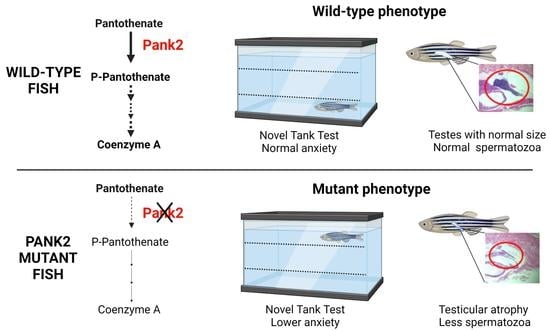
Bi-Allelic Mutations in Zebrafish pank2 Gene Lead to Testicular Atrophy and Perturbed Behavior without Signs of Neurodegeneration
Abstract
1. Introduction
2. Results
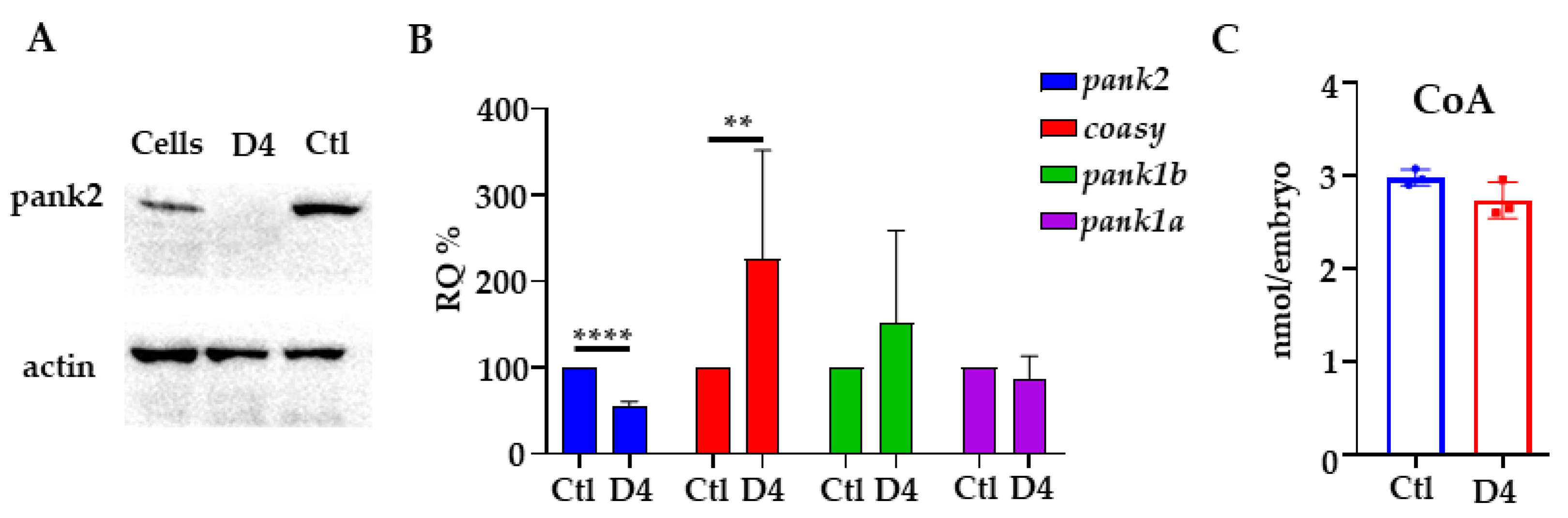
2.1. Generation and Validation of the pank2 Mutant Line
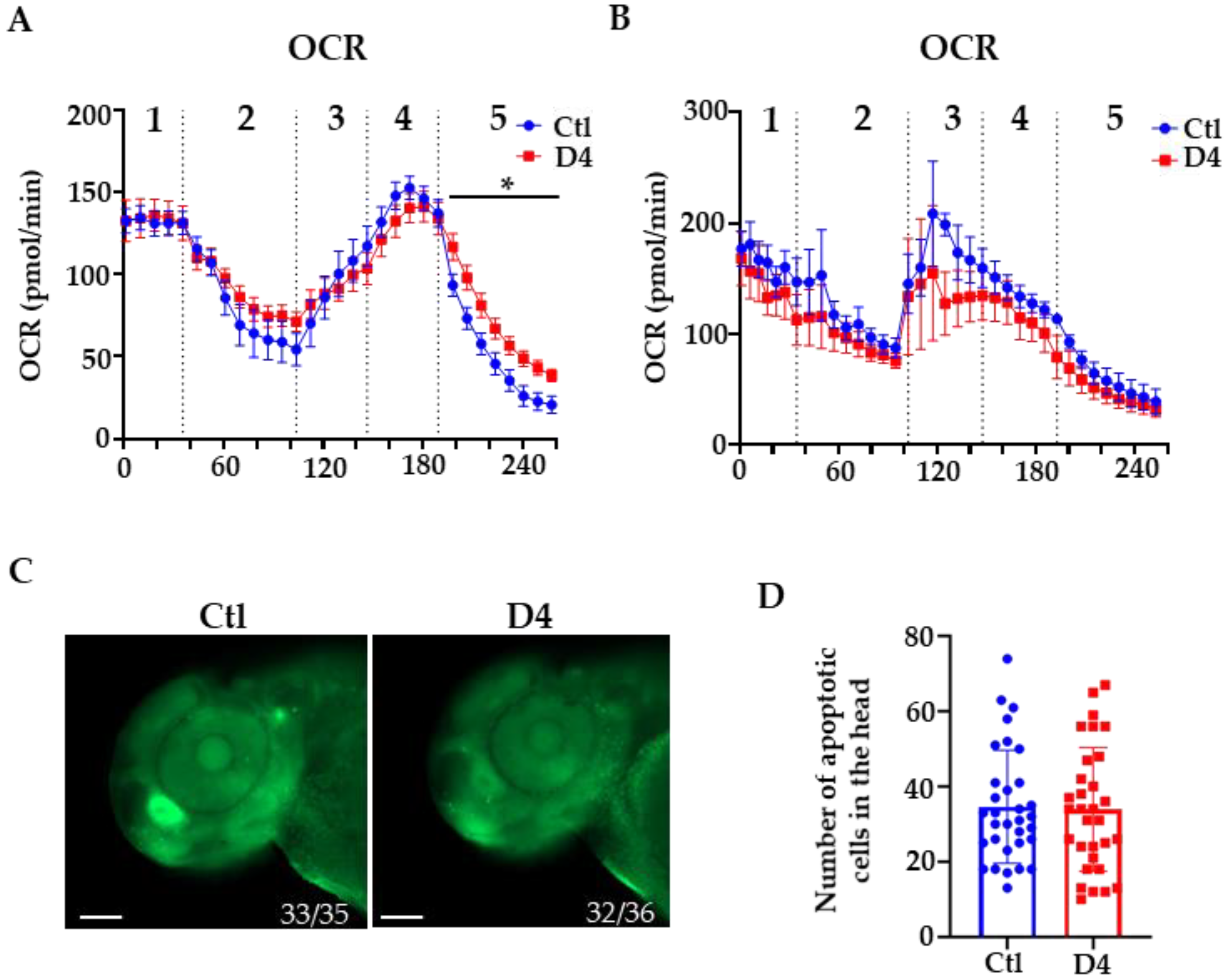
2.2. Evaluation of Mitochondrial Respiration and Cell Death
2.3. Assessment of Neural Development in the pank2 Mutant Line
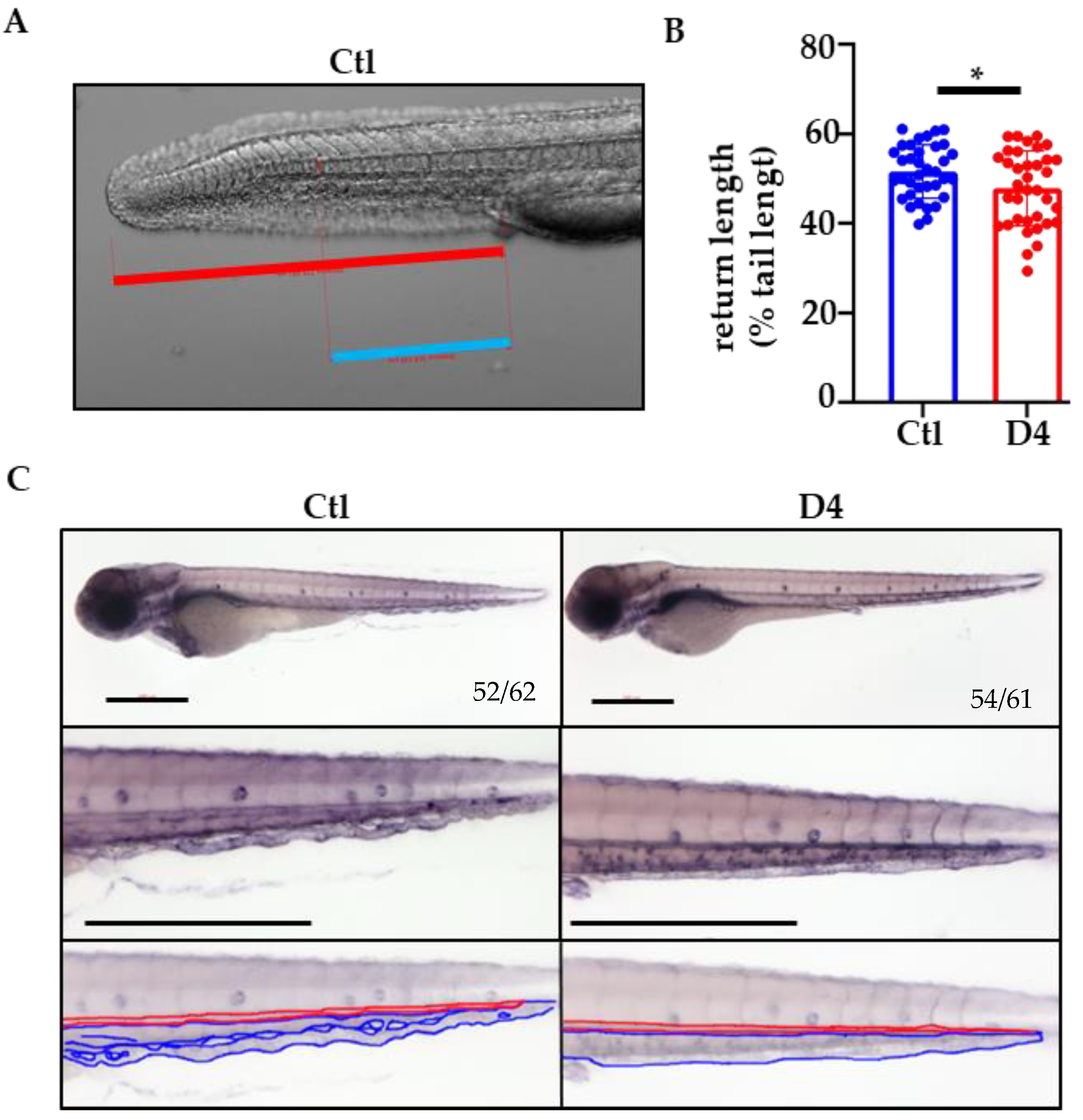
2.4. Vascular Development in pank2 D4 Mutant Embryos
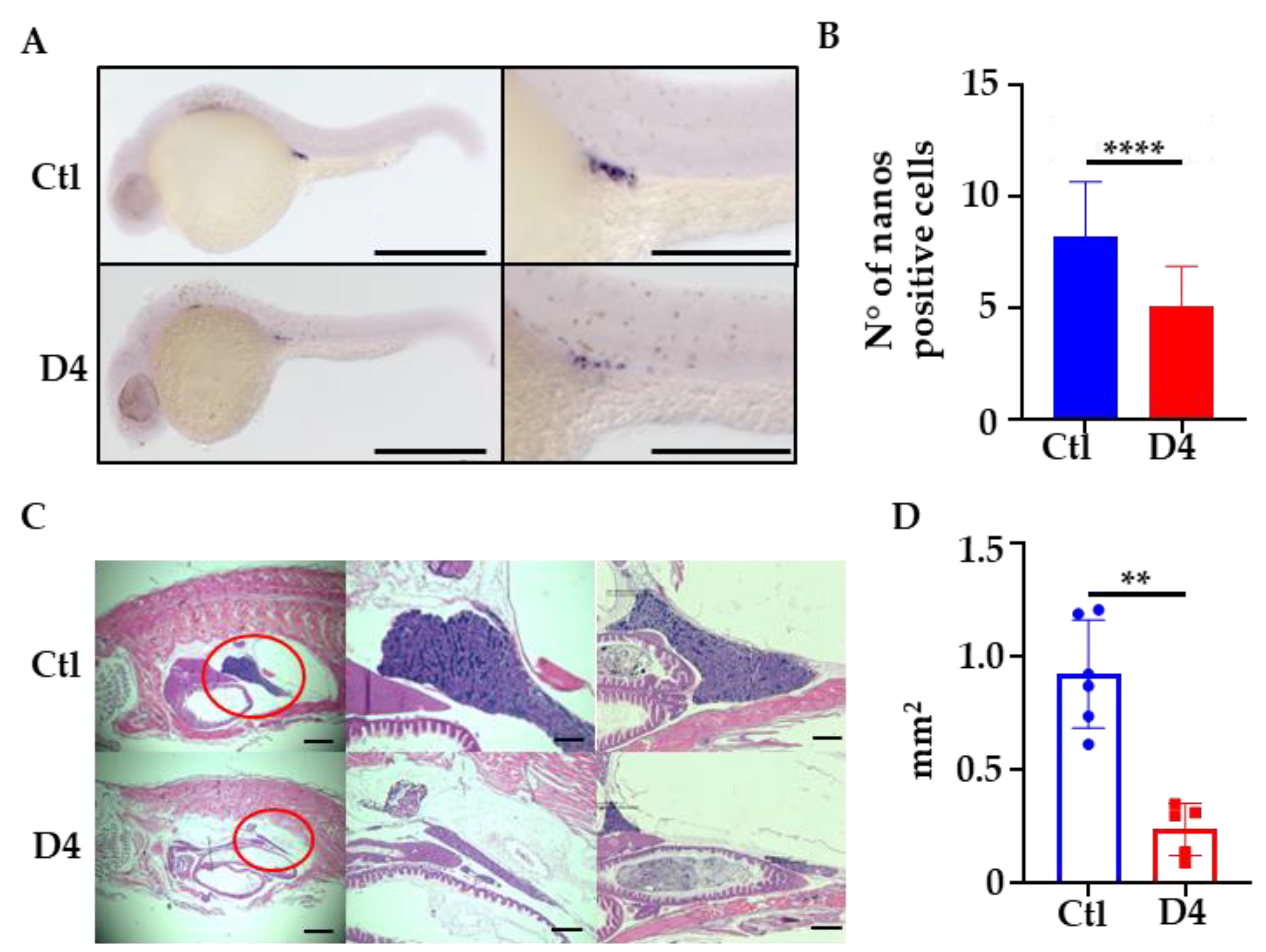
2.5. Analysis of Primordial Germ Cells Development in D4 Embryos
2.6. One-Year-Old D4 Mutants Display Testicular Atrophy
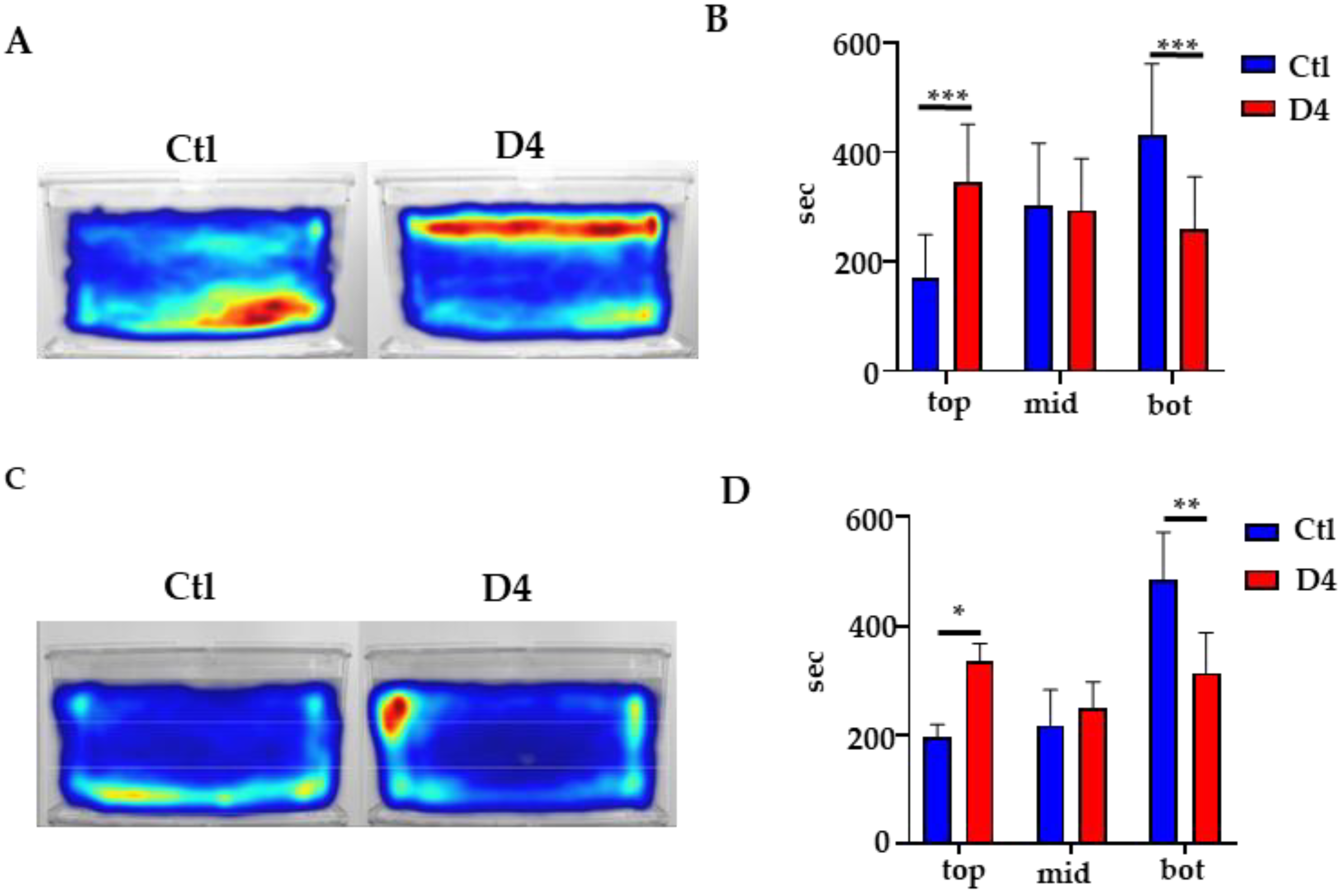
2.7. Behavioral Studies
3. Discussion
4. Materials and Methods
4.1. Fish Maintenance
4.2. Whole-Mount In Situ Hybridization (WISH)
4.3. DNA Extraction from a Single Embryo
4.4. Extraction of DNA from an Adult Fish
4.5. Heteroduplex Mobility Assay (HMA) Analysis
4.6. Western Blot
4.7. RNA-Extraction and qPCR
4.8. Coenzyme A Quantification
4.9. Microinjection
4.10. Venous Return Length Quantification
4.11. Phosphatase Assay
4.12. Oxygen Consumption Rate Measurement
4.13. Acridine Orange Staining
4.14. Immunofluorescence
4.15. Histological Section and Haematoxylin-Eosin Staining
4.16. Ejaculate Collection
4.17. Sperm Concentration
4.18. Sperm Viability
4.19. Iron Quantification
4.20. Larval Stages Behavioral Analysis
4.21. Adult Stages Behavioral Analysis
4.22. Testis Area Analysis
4.23. Adult Length Measurement
4.24. Microscopy
4.25. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hayflick, S.J. Unraveling the Hallervorden-Spatz syndrome: Pantothenate kinase-associated neurodegeneration is the name. Curr. Opin. Pediatr. 2003, 15, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Levi, S.; Tiranti, V. Neurodegeneration with Brain Iron Accumulation Disorders: Valuable Models Aimed at Understanding the Pathogenesis of Iron Deposition. Pharmaceuticals 2019, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Levi, S.; Finazzi, D. Neurodegeneration with brain iron accumulation: Update on pathogenic mechanisms. Front. Pharmacol. 2014, 5, 99. [Google Scholar] [CrossRef]
