Transcriptomic, Metabolomic and Ionomic Analyses Reveal Early Modulation of Leaf Mineral Content in Brassica napus under Mild or Severe Drought
Abstract
:1. Introduction
2. Results
2.1. General Effect of Water Deficit on Growth
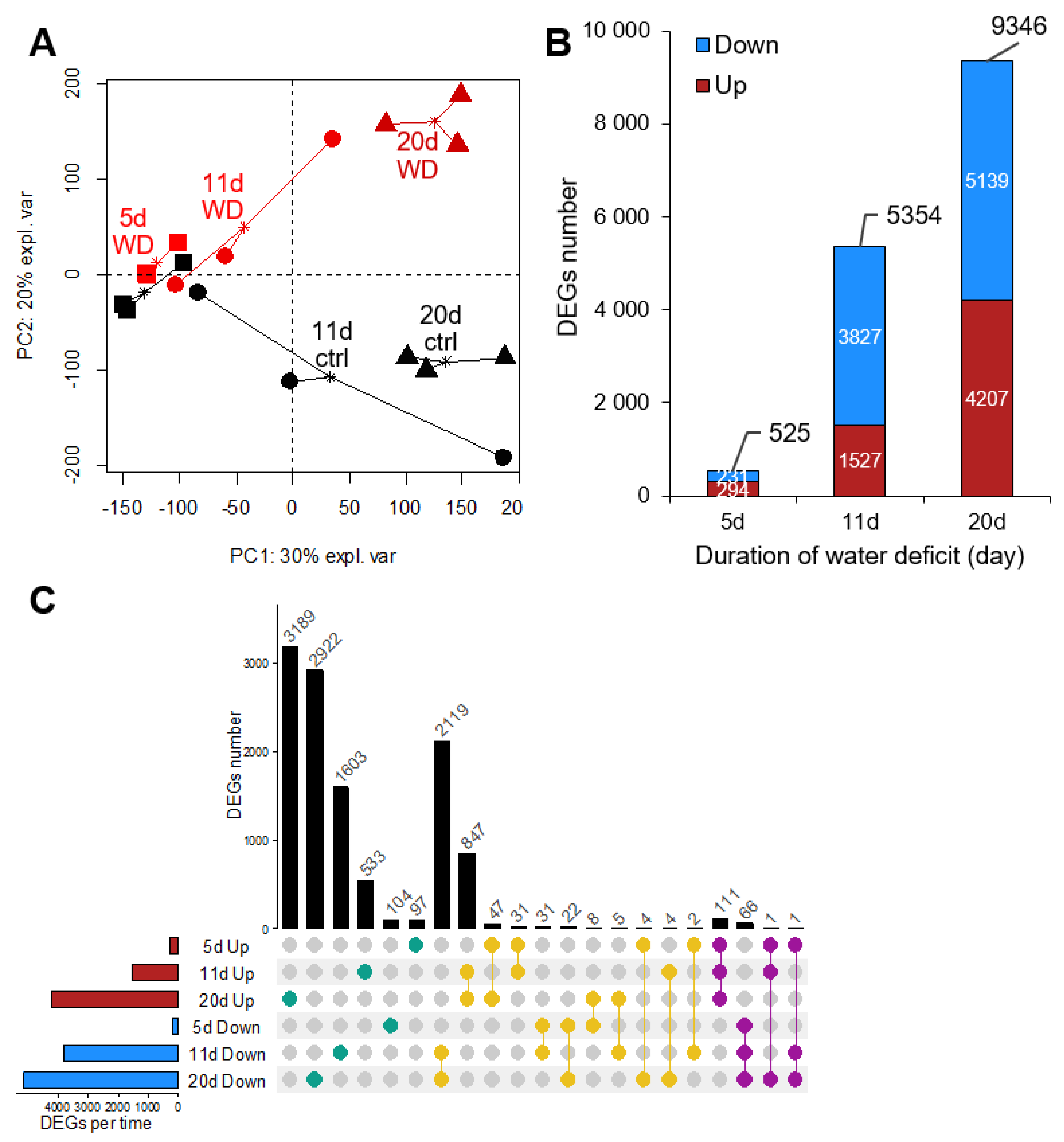
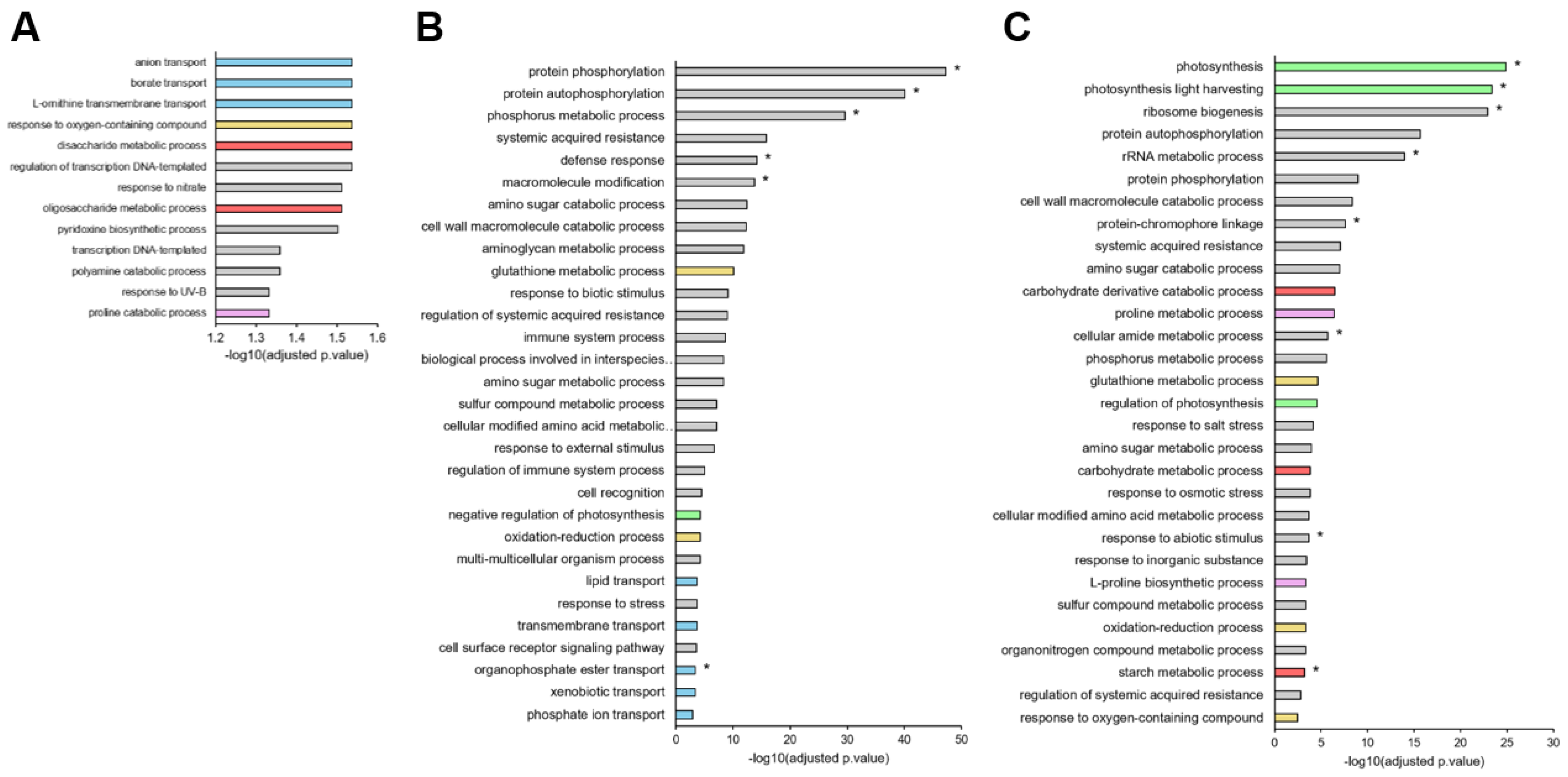
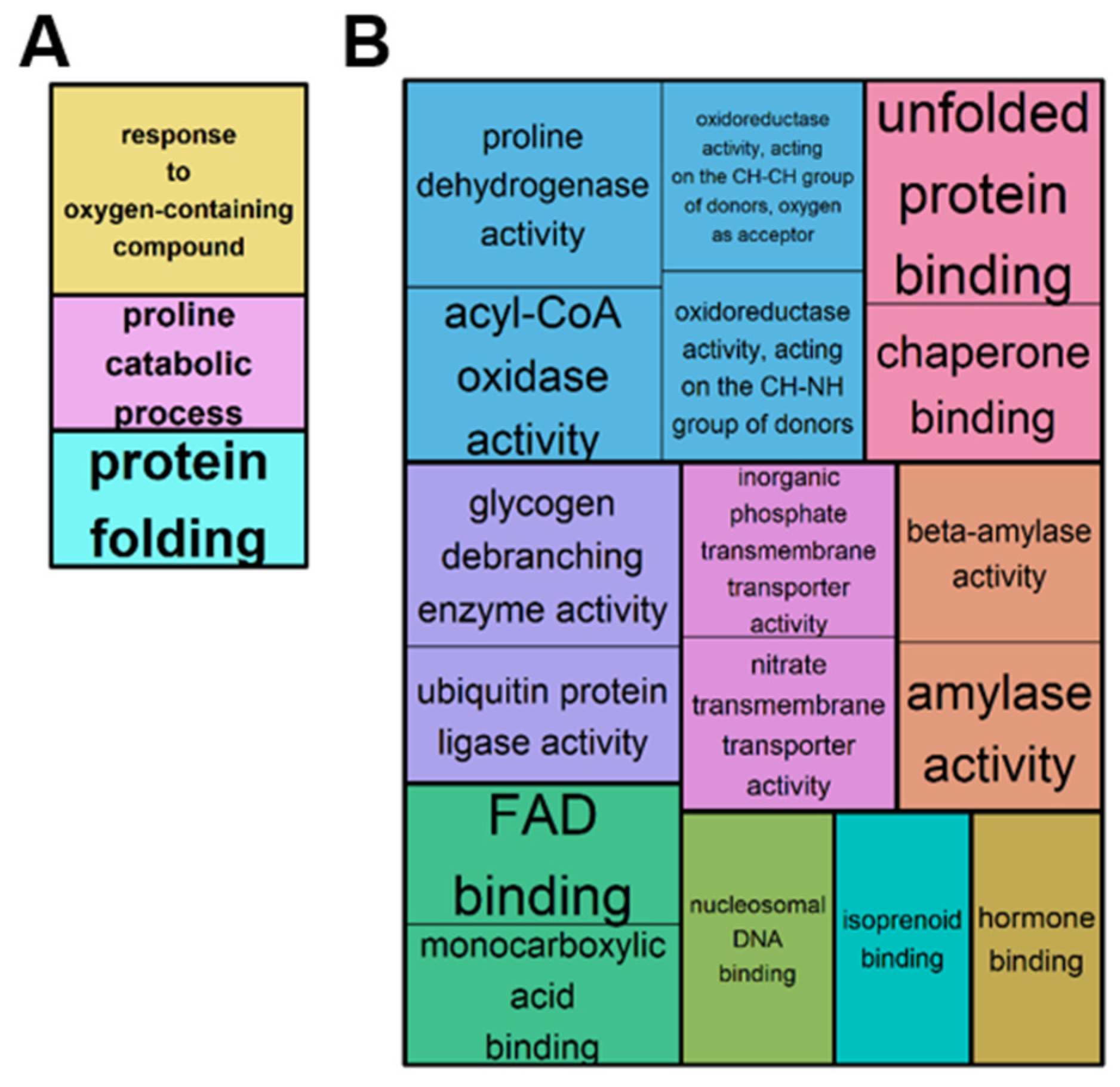
2.2. Overview of the Leaf Transcriptome and Metabolome of Brassica napus Exposed to Water Deficit
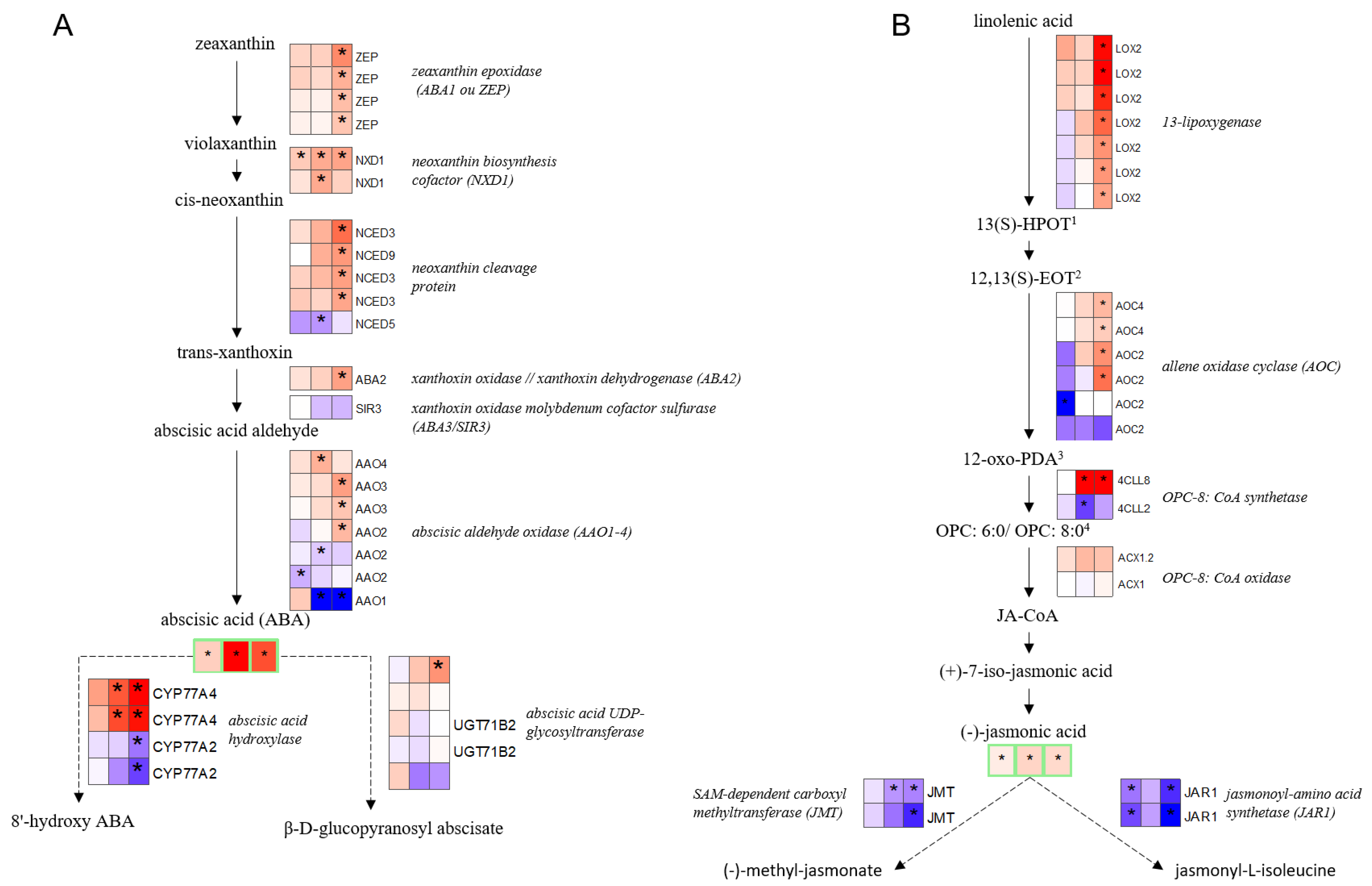
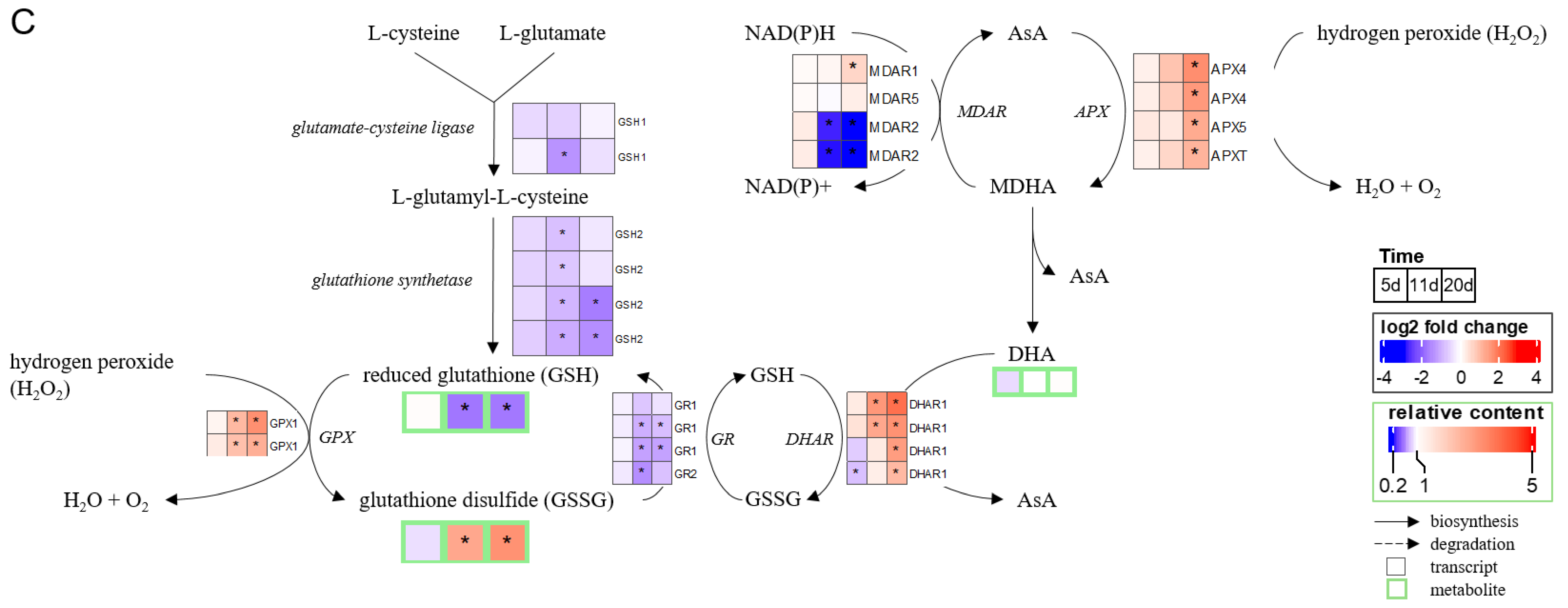
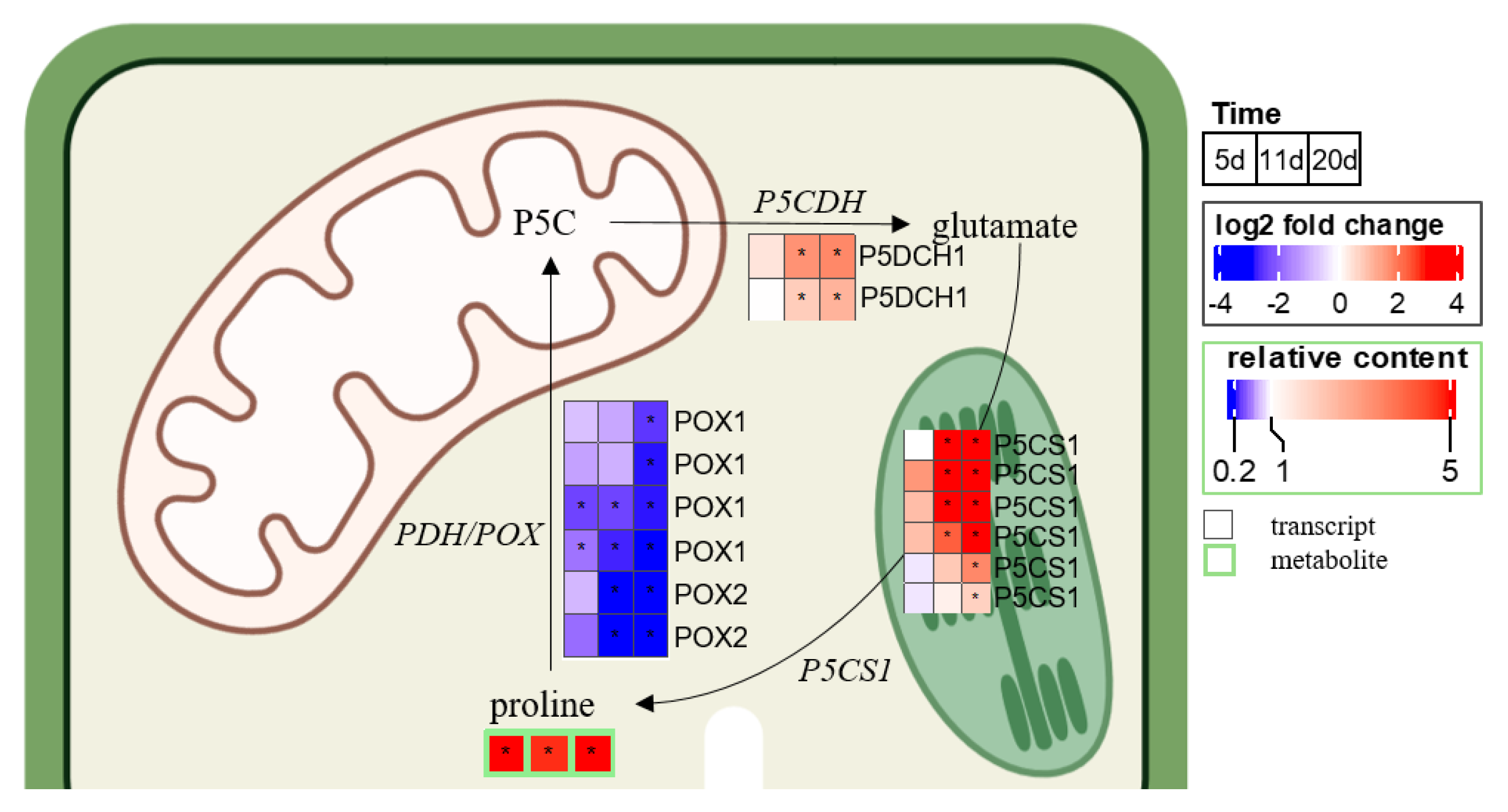
2.3. Chronology of Metabolic Events: The Case of ABA, JA, Glutathione, and Proline
2.4. Early Effects of Water Deficit on the Leaf Ionome
3. Discussion
3.1. Leaf Mineral Content Is Affected Early by Drought
3.2. At the Molecular Level, a Specific Down-Regulation of Genes Encoding Transporters Precedes the Modification of Leaf Ionomic Content
3.3. Global Transcriptomic Analysis Revealed That Downregulation of Mineral Nutrition Is Elicited before Phytohormone and Proline Metabolism or the Occurrence of Oxidative Stress
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. RNA Extraction, Reverse Transcription, and Q-PCR Analyses
4.3. Transcriptomic Analysis by RNA-Sequencing (RNA-Seq)
4.4. RNA-Seq Bioinformatic Analysis
4.5. Untargeted Metabolic Profiling Using UPLC-MS/MS
4.6. Phytohormone Analysis
4.7. Proline Quantitative Analysis
4.8. Element Content Analysis by Mass Spectrometry and X-ray Fluorescence
4.9. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dai, A. Increasing Drought under Global Warming in Observations and Models. Nat. Clim. Chang. 2013, 3, 52–58. [Google Scholar] [CrossRef]
- Qaderi, M.M.; Kurepin, L.V.; Reid, D.M. Growth and Physiological Responses of Canola (Brassica napus) to Three Components of Global Climate Change: Temperature, Carbon Dioxide and Drought. Physiol. Plant. 2006, 128, 710–721. [Google Scholar] [CrossRef]
- Champolivier, L.; Merrien, A. Effects of Water Stress Applied at Different Growth Stages to Brassica napus L. Var. Oleifera on Yield, Yield Components and Seed Quality. Eur. J. Agron. 1996, 5, 153–160. [Google Scholar] [CrossRef]
- Ghobadi, M.; Bakhshandeh, M.; Fathi, G.; Gharineh, M.H.; Alami-Said, K.; Naderi, A.; Ghobadi, M.E. Short and Long Periods of Water Stress during Different Growth Stages of Canola (Brassica napus L.): Effect on Yield, Yield Components, Seed Oil and Protein Contents. J. Agron 2006, 5, 336–341. [Google Scholar]
- Branka, K.; Bosko Andrija, G.; Angelina, T.; Goran, D. Yield and Chemical Composition of Soybean Seed under Different Irrigation Regimes in the Vojvodina Region. Plant Soil Environ. 2017, 63, 34–39. [Google Scholar] [CrossRef] [Green Version]
- Soares, J.C.; Santos, C.S.; Carvalho, S.M.P.; Pintado, M.M.; Vasconcelos, M.W. Preserving the Nutritional Quality of Crop Plants under a Changing Climate: Importance and Strategies. Plant Soil 2019, 443, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Myers, S.S.; Smith, M.R.; Guth, S.; Golden, C.D.; Vaitla, B.; Mueller, N.D.; Dangour, A.D.; Huybers, P. Climate Change and Global Food Systems: Potential Impacts on Food Security and Undernutrition. Annu. Rev. Public Health 2017, 38, 259–277. [Google Scholar] [CrossRef] [PubMed]
- Haberle, J.; Svoboda, P.; Raimanova, I. The Effect of Post-Anthesis Water Supply on Grain Nitrogen Concentration and Grain Nitrogen Yield of Winter Wheat. Plant Soil Environ. 2008, 54, 304–312. [Google Scholar] [CrossRef] [Green Version]
- Guzmán, C.; Autrique, J.E.; Mondal, S.; Singh, R.P.; Govindan, V.; Morales-Dorantes, A.; Posadas-Romano, G.; Crossa, J.; Ammar, K.; Peña, R.J. Response to Drought and Heat Stress on Wheat Quality, with Special Emphasis on Bread-Making Quality, in Durum Wheat. Field Crops Res. 2016, 186, 157–165. [Google Scholar] [CrossRef]
- Fan, M.-S.; Zhao, F.-J.; Fairweather-Tait, S.J.; Poulton, P.R.; Dunham, S.J.; McGrath, S.P. Evidence of Decreasing Mineral Density in Wheat Grain over the Last 160 Years. J. Trace Elem. Med. Biol. 2008, 22, 315–324. [Google Scholar] [CrossRef]
