Molecular Evolution of Tubulins in Diatoms
Abstract
1. Introduction
2. Results
2.1. Tubulin Identification
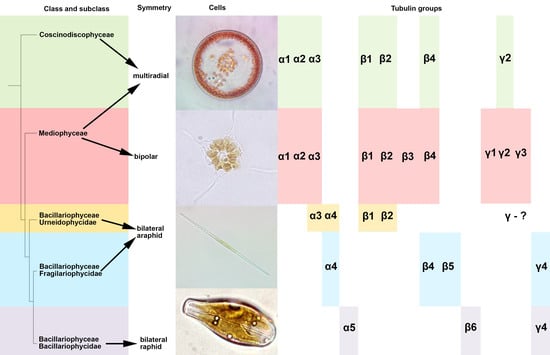
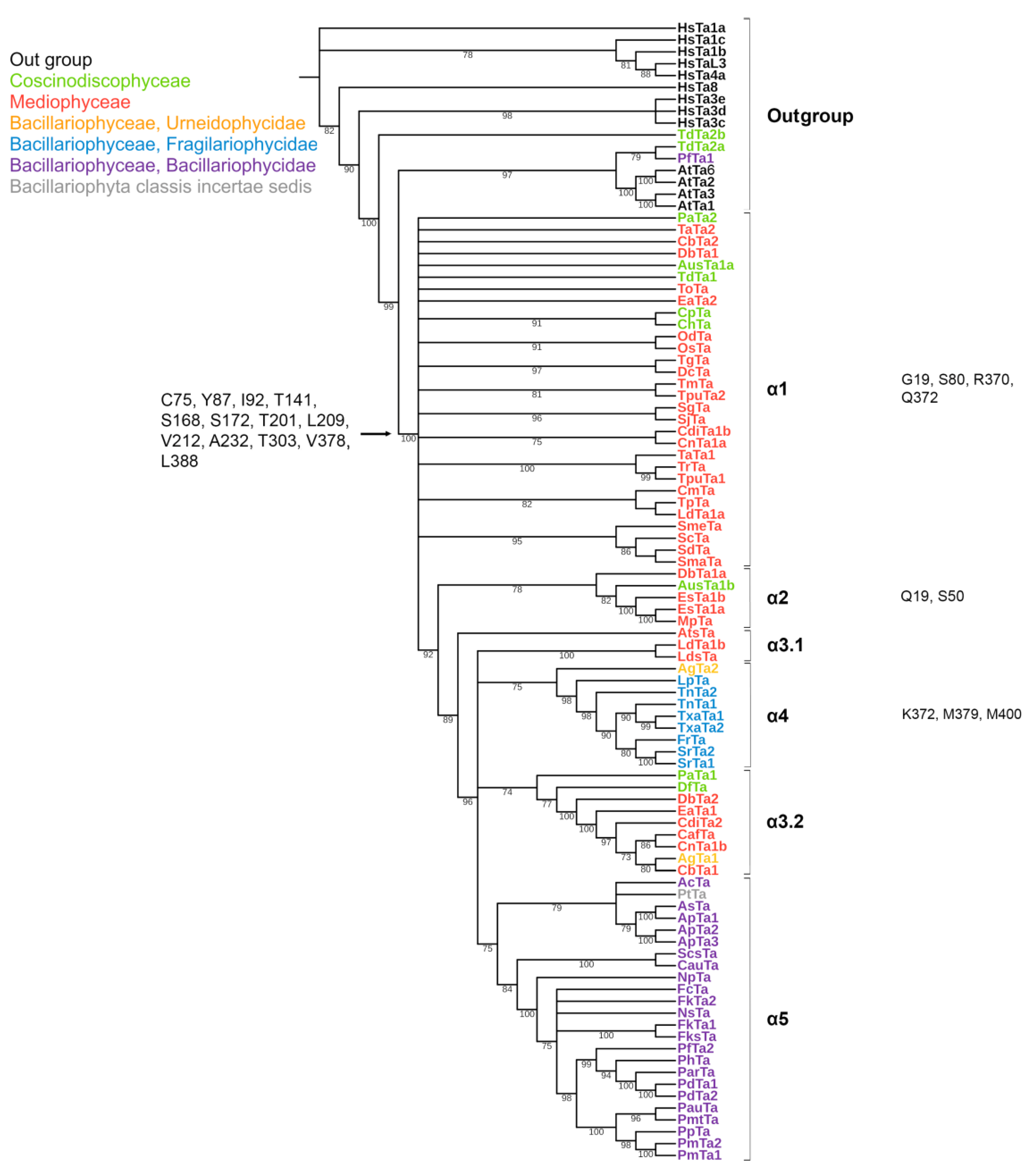
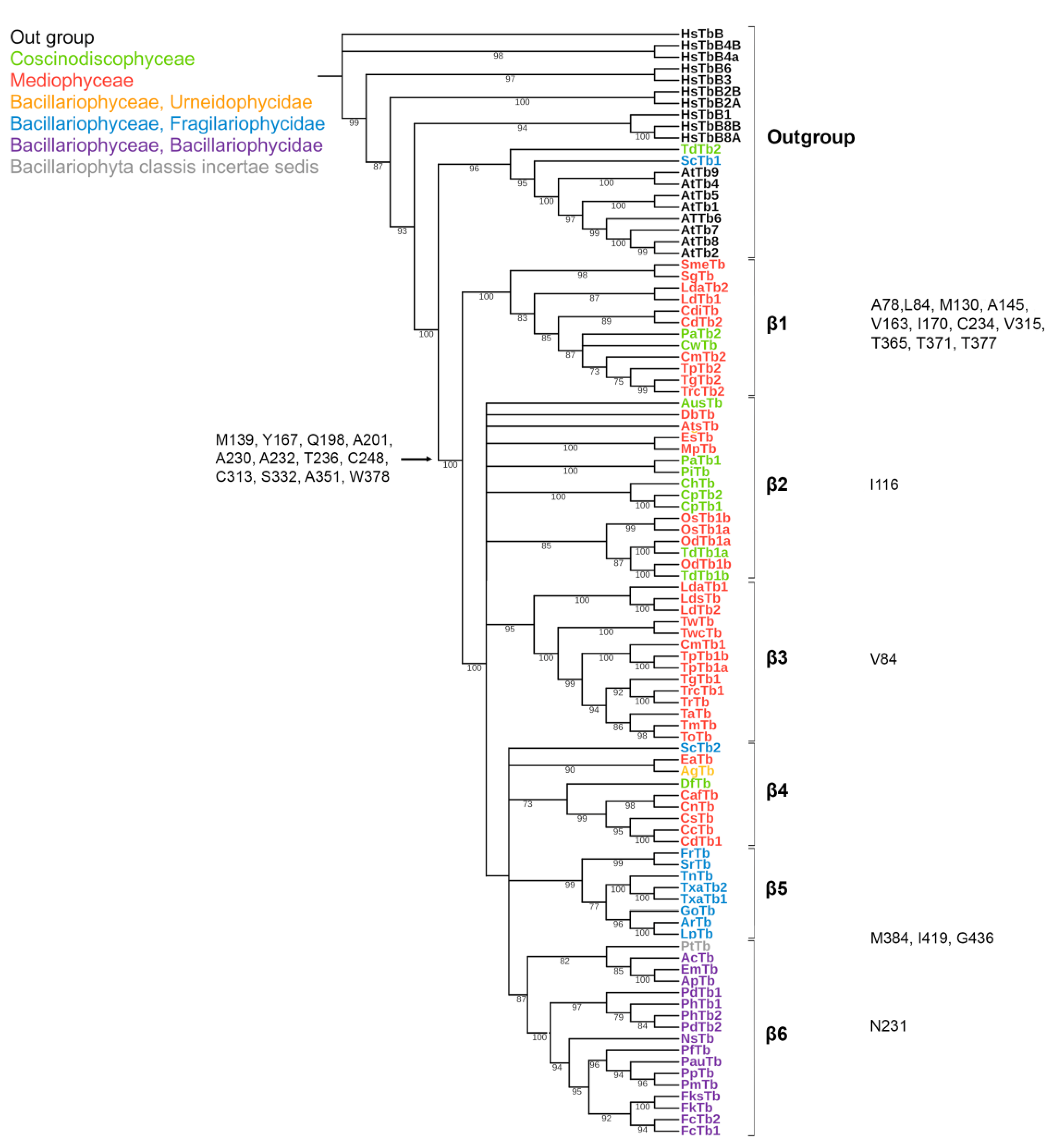