- Kolarova, H.; Tan, J.; Strom, T.M.; Meitinger, T.; Wagner, M.; Klopstock, T. Lifetime risk of autosomal recessive neurodegeneration with brain iron accumulation (NBIA) disorders calculated from genetic databases. EBioMedicine 2022, 77, 103869. [Google Scholar] [CrossRef]
- Hartig, M.B.; Hörtnagel, K.; Garavaglia, B.; Zorzi, G.; Kmiec, T.; Klopstock, T.; Rostasy, K.; Svetel, M.; Kostic, V.S.; Schuelke, M.; et al. Genotypic and phenotypic spectrum of PANK2 mutations in patients with neurodegeneration with brain iron accumulation. Ann. Neurol. 2006, 59, 248–256. [Google Scholar] [CrossRef] [PubMed]
- Hayflick, S.J.; Hartman, M.; Coryell, J.; Gitschier, J.; Rowley, H. Brain MRI in neurodegeneration with brain iron accumulation with and without PANK2 mutations. AJNR Am. J. Neuroradiol. 2006, 27, 1230–1233. [Google Scholar]
- Kruer, M.C.; Hiken, M.; Gregory, A.; Malandrini, A.; Clark, D.; Hogarth, P.; Grafe, M.; Hayflick, S.J.; Woltjer, R.L. Novel histopathologic findings in molecularly-confirmed pantothenate kinase-associated neurodegeneration. Brain 2011, 134, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Westaway, S.; Levinson, B.; Johnson, M.; Gitschier, J.; Hayflick, S. A novel pantothenate kinase gene (PANK2) is defective in Hallervorden-Spatz syndrome. Nat. Genet. 2001, 28, 345–349. [Google Scholar] [CrossRef]
- Mignani, L.; Gnutti, B.; Zizioli, D.; Finazzi, D. Coenzyme a Biochemistry: From Neurodevelopment to Neurodegeneration. Brain Sci. 2021, 11, 1031. [Google Scholar] [CrossRef]
- Sharma, L.K.; Subramanian, C.; Yun, M.K.; Frank, M.W.; White, S.W.; Rock, C.O.; Lee, R.E.; Jackowski, S. A therapeutic approach to pantothenate kinase associated neurodegeneration. Nat. Commun. 2018, 9, 4399. [Google Scholar] [CrossRef]
- Rana, A.; Seinen, E.; Siudeja, K.; Muntendam, R.; Srinivasan, B.; van der Want, J.J.; Hayflick, S.; Reijngoud, D.J.; Kayser, O.; Sibon, O.C. Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration. Proc. Natl. Acad. Sci. USA 2010, 107, 6988–6993. [Google Scholar] [CrossRef] [PubMed]
- Brunetti, D.; Dusi, S.; Morbin, M.; Uggetti, A.; Moda, F.; D’Amato, I.; Giordano, C.; d’Amati, G.; Cozzi, A.; Levi, S.; et al. Pantothenate kinase-associated neurodegeneration: Altered mitochondria membrane potential and defective respiration in Pank2 knock-out mouse model. Hum. Mol. Genet. 2012, 21, 5294–5305. [Google Scholar] [CrossRef] [PubMed]