- Velu, G.; Guzman, C.; Mondal, S.; Autrique, J.E.; Huerta, J.; Singh, R.P. Effect of Drought and Elevated Temperature on Grain Zinc and Iron Concentrations in CIMMYT Spring Wheat. J. Cereal Sci. 2016, 69, 182–186. [Google Scholar] [CrossRef]
- Rawat, N.; Neelam, K.; Tiwari, V.K.; Dhaliwal, H.S. Biofortification of Cereals to Overcome Hidden Hunger. Plant Breed. 2013, 132, 437–445. [Google Scholar] [CrossRef]
- Simova-Stoilova, L.; Vassileva, V.; Feller, U. Selection and Breeding of Suitable Crop Genotypes for Drought and Heat Periods in a Changing Climate: Which Morphological and Physiological Properties Should Be Considered? Agriculture 2016, 6, 26. [Google Scholar] [CrossRef] [Green Version]
- Tardieu, F. Any Trait or Trait-Related Allele Can Confer Drought Tolerance: Just Design the Right Drought Scenario. J. Exp. Bot. 2012, 63, 25–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, M.; Monroe, J.G.; Suhail, Y.; Villiers, F.; Mullen, J.; Pater, D.; Hauser, F.; Jeon, B.W.; Bader, J.S.; Kwak, J.M.; et al. Molecular and Systems Approaches towards Drought-Tolerant Canola Crops. New Phytol. 2016, 210, 1169–1189. [Google Scholar] [CrossRef] [Green Version]
- Araus, J.L.; Slafer, G.A.; Reynolds, M.P.; Royo, C. Plant Breeding and Drought in C3 Cereals: What Should We Breed For? Ann. Bot. 2002, 89, 925–940. [Google Scholar] [CrossRef]
- Lawlor, D.W. Genetic Engineering to Improve Plant Performance under Drought: Physiological Evaluation of Achievements, Limitations, and Possibilities. J. Exp. Bot. 2013, 64, 83–108. [Google Scholar] [CrossRef] [Green Version]
- Farooq, M.; Wahid, A.; Kobayashi, N.; Fujita, D.; Basra, S.M.A. Plant Drought Stress: Effects, Mechanisms and Management. Agron. Sustain. Dev. 2009, 29, 185–212. [Google Scholar] [CrossRef] [Green Version]
- Nadeem, M.; Li, J.; Yahya, M.; Sher, A.; Ma, C.; Wang, X.; Qiu, L. Research Progress and Perspective on Drought Stress in Legumes: A Review. Int. J. Mol. Sci. 2019, 20, 2541. [Google Scholar] [CrossRef] [Green Version]
- Raza, M.A.S.; Shahid, A.M.; Saleem, M.F.; Khan, I.H.; Ahmad, S.; Ali, M.; Iqbal, R. Effects and Management Strategies to Mitigate Drought Stress in Oilseed Rape (Brassica napus L.): A Review. Zemdirbyste 2017, 104, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Bodner, G.; Nakhforoosh, A.; Kaul, H.-P. Management of Crop Water under Drought: A Review. Agron. Sustain. Dev. 2015, 35, 401–442. [Google Scholar] [CrossRef]
- Rodríguez, M.; Canales, E.; Borrás-Hidalgo, O. Molecular Aspects of Abiotic Stress in Plants. Biotecnol. Apl. 2005, 22, 1–10. [Google Scholar]
- Zargar, S.M.; Gupta, N.; Nazir, M.; Mahajan, R.; Malik, F.A.; Sofi, N.R.; Shikari, A.B.; Salgotra, R.K. Impact of Drought on Photosynthesis: Molecular Perspective. Plant Gene 2017, 11, 154–159. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene Networks Involved in Drought Stress Response and Tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Ren, F.; Zhong, H.; Jiang, W.; Li, X. Identification and Expression Analysis of Genes in Response to High-Salinity and Drought Stresses in Brassica napus. Acta Biochim. Biophys. Sin. 2010, 42, 154–164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding Plant Responses to Drought—From Genes to the Whole Plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Park, S.-H.; Lee, B.-R.; La, V.H.; Mamun, M.A.; Bae, D.-W.; Kim, T.-H. Drought Intensity-Responsive Salicylic Acid and Abscisic Acid Crosstalk with the Sugar Signaling and Metabolic Pathway in Brassica Napus. Plants 2021, 10, 610. [Google Scholar] [CrossRef] [PubMed]
- Qaderi, M.M.; Kurepin, L.V.; Reid, D.M. Effects of Temperature and Watering Regime on Growth, Gas Exchange and Abscisic Acid Content of Canola (Brassica napus) Seedlings. Environ. Exp. Bot. 2012, 75, 107–113. [Google Scholar] [CrossRef]
