, 15776–15781. [Google Scholar] [CrossRef]
- Derveaux, S.; Vandesompele, J.; Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 2010, 50, 227–230. [Google Scholar] [CrossRef]
- Schena, M.; Shalon, D.; Davis, R.W.; Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 1995, 270, 467–470. [Google Scholar] [CrossRef]
- Garber, M.; Grabherr, M.G.; Guttman, M.; Trapnell, C. Computational methods for transcriptome annotation and quantification using RNA-seq. Nat. Methods 2011, 8, 469–477. [Google Scholar] [CrossRef]
- Wang, Z.; Gerstein, M.; Snyder, M. RNA-Seq: A revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009, 10, 57–63. [Google Scholar] [CrossRef]
- Rangan, P.; Furtado, A.; Henry, R.J. The transcriptome of the developing grain: A resource for understanding seed development and the molecular control of the functional and nutritional properties of wheat. BMC Genom. 2017, 18, 766. [Google Scholar] [CrossRef]
- Guan, Y.; Li, G.; Chu, Z.; Ru, Z.; Jiang, X.; Wen, Z.; Zhang, G.; Wang, Y.; Zhang, Y.; Wei, W. Transcriptome analysis reveals important candidate genes involved in grain-size formation at the stage of grain enlargement in common wheat cultivar “Bainong 4199”. PLoS ONE 2019, 14, e0214149. [Google Scholar] [CrossRef]
- Qu, J.; Ma, C.; Feng, J.; Xu, S.; Wang, L.; Li, F.; Li, Y.; Zhang, R.; Zhang, X.; Xue, J.; et al. Transcriptome dynamics during maize endosperm development. PLoS ONE 2016, 11, e0163814. [Google Scholar] [CrossRef]
- Kandpal, M.; Vishwakarma, C.; Krishnan, K.; Chinnusamy, V.; Pareek, A.; Sharma, M.K.; Sharma, R. Gene expression dynamics in rice peduncles at the heading stage. Front. Genet. 2020, 11, 584678. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wang, D.; Li, J.; Lei, Y.; Li, G.; Cai, W.; Zhao, X.; Liang, W.; Zhang, D. Transcriptome analysis reveals photoperiod-associated genes expressed in rice anthers. Front. Plant Sci. 2021, 12, 621561. [Google Scholar] [CrossRef] [PubMed]
- Cheng, A.-P.; Chen, S.-Y.; Lai, M.-H.; Wu, D.-H.; Lin, S.-S.; Chen, C.-Y.; Chung, C.-L. Transcriptome analysis of early defenses in rice against fusarium fujikuroi. Rice 2020, 13, 65. [Google Scholar] [CrossRef]
- Liang, Y.; Tabien, R.E.; Tarpley, L.; Mohammed, A.R.; Septiningsih, E.M. Transcriptome profiling of two rice genotypes under mild field drought stress during grain-filling stage. AoB Plants 2021, 13, plab043. [Google Scholar] [CrossRef]
- Sun, L.; Wang, J.; Song, K.; Sun, Y.; Qin, Q.; Xue, Y. Transcriptome analysis of rice (Oryza sativa L.) shoots responsive to cadmium stress. Sci. Rep. 2019, 9, 10177. [Google Scholar] [CrossRef]
- Zhu, T.; Budworth, P.; Chen, W.; Provart, N.; Chang, H.S.; Guimil, S.; Su, W.; Estes, B.; Zou, G.; Wang, X. Transcriptional control of nutrient partitioning during rice grain filling. Plant Biotechnol. J. 2003, 1, 59–70. [Google Scholar] [CrossRef]
- Ohdan, T.; Francisco, P.B., Jr.; Sawada, T.; Hirose, T.; Terao, T.; Satoh, H.; Nakamura, Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. J. Exp. Bot. 2005, 56, 3229–3244. [Google Scholar] [CrossRef]
- Zhang, J.; Chen, L.; Fu, C.; Wang, L.; Liu, H.; Cheng, Y.; Li, S.; Deng, Q.; Wang, S.; Zhu, J.; et al. Comparative transcriptome analyses of gene expression changes triggered by rhizoctonia solani ag1 ia infection in resistant and susceptible rice varieties. Front. Plant Sci. 2017, 8, 1422. [Google Scholar] [CrossRef]
- Moner, A.M.; Furtado, A.; Chivers, I.; Fox, G.; Crayn, D.; Henry, R.J. Diversity and evolution of rice progenitors in Australia. Ecol. Evol. 2018, 8, 4360–4366. [Google Scholar] [CrossRef]
- Kawahara, Y.; de la Bastide, M.; Hamilton, J.P.; Kanamori, H.; McCombie, W.R.; Ouyang, S.; Schwartz, D.C.; Tanaka, T.; Wu, J.; Zhou, S.; et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice 2013, 6, 4. [Google Scholar] [CrossRef] [PubMed]
- Kukurba, K.R.; Montgomery, S.B. RNA sequencing and analysis. Cold Spring Harb. Protoc. 2015, 2015, 951–969. [Google Scholar] [CrossRef]
- Maekawa, M. Geographical distribution of the genes for black hull coloration. Rice Genet. Newl. 1984, 1, 104–105. [Google Scholar]
- Sweeney, M.T.; Thomson, M.J.; Pfeil, B.E.; McCouch, S. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 2006, 18, 283–294. [Google Scholar] [CrossRef]
- Gan, L.; Huang, B.; Song, Z.; Zhang, Y.; Zhang, Y.; Chen, S.; Tong, L.; Wei, Z.; Yu, L.; Luo, X.; et al. Unique glutelin expression patterns and seed endosperm structure facilitate glutelin accumulation in polyploid rice seed. Rice 2021, 14, 61. [Google Scholar] [CrossRef]