- Campanella, A.; Privitera, D.; Guaraldo, M.; Rovelli, E.; Barzaghi, C.; Garavaglia, B.; Santambrogio, P.; Cozzi, A.; Levi, S. Skin fibroblasts from pantothenate kinase-associated neurodegeneration patients show altered cellular oxidative status and have defective iron-handling properties. Hum. Mol. Genet. 2012, 21, 4049–4059. [Google Scholar] [CrossRef] [PubMed]
- Santambrogio, P.; Dusi, S.; Guaraldo, M.; Rotundo, L.I.; Broccoli, V.; Garavaglia, B.; Tiranti, V.; Levi, S. Mitochondrial iron and energetic dysfunction distinguish fibroblasts and induced neurons from pantothenate kinase-associated neurodegeneration patients. Neurobiol. Dis. 2015, 81, 144–153. [Google Scholar] [CrossRef]
- Álvarez-Córdoba, M.; Fernández Khoury, A.; Villanueva-Paz, M.; Gómez-Navarro, C.; Villalón-García, I.; Suárez-Rivero, J.M.; Povea-Cabello, S.; de la Mata, M.; Cotán, D.; Talaverón-Rey, M.; et al. Pantothenate Rescues Iron Accumulation in Pantothenate Kinase-Associated Neurodegeneration Depending on the Type of Mutation. Mol. Neurobiol. 2018, 56, 3638–3656. [Google Scholar] [CrossRef]
- Arber, C.; Angelova, P.R.; Wiethoff, S.; Tsuchiya, Y.; Mazzacuva, F.; Preza, E.; Bhatia, K.P.; Mills, K.; Gout, I.; Abramov, A.Y.; et al. iPSC-derived neuronal models of PANK2-associated neurodegeneration reveal mitochondrial dysfunction contributing to early disease. PLoS ONE 2017, 12, e0184104. [Google Scholar] [CrossRef]
- Orellana, D.I.; Santambrogio, P.; Rubio, A.; Yekhlef, L.; Cancellieri, C.; Dusi, S.; Giannelli, S.G.; Venco, P.; Mazzara, P.G.; Cozzi, A.; et al. Coenzyme A corrects pathological defects in human neurons of PANK2-associated neurodegeneration. EMBO Mol. Med. 2016, 8, 1197–1211. [Google Scholar] [CrossRef]
- Elbaum, D.; Beconi, M.G.; Monteagudo, E.; Di Marco, A.; Quinton, M.S.; Lyons, K.A.; Vaino, A.; Harper, S. Fosmetpantotenate (RE-024), a phosphopantothenate replacement therapy for pantothenate kinase-associated neurodegeneration: Mechanism of action and efficacy in nonclinical models. PLoS ONE 2018, 13, e0192028. [Google Scholar] [CrossRef]
- Srinivasan, B.; Baratashvili, M.; van der Zwaag, M.; Kanon, B.; Colombelli, C.; Lambrechts, R.A.; Schaap, O.; Nollen, E.A.; Podgoršek, A.; Kosec, G.; et al. Extracellular 4’-phosphopantetheine is a source for intracellular coenzyme A synthesis. Nat. Chem. Biol. 2015, 11, 784–792. [Google Scholar] [CrossRef]