- Liu, S.; Lv, Z.; Liu, Y.; Li, L.; Zhang, L. Network Analysis of ABA-Dependent and ABA-Independent Drought Responsive Genes in Arabidopsis Thaliana. Genet. Mol. Biol. 2018, 41, 624–637. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chhaya; Yadav, B.; Jogawat, A.; Gnanasekaran, P.; Kumari, P.; Lakra, N.; Lal, S.K.; Pawar, J.; Narayan, O.P. An Overview of Recent Advancement in Phytohormones-Mediated Stress Management and Drought Tolerance in Crop Plants. Plant Gene 2021, 25, 100264. [Google Scholar] [CrossRef]
- Gill, S.S.; Tuteja, N. Reactive Oxygen Species and Antioxidant Machinery in Abiotic Stress Tolerance in Crop Plants. Plant Physiol. Biochem. 2010, 48, 909–930. [Google Scholar] [CrossRef]
- La, V.H.; Lee, B.-R.; Islam, M.T.; Park, S.-H.; Jung, H.; Bae, D.-W.; Kim, T.-H. Characterization of Salicylic Acid-Mediated Modulation of the Drought Stress Responses: Reactive Oxygen Species, Proline, and Redox State in Brassica napus. Environ. Exp. Bot. 2019, 157, 1–10. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Anee, T.I.; Parvin, K.; Nahar, K.; Mahmud, J.A.; Fujita, M. Regulation of Ascorbate-Glutathione Pathway in Mitigating Oxidative Damage in Plants under Abiotic Stress. Antioxidants 2019, 8, 384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashraf, M.; Shahbaz, M.; Ali, Q. Drought-Induced Modulation in Growth and Mineral Nutrients in Canola (Brassica napus L.). Pak. J. Bot 2013, 45, 93–98. [Google Scholar]
- Hu, Y.; Schmidhalter, U. Drought and Salinity: A Comparison of Their Effects on Mineral Nutrition of Plants. Z. Pflanzenernähr. Bodenk. 2005, 168, 541–549. [Google Scholar] [CrossRef]
- Bagherikia, S.; Pahlevani, M.; Yamchi, A.; Zaynalinezhad, K.; Mostafaie, A. Transcript Profiling of Genes Encoding Fructan and Sucrose Metabolism in Wheat Under Terminal Drought Stress. J. Plant Growth Regul. 2019, 38, 148–163. [Google Scholar] [CrossRef]
- Durand, M.; Porcheron, B.; Hennion, N.; Maurousset, L.; Lemoine, R.; Pourtau, N. Water Deficit Enhances C Export to the Roots in Arabidopsis Thaliana Plants with Contribution of Sucrose Transporters in Both Shoot and Roots. Plant Physiol. 2016, 170, 1460–1479. [Google Scholar] [CrossRef] [Green Version]
- Hou, J.; Huang, X.; Sun, W.; Du, C.; Wang, C.; Xie, Y.; Ma, Y.; Ma, D. Accumulation of Water-Soluble Carbohydrates and Gene Expression in Wheat Stems Correlates with Drought Resistance. J. Plant Physiol. 2018, 231, 182–191. [Google Scholar] [CrossRef]
- Fàbregas, N.; Fernie, A.R. The Metabolic Response to Drought. J. Exp. Bot. 2019, 70, 1077–1085. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michaletti, A.; Naghavi, M.R.; Toorchi, M.; Zolla, L.; Rinalducci, S. Metabolomics and Proteomics Reveal Drought-Stress Responses of Leaf Tissues from Spring-Wheat. Sci. Rep. 2018, 8, 5710. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zhang, X.; Zhang, K.; An, H.; Hu, K.; Wen, J.; Shen, J.; Ma, C.; Yi, B.; Tu, J.; et al. Comparative Analysis of the Brassica napus Root and Leaf Transcript Profiling in Response to Drought Stress. Int. J. Mol. Sci. 2015, 16, 18752–18777. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rasheed, S.; Bashir, K.; Matsui, A.; Tanaka, M.; Seki, M. Transcriptomic Analysis of Soil-Grown Arabidopsis Thaliana Roots and Shoots in Response to a Drought Stress. Front. Plant Sci. 2016, 7, 180. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Mason, A.S.; Wu, J.; Liu, S.; Zhang, X.; Luo, T.; Redden, R.; Batley, J.; Hu, L.; Yan, G. Identification of Putative Candidate Genes for Water Stress Tolerance in Canola (Brassica napus). Front. Plant Sci. 2015, 6, 1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yong, H.-Y.; Zou, Z.; Kok, E.-P.; Kwan, B.-H.; Chow, K.; Nasu, S.; Nanzyo, M.; Kitashiba, H.; Nishio, T. Comparative Transcriptome Analysis of Leaves and Roots in Response to Sudden Increase in Salinity in Brassica napus by RNA-Seq. BioMed Res. Int. 2014, 2014, e467395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, P.; Yang, C.; Chen, H.; Song, C.; Zhang, X.; Wang, D. Transcriptomic Basis for Drought-Resistance in Brassica napus L. Sci. Rep. 2017, 7, 40532. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gargallo-Garriga, A.; Sardans, J.; Pérez-Trujillo, M.; Rivas-Ubach, A.; Oravec, M.; Vecerova, K.; Urban, O.; Jentsch, A.; Kreyling, J.; Beierkuhnlein, C.; et al. Opposite Metabolic Responses of Shoots and Roots to Drought. Sci. Rep. 2015, 4, 6829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mohammadi, P.P.; Moieni, A.; Komatsu, S. Comparative Proteome Analysis of Drought-Sensitive and Drought-Tolerant Rapeseed Roots and Their Hybrid F1 Line under Drought Stress. Amino Acids 2012, 43, 2137–2152. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, L.; Hu, S.; Zhang, J.; Wang, L.; Ping, X.; Wang, J.; Li, J.; Lu, K.; Tang, Z.; et al. Transcriptome and Proteome Analyses of the Molecular Mechanisms Underlying Changes in Oil Storage under Drought Stress in Brassica napus L. GCB Bioenergy 2021, 13, 1071–1086. [Google Scholar] [CrossRef]
- Waraich, E.A.; Ahmad, R.; Ashraf, M.Y. Role of Mineral Nutrition in Alleviation of Drought Stress in Plants. Aust. J. Crop Sci. 2011, 5, 764. [Google Scholar]
- Canales, J.; Uribe, F.; Henríquez-Valencia, C.; Lovazzano, C.; Medina, J.; Vidal, E.A. Transcriptomic Analysis at Organ and Time Scale Reveals Gene Regulatory Networks Controlling the Sulfate Starvation Response of Solanum Lycopersicum. BMC Plant Biol. 2020, 20, 385. [Google Scholar] [CrossRef]
- Forieri, I.; Wirtz, M.; Hell, R. Toward New Perspectives on the Interaction of Iron and Sulfur Metabolism in Plants. Front. Plant Sci. 2013, 4, 357. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Yadav, B.S.; Raj, U.; Freilich, S.; Varadwaj, P.K. Transcriptomic Analysis of Soil Grown T. Aestivum Cv. Root to Reveal the Changes in Expression of Genes in Response to Multiple Nutrients Deficiency. Front. Plant Sci. 2017, 8, 1025. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Courbet, G.; D’Oria, A.; Maillard, A.; Jing, L.; Pluchon, S.; Arkoun, M.; Pateyron, S.; Paysant Le Roux, C.; Diquélou, S.; Ourry, A.; et al. Comparative Omics Analysis of Brassica napus Roots Subjected to Six Individual Macronutrient Deprivations Reveals Deficiency-Specific Genes and Metabolomic Profiles. Int. J. Mol. Sci. 2021, 22, 11679. [Google Scholar] [CrossRef]
- White, P.J.; Bowen, H.C.; Parmaguru, P.; Fritz, M.; Spracklen, W.P.; Spiby, R.E.; Meacham, M.C.; Mead, A.; Harriman, M.; Trueman, L.J.; et al. Interactions between Selenium and Sulphur Nutrition in Arabidopsis thaliana. J. Exp. Bot. 2004, 55, 1927–1937. [Google Scholar] [CrossRef] [Green Version]
- Salt, D.E.; Baxter, I.; Lahner, B. Ionomics and the Study of the Plant Ionome. Annu. Rev. Plant Biol. 2008, 59, 709–733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baxter, I.R.; Vitek, O.; Lahner, B.; Muthukumar, B.; Borghi, M.; Morrissey, J.; Guerinot, M.L.; Salt, D.E. The Leaf Ionome as a Multivariable System to Detect a Plant’s Physiological Status. Proc. Natl. Acad. Sci. USA 2008, 105, 12081–12086. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stich, B.; Benke, A.; Schmidt, M.; Urbany, C.; Shi, R.; von Wirén, N. The Maize Shoot Ionome: Its Interaction Partners, Predictive Power, and Genetic Determinants. Plant Cell Environ. 2020, 43, 2095–2111. [Google Scholar] [CrossRef]
- Cao, Y.; Ye, S.; Yao, X. Leaf Ionome to Predict the Physiological Status of Nitrogen, Phosphorus, and Potassium in Camellia oleifera. Pak. J. Bot 2019, 51, 1349–1355. [Google Scholar] [CrossRef]
- Campos, A.C.A.; Kruijer, W.; Alexander, R.; Akkers, R.C.; Danku, J.; Salt, D.E.; Aarts, M.G. Natural Variation in Arabidopsis Thaliana Reveals Shoot Ionome, Biomass, and Gene Expression Changes as Biomarkers for Zinc Deficiency Tolerance. J. Exp. Bot. 2017, 68, 3643–3656. [Google Scholar] [CrossRef] [Green Version]
- Pii, Y.; Cesco, S.; Mimmo, T. Shoot Ionome to Predict the Synergism and Antagonism between Nutrients as Affected by Substrate and Physiological Status. Plant Physiol. Biochem. 2015, 94, 48–56. [Google Scholar] [CrossRef]