- Konishi, S.; Izawa, T.; Lin, S.Y.; Ebana, K.; Fukuta, Y.; Sasaki, T.; Yano, M. An SNP caused loss of seed shattering during rice domestication. Science 2006, 312, 1392–1396. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhou, A.; Sang, T. Rice domestication by reducing shattering. Science 2006, 311, 1936–1939. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Komatsu, A.; Ohtake, M.; Eun, H.; Shimizu, A.; Kato, H. Direct identification of a mutation in OsSh1 causing non-shattering in a rice (Oryza sativa L.) mutant cultivar using whole-genome resequencing. Sci. Rep. 2020, 10, 14936. [Google Scholar] [CrossRef]
- Li, N.; Xu, R.; Duan, P.; Li, Y. Control of grain size in rice. Plant Reprod. 2018, 31, 237–251. [Google Scholar] [CrossRef]
- Tikapunya, T.; Fox, G.; Furtado, A.; Henry, R. Grain physical characteristic of the Australian wild rices. Plant Genet. Resour. -Charact. Util. 2017, 15, 409–420. [Google Scholar] [CrossRef]
- Mao, H.; Sun, S.; Yao, J.; Wang, C.; Yu, S.; Xu, C.; Li, X.; Zhang, Q. Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. USA 2010, 107, 19579–19584. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Fan, C.; Xing, Y.; Jiang, Y.; Luo, L.; Sun, L.; Shao, D.; Xu, C.; Li, X.; Xiao, J.; et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 2011, 43, 1266–1269. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.-F.; Si, L.; Wang, Z.; Zhou, Y.; Zhu, J.; Shangguan, Y.; Lu, D.; Fan, D.; Li, C.; Lin, H.; et al. Genetic control of a transition from black to straw-white seed hull in rice domestication. Plant Physiol. 2011, 155, 1301–1311. [Google Scholar] [CrossRef] [PubMed]
- Yu, G.; Olsen, K.M.; Schaal, B.A. Molecular evolution of the endosperm starch synthesis pathway genes in rice (Oryza sativa L.) and its wild ancestor, O. rufipogon L. Mol. Biol. Evol. 2011, 28, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Juliano, B.O. Free sugars in relation to starch accumulation in developing rice grain. Plant Physiol. 1977, 59, 417–421. [Google Scholar] [CrossRef]
- Odegard, W.; Liu, J.J.; de Lumen, B.O. Cloning and expression of rice (Oryza sativa) sucrose synthase 1 (RSs1) in developing seed endosperm. Plant Sci. 1996, 113, 67–78. [Google Scholar] [CrossRef]
- Kawakatsu, T.; Yamamoto, M.P.; Hirose, S.; Yano, M.; Takaiwa, F. Characterization of a new rice glutelin gene GluD-1 expressed in the starchy endosperm. J. Exp. Bot. 2008, 59, 4233–4245. [Google Scholar] [CrossRef]
- Takaiwa, F.; Oono, K. Genomic DNA sequences of two new genes for new storage protein glutelin in rice. Jpn. J. Genet. 1991, 66, 161–171. [Google Scholar] [CrossRef][Green Version]
- Sotowa, M.; Ootsuka, K.; Kobayashi, Y.; Hao, Y.; Tanaka, K.; Ichitani, K.; Flowers, J.M.; Purugganan, M.D.; Nakamura, I.; Sato, Y.; et al. Molecular relationships between Australian annual wild rice, Oryza meridionalis, and two related perennial forms. Rice 2013, 6, 26. [Google Scholar] [CrossRef]
- Singh, G.; Kumar, S.; Singh, P. A quick method to isolate RNA from wheat and other carbohydrate-rich seeds. Plant Molecular Biol. Report. 2003, 21, 93. [Google Scholar] [CrossRef]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Liu, J.; Wang, H.; Wong, L.; Chua, N.-H. PLncDB: Plant long non-coding RNA database. Bioinformatics 2013, 29, 1068–1071. [Google Scholar] [CrossRef]
- Götz, S.; García-Gómez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talón, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.G.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M.; et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hasan, S.; Furtado, A.; Henry, R. Gene Expression in the Developing Seed of Wild and Domesticated Rice. Int. J. Mol. Sci. 2022, 23, 13351. https://doi.org/10.3390/ijms232113351
Hasan S, Furtado A, Henry R. Gene Expression in the Developing Seed of Wild and Domesticated Rice. International Journal of Molecular Sciences. 2022; 23(21):13351. https://doi.org/10.3390/ijms232113351
Chicago/Turabian StyleHasan, Sharmin, Agnelo Furtado, and Robert Henry. 2022. "Gene Expression in the Developing Seed of Wild and Domesticated Rice" International Journal of Molecular Sciences 23, no. 21: 13351. https://doi.org/10.3390/ijms232113351
APA StyleHasan, S., Furtado, A., & Henry, R. (2022). Gene Expression in the Developing Seed of Wild and Domesticated Rice. International Journal of Molecular Sciences, 23(21), 13351. https://doi.org/10.3390/ijms232113351