- Jeong, S.Y.; Hogarth, P.; Placzek, A.; Gregory, A.M.; Fox, R.; Zhen, D.; Hamada, J.; van der Zwaag, M.; Lambrechts, R.; Jin, H.; et al. 4’-Phosphopantetheine corrects CoA, iron, and dopamine metabolic defects in mammalian models of PKAN. EMBO Mol. Med. 2019, 11, e10489. [Google Scholar] [CrossRef]
- Huang, Y.; Wan, Z.; Tang, Y.; Xu, J.; Laboret, B.; Nallamothu, S.; Yang, C.; Liu, B.; Lu, R.O.; Lu, B.; et al. Pantothenate kinase 2 interacts with PINK1 to regulate mitochondrial quality control via acetyl-CoA metabolism. Nat. Commun. 2022, 13, 2412. [Google Scholar] [CrossRef] [PubMed]
- Poli, M.; Derosas, M.; Luscieti, S.; Cavadini, P.; Campanella, A.; Verardi, R.; Finazzi, D.; Arosio, P. Pantothenate kinase-2 (Pank2) silencing causes cell growth reduction, cell-specific ferroportin upregulation and iron deregulation. Neurobiol. Dis. 2010, 39, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Santambrogio, P.; Ripamonti, M.; Paolizzi, C.; Panteghini, C.; Carecchio, M.; Chiapparini, L.; Raimondi, M.; Rubio, A.; Di Meo, I.; Cozzi, A.; et al. Harmful Iron-Calcium Relationship in Pantothenate kinase Associated Neurodegeneration. Int. J. Mol. Sci. 2020, 21, 3664. [Google Scholar] [CrossRef] [PubMed]
- Santambrogio, P.; Ripamonti, M.; Cozzi, A.; Raimondi, M.; Cavestro, C.; Di Meo, I.; Rubio, A.; Taverna, S.; Tiranti, V.; Levi, S. Massive iron accumulation in PKAN-derived neurons and astrocytes: Light on the human pathological phenotype. Cell Death Dis. 2022, 13, 185. [Google Scholar] [CrossRef]
- Bosveld, F.; Rana, A.; van der Wouden, P.; Lemstra, W.; Ritsema, M.; Kampinga, H.; Sibon, O. De novo CoA biosynthesis is required to maintain DNA integrity during development of the Drosophila nervous system. Hum. Mol. Genet. 2008, 17, 2058–2069. [Google Scholar] [CrossRef]
- Kuo, Y.; Duncan, J.; Westaway, S.; Yang, H.; Nune, G.; Xu, E.; Hayflick, S.; Gitschier, J. Deficiency of pantothenate kinase 2 (Pank2) in mice leads to retinal degeneration and azoospermia. Hum. Mol. Genet. 2005, 14, 49–57. [Google Scholar] [CrossRef]
- Lambrechts, R.A.; Schepers, H.; Yu, Y.; van der Zwaag, M.; Autio, K.J.; Vieira-Lara, M.A.; Bakker, B.M.; Tijssen, M.A.; Hayflick, S.J.; Grzeschik, N.A.; et al. CoA-dependent activation of mitochondrial acyl carrier protein links four neurodegenerative diseases. EMBO Mol. Med. 2019, 11, e10488. [Google Scholar] [CrossRef]