- D’Oria, A.; Courbet, G.; Lornac, A.; Pluchon, S.; Arkoun, M.; Maillard, A.; Etienne, P.; Diquélou, S.; Ourry, A. Specificity and Plasticity of the Functional Ionome of Brassica napus and Triticum Aestivum Exposed to Micronutrient or Beneficial Nutrient Deprivation and Predictive Sensitivity of the Ionomic Signatures. Front. Plant Sci. 2021, 12, 79. [Google Scholar] [CrossRef]
- Acosta-Gamboa, L.M.; Liu, S.; Langley, E.; Campbell, Z.; Castro-Guerrero, N.; Mendoza-Cozatl, D.; Lorence, A. Moderate to Severe Water Limitation Differentially Affects the Phenome and Ionome of Arabidopsis. Funct. Plant Biol. 2017, 44, 94. [Google Scholar] [CrossRef]
- Sánchez-Rodríguez, E.; del Mar Rubio-Wilhelmi, M.; Cervilla, L.M.; Blasco, B.; Rios, J.J.; Leyva, R.; Romero, L.; Ruiz, J.M. Study of the Ionome and Uptake Fluxes in Cherry Tomato Plants under Moderate Water Stress Conditions. Plant Soil 2010, 335, 339–347. [Google Scholar] [CrossRef]
- Fischer, S.; Hilger, T.; Piepho, H.-P.; Jordan, I.; Cadisch, G. Do We Need More Drought for Better Nutrition? The Effect of Precipitation on Nutrient Concentration in East African Food Crops. Sci. Total Environ. 2019, 658, 405–415. [Google Scholar] [CrossRef]
- D’Oria, A.; Courbet, G.; Billiot, B.; Jing, L.; Pluchon, S.; Arkoun, M.; Paysant-Le Roux, C.; Trouverie, J.; Etienne, P.; Diquélou, S.; et al. Drought Specifically Downregulates Mineral Nutrition: Plant Ionomic Content and Associated Gene Expression. Plant Direct 2022. submitted. [Google Scholar]
- Whitt, L.; Ricachenevsky, F.K.; Ziegler, G.Z.; Clemens, S.; Walker, E.; Maathuis, F.J.M.; Kear, P.; Baxter, I. A Curated List of Genes That Affect the Plant Ionome. Plant Direct 2020, 4, e00272. [Google Scholar] [CrossRef]
- Akmouche, Y.; Cheneby, J.; Lamboeuf, M.; Elie, N.; Laperche, A.; Bertheloot, J.; D’Hooghe, P.; Trouverie, J.; Avice, J.-C.; Etienne, P.; et al. Do Nitrogen- and Sulphur-Remobilization-Related Parameters Measured at the Onset of the Reproductive Stage Provide Early Indicators to Adjust N and S Fertilization in Oilseed Rape (Brassica napus L.) Grown under N- and/or S-Limiting Supplies? Planta 2019, 250, 2047–2062. [Google Scholar] [CrossRef]
- Grant, C.A.; Bailey, L.D. Fertility Management in Canola Production. Can. J. Plant Sci. 1993, 73, 651–670. [Google Scholar] [CrossRef]
- Poisson, E.; Trouverie, J.; Brunel-Muguet, S.; Akmouche, Y.; Pontet, C.; Pinochet, X.; Avice, J.-C. Seed Yield Components and Seed Quality of Oilseed Rape Are Impacted by Sulfur Fertilization and Its Interactions With Nitrogen Fertilization. Front. Plant Sci. 2019, 10, 458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girondé, A.; Dubousset, L.; Trouverie, J.; Etienne, P.; Avice, J.-C. The Impact of Sulfate Restriction on Seed Yield and Quality of Winter Oilseed Rape Depends on the Ability to Remobilize Sulfate from Vegetative Tissues to Reproductive Organs. Front. Plant Sci. 2014, 5, 695. [Google Scholar] [CrossRef] [Green Version]
- Rathke, G.-W.; Christen, O.; Diepenbrock, W. Effects of Nitrogen Source and Rate on Productivity and Quality of Winter Oilseed Rape (Brassica napus L.) Grown in Different Crop Rotations. Field Crops Res. 2005, 94, 103–113. [Google Scholar] [CrossRef]
- Magno Massuia de Almeida, L.; Avice, J.-C.; Morvan-Bertrand, A.; Wagner, M.-H.; González-Centeno, M.R.; Teissedre, P.-L.; Bessoule, J.-J.; Le Guédard, M.; Kim, T.H.; Mollier, A.; et al. High Temperature Patterns at the Onset of Seed Maturation Determine Seed Yield and Quality in Oilseed Rape (Brassica napus L.) in Relation to Sulphur Nutrition. Environ. Exp. Bot. 2021, 185, 104400. [Google Scholar] [CrossRef]
- Baux, A.; Colbach, N.; Allirand, J.M.; Jullien, A.; Ney, B.; Pellet, D. Insights into Temperature Effects on the Fatty Acid Composition of Oilseed Rape Varieties. Eur. J. Agron. 2013, 49, 12–19. [Google Scholar] [CrossRef]
- Loladze, I. Hidden Shift of the Ionome of Plants Exposed to Elevated CO2 Depletes Minerals at the Base of Human Nutrition. eLife 2014, 3, e02245. [Google Scholar] [CrossRef] [PubMed]
- Bouchereau, A.; Clossais-Besnard, N.; Bensaoud, A.; Leport, L.; Renard, M. Water Stress Effects on Rapeseed Quality. Eur. J. Agron. 1996, 5, 19–30. [Google Scholar] [CrossRef]