- Petit, F.; Drecourt, A.; Dussiot, M.; Zangarelli, C.; Hermine, O.; Munnich, A.; Rötig, A. Defective palmitoylation of transferrin receptor triggers iron overload in Friedreich ataxia fibroblasts. Blood 2021, 137, 2090–2102. [Google Scholar] [CrossRef]
- Klopstock, T.; Videnovic, A.; Bischoff, A.T.; Bonnet, C.; Cif, L.; Comella, C.; Correa-Vela, M.; Escolar, M.L.; Fraser, J.L.; Gonzalez, V.; et al. Fosmetpantotenate Randomized Controlled Trial in Pantothenate Kinase-Associated Neurodegeneration. Mov. Disord. 2020, 36, 1342–1352. [Google Scholar] [CrossRef]
- Chang, X.; Zhang, J.; Jiang, Y.; Yao, B.; Wang, J.; Wu, Y. Pilot trial on the efficacy and safety of pantethine in children with pantothenate kinase-associated neurodegeneration: A single-arm, open-label study. Orphanet J. Rare Dis. 2020, 15, 248. [Google Scholar] [CrossRef]
- Spence, R.; Gerlach, G.; Lawrence, C.; Smith, C. The behaviour and ecology of the zebrafish, Danio rerio. Biol. Rev. Camb. Philos. Soc. 2008, 83, 13–34. [Google Scholar] [CrossRef] [PubMed]
- Zizioli, D.; Tiso, N.; Guglielmi, A.; Saraceno, C.; Busolin, G.; Giuliani, R.; Khatri, D.; Monti, E.; Borsani, G.; Argenton, F.; et al. Knock-down of pantothenate kinase 2 severely affects the development of the nervous and vascular system in zebrafish, providing new insights into PKAN disease. Neurobiol. Dis. 2016, 85, 35–48. [Google Scholar] [CrossRef] [PubMed]
- Khatri, D.; Zizioli, D.; Trivedi, A.; Borsani, G.; Monti, E.; Finazzi, D. Overexpression of Human Mutant PANK2 Proteins Affects Development and Motor Behavior of Zebrafish Embryos. Neuromol. Med. 2018, 21, 120–131. [Google Scholar] [CrossRef]
- Pagani, F.; Trivedi, A.; Khatri, D.; Zizioli, D.; Garrafa, E.; Mitola, S.; Finazzi, D. Silencing of pantothenate kinase 2 reduces endothelial cell angiogenesis. Mol. Med. Rep. 2018, 18, 4739–4746. [Google Scholar] [CrossRef] [PubMed]
- Gagnon, J.A.; Valen, E.; Thyme, S.B.; Huang, P.; Ahkmetova, L.; Pauli, A.; Montague, T.G.; Zimmerman, S.; Richter, C.; Schier, A.F. Efficient mutagenesis by Cas9 protein-mediated oligonucleotide insertion and large-scale assessment of single-guide RNAs. PLoS ONE 2014, 9, e98186. [Google Scholar] [CrossRef]
- Khatri, D.; Mignani, L.; Zizioli, D.; Ritelli, M.; Monti, E.; Finazzi, D. Abnormal Vasculature Development in Zebrafish Embryos with Reduced Expression of Pantothenate Kinase 2 Gene. Bull. Exp. Biol. Med. 2020, 170, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Korzh, V.; Sleptsova, I.; Liao, J.; He, J.; Gong, Z. Expression of zebrafish bHLH genes ngn1 and nrd defines distinct stages of neural differentiation. Dev. Dyn. 1998, 213, 92–104. [Google Scholar] [CrossRef]
- Mueller, T.; Wullimann, M.F. Expression domains of neuroD (nrd) in the early postembryonic zebrafish brain. Brain Res. Bull. 2002, 57, 377–379. [Google Scholar] [CrossRef]
- Ganz, J.; Kaslin, J.; Freudenreich, D.; Machate, A.; Geffarth, M.; Brand, M. Subdivisions of the adult zebrafish subpallium by molecular marker analysis. J. Comp. Neurol. 2012, 520, 633–655. [Google Scholar] [CrossRef]
- Yu, M.; Xi, Y.; Pollack, J.; Debiais-Thibaud, M.; Macdonald, R.B.; Ekker, M. Activity of dlx5a/dlx6a regulatory elements during zebrafish GABAergic neuron development. Int. J. Dev. Neurosci. 2011, 29, 681–691. [Google Scholar] [CrossRef]
- Köprunner, M.; Thisse, C.; Thisse, B.; Raz, E. A zebrafish nanos-related gene is essential for the development of primordial germ cells. Genes Dev. 2001, 15, 2877–2885. [Google Scholar] [CrossRef] [PubMed]
- Basnet, R.M.; Guarienti, M.; Memo, M. Zebrafish Embryo as an In Vivo Model for Behavioral and Pharmacological Characterization of Methylxanthine Drugs. Int. J. Mol. Sci. 2017, 18, 596. [Google Scholar] [CrossRef]
- Bencan, Z.; Sledge, D.; Levin, E.D. Buspirone, chlordiazepoxide and diazepam effects in a zebrafish model of anxiety. Pharmacol. Biochem. Behav. 2009, 94, 75–80. [Google Scholar] [CrossRef]
- Levin, E.D. Zebrafish assessment of cognitive improvement and anxiolysis: Filling the gap between in vitro and rodent models for drug development. Rev. Neurosci. 2011, 22, 75–84. [Google Scholar] [CrossRef] [PubMed]
- Xi, Y.; Noble, S.; Ekker, M. Modeling neurodegeneration in zebrafish. Curr. Neurol. Neurosci. Rep. 2011, 11, 274–282. [Google Scholar] [CrossRef] [PubMed]
- MacDonald, R.B.; Debiais-Thibaud, M.; Talbot, J.C.; Ekker, M. The relationship between dlx and gad1 expression indicates highly conserved genetic pathways in the zebrafish forebrain. Dev. Dyn. 2010, 239, 2298–2306. [Google Scholar] [CrossRef]