- He, M.; Dijkstra, F.A. Drought Effect on Plant Nitrogen and Phosphorus: A Meta-Analysis. New Phytol. 2014, 204, 924–931. [Google Scholar] [CrossRef]
- Cakmak, I. The Role of Potassium in Alleviating Detrimental Effects of Abiotic Stresses in Plants. J. Plant Nutr. Soil Sci. 2005, 168, 521–530. [Google Scholar] [CrossRef]
- Wang, M.; Zheng, Q.; Shen, Q.; Guo, S. The Critical Role of Potassium in Plant Stress Response. Int. J. Mol. Sci. 2013, 14, 7370–7390. [Google Scholar] [CrossRef] [Green Version]
- Erel, R.; Ben-Gal, A.; Dag, A.; Schwartz, A.; Yermiyahu, U. Sodium Replacement of Potassium in Physiological Processes of Olive Trees (Var. Barnea) as Affected by Drought. Tree Physiol. 2014, 34, 1102–1117. [Google Scholar] [CrossRef] [Green Version]
- Malvi, U.R. Interaction of Micronutrients with Major Nutrients with Special Reference to Potassium. Karnataka J. Agric. Sci. 2011, 24, 1102–1117. [Google Scholar]
- Etienne, P.; Diquelou, S.; Prudent, M.; Salon, C.; Maillard, A.; Ourry, A. Macro and Micronutrient Storage in Plants and Their Remobilization When Facing Scarcity: The Case of Drought. Agriculture 2018, 8, 14. [Google Scholar] [CrossRef] [Green Version]
- Baxter, I.; Muthukumar, B.; Park, H.C.; Buchner, P.; Lahner, B.; Danku, J.; Zhao, K.; Lee, J.; Hawkesford, M.J.; Guerinot, M.L. Variation in Molybdenum Content across Broadly Distributed Populations of Arabidopsis thaliana Is Controlled by a Mitochondrial Molybdenum Transporter (MOT1). PLoS Genet. 2008, 4, e1000004. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tejada-Jiménez, M.; Llamas, Á.; Sanz-Luque, E.; Galván, A.; Fernández, E. A High-Affinity Molybdate Transporter in Eukaryotes. Proc. Natl. Acad. Sci. USA 2007, 104, 20126–20130. [Google Scholar] [CrossRef] [Green Version]
- Puig, S.; Andrés-Colás, N.; García-Molina, A.; Peñarrubia, L. Copper and Iron Homeostasis in Arabidopsis: Responses to Metal Deficiencies, Interactions and Biotechnological Applications. Plant Cell Environ. 2007, 30, 271–290. [Google Scholar] [CrossRef]
- Cailliatte, R.; Schikora, A.; Briat, J.-F.; Mari, S.; Curie, C. High-Affinity Manganese Uptake by the Metal Transporter NRAMP1 Is Essential for Arabidopsis Growth in Low Manganese Conditions. Plant Cell 2010, 22, 904–917. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Itai, R.N.; Ogo, Y.; Kakei, Y.; Nakanishi, H.; Takahashi, M.; Nishizawa, N.K. The Rice Transcription Factor IDEF1 Is Essential for the Early Response to Iron Deficiency, and Induces Vegetative Expression of Late Embryogenesis Abundant Genes. Plant J. 2009, 60, 948–961. [Google Scholar] [CrossRef]
- Lei, G.J.; Zhu, X.F.; Wang, Z.W.; Dong, F.; Dong, N.Y.; Zheng, S.J. Abscisic Acid Alleviates Iron Deficiency by Promoting Root Iron Reutilization and Transport from Root to Shoot in Arabidopsis. Plant Cell Environ. 2014, 37, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Dugasa, M.T.; Feng, X.; Wang, N.-H.; Wang, J.; Wu, F. Comparative Transcriptome and Tolerance Mechanism Analysis in the Two Contrasting Wheat (Triticum aestivum L.) Cultivars in Response to Drought and Salinity Stresses. Plant Growth Regul. 2021, 94, 101–114. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Yi, J.; Yang, Y.; Lei, C.; Gong, M. Transcriptome Response to Drought, Rehydration and Re-Dehydration in Potato. Int. J. Mol. Sci. 2020, 21, 159. [Google Scholar] [CrossRef] [Green Version]
- Gong, L.; Zhang, H.; Gan, X.; Zhang, L.; Chen, Y.; Nie, F.; Shi, L.; Li, M.; Guo, Z.; Zhang, G.; et al. Transcriptome Profiling of the Potato (Solanum Tuberosum L.) Plant under Drought Stress and Water-Stimulus Conditions. PLoS ONE 2015, 10, e0128041. [Google Scholar] [CrossRef]
- Mar, C. Transcriptomic Response to Water Deficit Reveals a Crucial Role of Phosphate Acquisition in a Drought-Tolerant Common Bean Landrace. Plants 2020, 9, 445. [Google Scholar]