- Solek, C.M.; Feng, S.; Perin, S.; Weinschutz Mendes, H.; Ekker, M. Lineage tracing of dlx1a/2a and dlx5a/6a expressing cells in the developing zebrafish brain. Dev. Biol. 2017, 427, 131–147. [Google Scholar] [CrossRef]
- Garcia, M.; Leonardi, R.; Zhang, Y.M.; Rehg, J.E.; Jackowski, S. Germline deletion of pantothenate kinases 1 and 2 reveals the key roles for CoA in postnatal metabolism. PLoS ONE 2012, 7, e40871. [Google Scholar] [CrossRef]
- El-Brolosy, M.A.; Kontarakis, Z.; Rossi, A.; Kuenne, C.; Günther, S.; Fukuda, N.; Kikhi, K.; Boezio, G.L.M.; Takacs, C.M.; Lai, S.L.; et al. Genetic compensation triggered by mutant mRNA degradation. Nature 2019, 568, 193–197. [Google Scholar] [CrossRef]
- Yu, Y.; van der Zwaag, M.; Wedman, J.J.; Permentier, H.; Plomp, N.; Jia, X.; Kanon, B.; Eggens-Meijer, E.; Buist, G.; Harmsen, H.; et al. Coenzyme A precursors flow from mother to zygote and from microbiome to host. Mol. Cell. 2022, 82, 2650–2665.e12. [Google Scholar] [CrossRef]
- Choi, J.; Mouillesseaux, K.; Wang, Z.; Fiji, H.D.; Kinderman, S.S.; Otto, G.W.; Geisler, R.; Kwon, O.; Chen, J.N. Aplexone targets the HMG-CoA reductase pathway and differentially regulates arteriovenous angiogenesis. Development 2011, 138, 1173–1181. [Google Scholar] [CrossRef] [PubMed]
- Siudeja, K.; Srinivasan, B.; Xu, L.; Rana, A.; de Jong, J.; Nollen, E.A.; Jackowski, S.; Sanford, L.; Hayflick, S.; Sibon, O.C. Impaired Coenzyme A metabolism affects histone and tubulin acetylation in Drosophila and human cell models of pantothenate kinase associated neurodegeneration. EMBO Mol. Med. 2011, 3, 755–766. [Google Scholar] [CrossRef] [PubMed]
- Drecourt, A.; Babdor, J.; Dussiot, M.; Petit, F.; Goudin, N.; Garfa-Traoré, M.; Habarou, F.; Bole-Feysot, C.; Nitschké, P.; Ottolenghi, C.; et al. Impaired Transferrin Receptor Palmitoylation and Recycling in Neurodegeneration with Brain Iron Accumulation. Am. J. Hum. Genet. 2018, 102, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Leoni, V.; Strittmatter, L.; Zorzi, G.; Zibordi, F.; Dusi, S.; Garavaglia, B.; Venco, P.; Caccia, C.; Souza, A.L.; Deik, A.; et al. Metabolic consequences of mitochondrial coenzyme A deficiency in patients with PANK2 mutations. Mol. Genet. Metab. 2012, 105, 463–471. [Google Scholar] [CrossRef]
- Wu, Z.; Li, C.; Lv, S.; Zhou, B. Pantothenate kinase-associated neurodegeneration: Insights from a Drosophila model. Hum. Mol. Genet. 2009, 18, 3659–3672. [Google Scholar] [CrossRef]
- Möhler, H. The GABA system in anxiety and depression and its therapeutic potential. Neuropharmacology 2012, 62, 42–53. [Google Scholar] [CrossRef]
- Baumann, B.; Danos, P.; Krell, D.; Diekmann, S.; Leschinger, A.; Stauch, R.; Wurthmann, C.; Bernstein, H.G.; Bogerts, B. Reduced volume of limbic system-affiliated basal ganglia in mood. J. Neuropsychiatry Clin. Neurosci. 1999, 11, 71–78. [Google Scholar] [CrossRef]
- Egan, R.J.; Bergner, C.L.; Hart, P.C.; Cachat, J.M.; Canavello, P.R.; Elegante, M.F.; Elkhayat, S.I.; Bartels, B.K.; Tien, A.K.; Tien, D.H.; et al. Understanding behavioral and physiological phenotypes of stress and anxiety in zebrafish. Behav. Brain Res. 2009, 205, 38–44. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mignani, L.; Zizioli, D.; Khatri, D.; Facchinello, N.; Schiavone, M.; De Palma, G.; Finazzi, D. Bi-Allelic Mutations in Zebrafish pank2 Gene Lead to Testicular Atrophy and Perturbed Behavior without Signs of Neurodegeneration. Int. J. Mol. Sci. 2022, 23, 12914. https://doi.org/10.3390/ijms232112914
Mignani L, Zizioli D, Khatri D, Facchinello N, Schiavone M, De Palma G, Finazzi D. Bi-Allelic Mutations in Zebrafish pank2 Gene Lead to Testicular Atrophy and Perturbed Behavior without Signs of Neurodegeneration. International Journal of Molecular Sciences. 2022; 23(21):12914. https://doi.org/10.3390/ijms232112914
Chicago/Turabian StyleMignani, Luca, Daniela Zizioli, Deepak Khatri, Nicola Facchinello, Marco Schiavone, Giuseppe De Palma, and Dario Finazzi. 2022. "Bi-Allelic Mutations in Zebrafish pank2 Gene Lead to Testicular Atrophy and Perturbed Behavior without Signs of Neurodegeneration" International Journal of Molecular Sciences 23, no. 21: 12914. https://doi.org/10.3390/ijms232112914
APA StyleMignani, L., Zizioli, D., Khatri, D., Facchinello, N., Schiavone, M., De Palma, G., & Finazzi, D. (2022). Bi-Allelic Mutations in Zebrafish pank2 Gene Lead to Testicular Atrophy and Perturbed Behavior without Signs of Neurodegeneration. International Journal of Molecular Sciences, 23(21), 12914. https://doi.org/10.3390/ijms232112914