- Padmalatha, K.V.; Dhandapani, G.; Kanakachari, M.; Kumar, S.; Dass, A.; Patil, D.P.; Rajamani, V.; Kumar, K.; Pathak, R.; Rawat, B.; et al. Genome-Wide Transcriptomic Analysis of Cotton under Drought Stress Reveal Significant down-Regulation of Genes and Pathways Involved in Fibre Elongation and up-Regulation of Defense Responsive Genes. Plant Mol. Biol. 2012, 78, 223–246. [Google Scholar] [CrossRef] [PubMed]
- Ranjan, A.; Pandey, N.; Lakhwani, D.; Dubey, N.K.; Pathre, U.V.; Sawant, S.V. Comparative Transcriptomic Analysis of Roots of Contrasting Gossypium Herbaceum Genotypes Revealing Adaptation to Drought. BMC Genom. 2012, 13, 680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iovieno, P.; Punzo, P.; Guida, G.; Mistretta, C.; Van Oosten, M.J.; Nurcato, R.; Bostan, H.; Colantuono, C.; Costa, A.; Bagnaresi, P.; et al. Transcriptomic Changes Drive Physiological Responses to Progressive Drought Stress and Rehydration in Tomato. Front. Plant Sci. 2016, 7, 371. [Google Scholar] [CrossRef] [Green Version]
- Duncan, O.; Trösch, J.; Fenske, R.; Taylor, N.L.; Millar, A.H. Resource: Mapping the Triticum aestivum Proteome. Plant J. 2017, 89, 601–616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verslues, P.E.; Agarwal, M.; Katiyar-Agarwal, S.; Zhu, J.; Zhu, J.-K. Methods and Concepts in Quantifying Resistance to Drought, Salt and Freezing, Abiotic Stresses That Affect Plant Water Status. Plant J. 2006, 45, 523–539. [Google Scholar] [CrossRef] [PubMed]
- Bhaskara, G.B.; Yang, T.-H.; Verslues, P.E. Dynamic Proline Metabolism: Importance and Regulation in Water Limited Environments. Front. Plant Sci. 2015, 6, 484. [Google Scholar] [CrossRef] [Green Version]
- Szabados, L.; Savouré, A. Proline: A Multifunctional Amino Acid. Trends Plant Sci. 2010, 15, 89–97. [Google Scholar] [CrossRef]
- Zhang, J.; Jia, W.; Yang, J.; Ismail, A.M. Role of ABA in Integrating Plant Responses to Drought and Salt Stresses. Field Crops Res. 2006, 97, 111–119. [Google Scholar] [CrossRef]
- Liu, H.; Ren, X.; Zhu, J.; Wu, X.; Liang, C. Effect of Exogenous Abscisic Acid on Morphology, Growth and Nutrient Uptake of Rice (Oryza sativa) Roots under Simulated Acid Rain Stress. Planta 2018, 248, 647–659. [Google Scholar] [CrossRef]
- Sorin, E.; Etienne, P.; Maillard, A.; Zamarreño, A.-M.; Garcia-Mina, J.-M.; Arkoun, M.; Jamois, F.; Cruz, F.; Yvin, J.-C.; Ourry, A. Effect of Sulphur Deprivation on Osmotic Potential Components and Nitrogen Metabolism in Oilseed Rape Leaves: Identification of a New Early Indicator. J. Exp. Bot. 2015, 66, 6175–6189. [Google Scholar] [CrossRef] [Green Version]
- Maillard, A.; Etienne, P.; Diquélou, S.; Trouverie, J.; Billard, V.; Yvin, J.-C.; Ourry, A. Nutrient Deficiencies Modify the Ionomic Composition of Plant Tissues: A Focus on Cross-Talk between Molybdenum and Other Nutrients in Brassica napus. J. Exp. Bot. 2016, 67, 5631–5641. [Google Scholar] [CrossRef] [Green Version]
- Haddad, C.; Arkoun, M.; Jamois, F.; Schwarzenberg, A.; Yvin, J.-C.; Etienne, P.; Laîné, P. Silicon Promotes Growth of Brassica napus L. and Delays Leaf Senescence Induced by Nitrogen Starvation. Front. Plant Sci. 2018, 9, 516. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kopylova, E.; Noé, L.; Touzet, H. SortMeRNA: Fast and Accurate Filtering of Ribosomal RNAs in Metatranscriptomic Data. Bioinformatics 2012, 28, 3211–3217. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor Package for Differential Expression Analysis of Digital Gene Expression Data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- Rigaill, G.; Balzergue, S.; Brunaud, V.; Blondet, E.; Rau, A.; Rogier, O.; Caius, J.; Maugis-Rabusseau, C.; Soubigou-Taconnat, L.; Aubourg, S.; et al. Synthetic Data Sets for the Identification of Key Ingredients for RNA-Seq Differential Analysis. Brief. Bioinform. 2018, 19, 65–76. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. G:Profiler: A Web Server for Functional Enrichment Analysis and Conversions of Gene Lists (2019 Update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [Green Version]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO Summarizes and Visualizes Long Lists of Gene Ontology Terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Usadel, B.; Poree, F.; Nagel, A.; Lohse, M.; Czedik-Eysenberg, A.; Stitt, M. A Guide to Using MapMan to Visualize and Compare Omics Data in Plants: A Case Study in the Crop Species, Maize. Plant Cell Environ. 2009, 32, 1211–1229. [Google Scholar] [CrossRef] [PubMed]
- Thimm, O.; Bläsing, O.; Gibon, Y.; Nagel, A.; Meyer, S.; Krüger, P.; Selbig, J.; Müller, L.A.; Rhee, S.Y.; Stitt, M. Mapman: A User-Driven Tool to Display Genomics Data Sets onto Diagrams of Metabolic Pathways and Other Biological Processes. Plant J. 2004, 37, 914–939. [Google Scholar] [CrossRef]
- Hawkins, C.; Ginzburg, D.; Zhao, K.; Dwyer, W.; Xue, B.; Xu, A.; Rice, S.; Cole, B.; Paley, S.; Karp, P.; et al. Plant Metabolic Network 15: A Resource of Genome-Wide Metabolism Databases for 126 Plants and Algae. J. Integr. Plant Biol. 2021, 63, 1888–1905. [Google Scholar] [CrossRef] [PubMed]
- Schläpfer, P.; Zhang, P.; Wang, C.; Kim, T.; Banf, M.; Chae, L.; Dreher, K.; Chavali, A.K.; Nilo-Poyanco, R.; Bernard, T.; et al. Genome-Wide Prediction of Metabolic Enzymes, Pathways, and Gene Clusters in Plants. Plant Physiol. 2017, 173, 2041–2059. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsugawa, H.; Cajka, T.; Kind, T.; Ma, Y.; Higgins, B.; Ikeda, K.; Kanazawa, M.; VanderGheynst, J.; Fiehn, O.; Arita, M. MS-DIAL: Data-Independent MS/MS Deconvolution for Comprehensive Metabolome Analysis. Nat Methods 2015, 12, 523–526. [Google Scholar] [CrossRef] [PubMed]
- van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, Scaling, and Transformations: Improving the Biological Information Content of Metabolomics Data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, L. Introduction to Multi-and Megavariate Data Analysis Using Projection Methods (PCA & PLS); Umetrics AB: Umeä, Sweden, 1999. [Google Scholar]
- Trygg, J.; Wold, S. Orthogonal projections to latent structures (O-PLS). J. Chemom. 2002, 16, 119–128. [Google Scholar] [CrossRef]
- Pang, Z.; Chong, J.; Zhou, G.; de Lima Morais, D.A.; Chang, L.; Barrette, M.; Gauthier, C.; Jacques, P.-É.; Li, S.; Xia, J. MetaboAnalyst 5.0: Narrowing the Gap between Raw Spectra and Functional Insights. Nucleic Acids Res. 2021, 49, W388–W396. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New Perspectives on Genomes, Pathways, Diseases and Drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Troll, W.; Lindsley, J. A Photometric Method for the Determination of Proline. J. Biol. Chem. 1955, 215, 655–660. [Google Scholar] [CrossRef]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
D’Oria, A.; Jing, L.; Arkoun, M.; Pluchon, S.; Pateyron, S.; Trouverie, J.; Etienne, P.; Diquélou, S.; Ourry, A. Transcriptomic, Metabolomic and Ionomic Analyses Reveal Early Modulation of Leaf Mineral Content in Brassica napus under Mild or Severe Drought. Int. J. Mol. Sci. 2022, 23, 781. https://doi.org/10.3390/ijms23020781
D’Oria A, Jing L, Arkoun M, Pluchon S, Pateyron S, Trouverie J, Etienne P, Diquélou S, Ourry A. Transcriptomic, Metabolomic and Ionomic Analyses Reveal Early Modulation of Leaf Mineral Content in Brassica napus under Mild or Severe Drought. International Journal of Molecular Sciences. 2022; 23(2):781. https://doi.org/10.3390/ijms23020781
Chicago/Turabian StyleD’Oria, Aurélien, Lun Jing, Mustapha Arkoun, Sylvain Pluchon, Stéphanie Pateyron, Jacques Trouverie, Philippe Etienne, Sylvain Diquélou, and Alain Ourry. 2022. "Transcriptomic, Metabolomic and Ionomic Analyses Reveal Early Modulation of Leaf Mineral Content in Brassica napus under Mild or Severe Drought" International Journal of Molecular Sciences 23, no. 2: 781. https://doi.org/10.3390/ijms23020781
APA StyleD’Oria, A., Jing, L., Arkoun, M., Pluchon, S., Pateyron, S., Trouverie, J., Etienne, P., Diquélou, S., & Ourry, A. (2022). Transcriptomic, Metabolomic and Ionomic Analyses Reveal Early Modulation of Leaf Mineral Content in Brassica napus under Mild or Severe Drought. International Journal of Molecular Sciences, 23(2), 781. https://doi.org/10.3390/ijms23020781