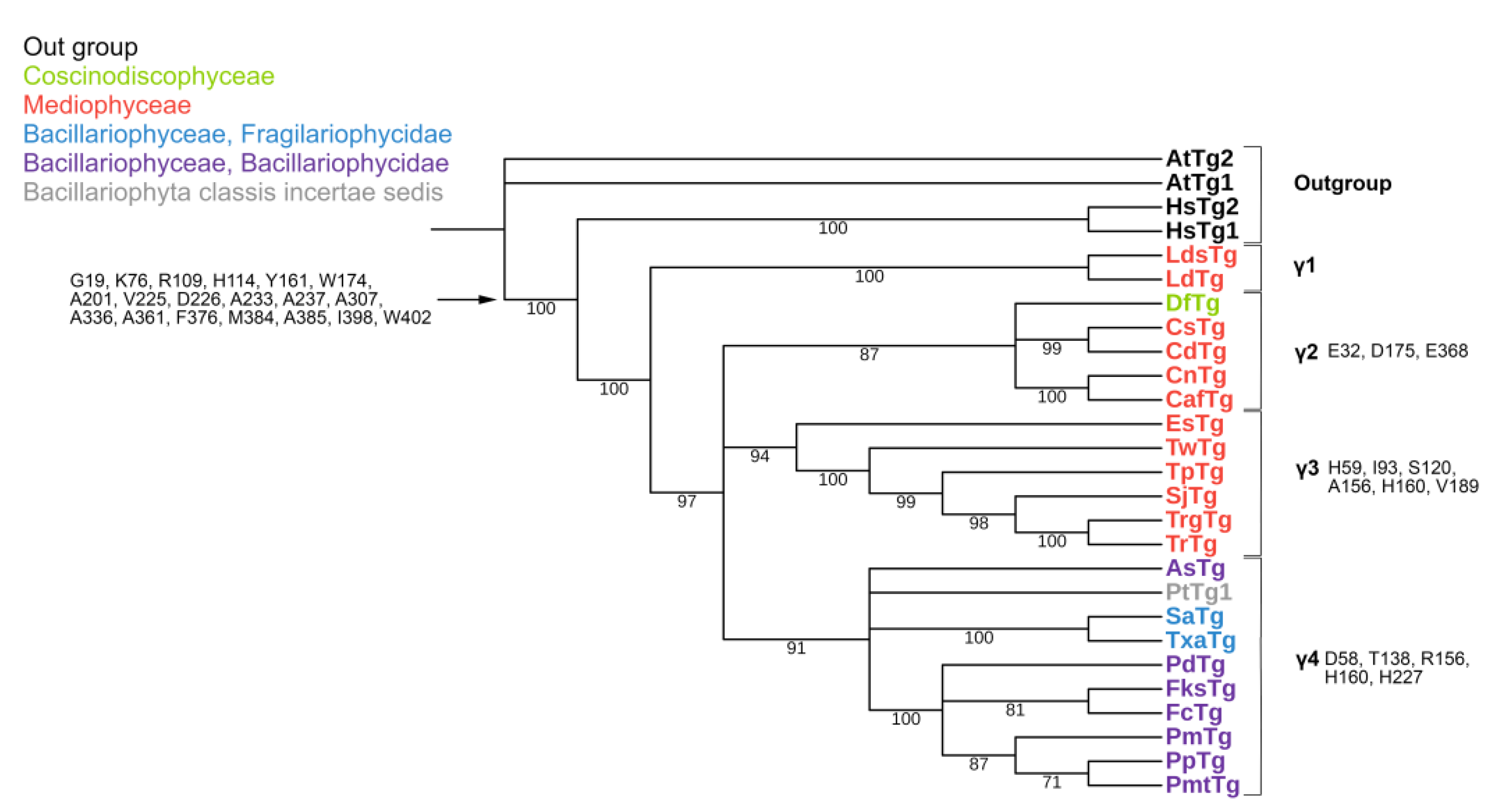
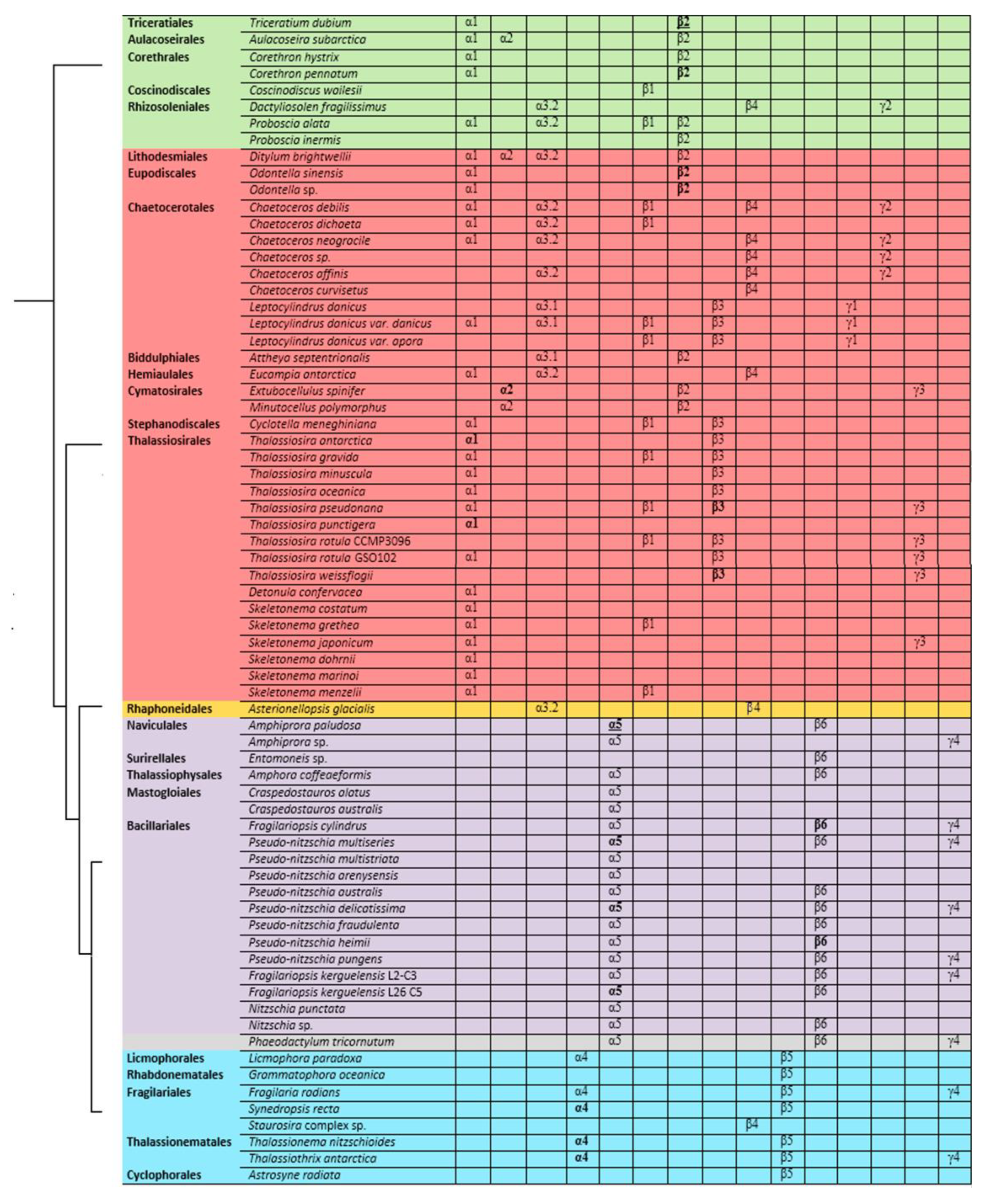
2.2. Phylogeny
2.3. Analysis of Specific Amino Acids in Different Groups
2.4. Analysis of Posttranslational Modification Sites
3. Discussion
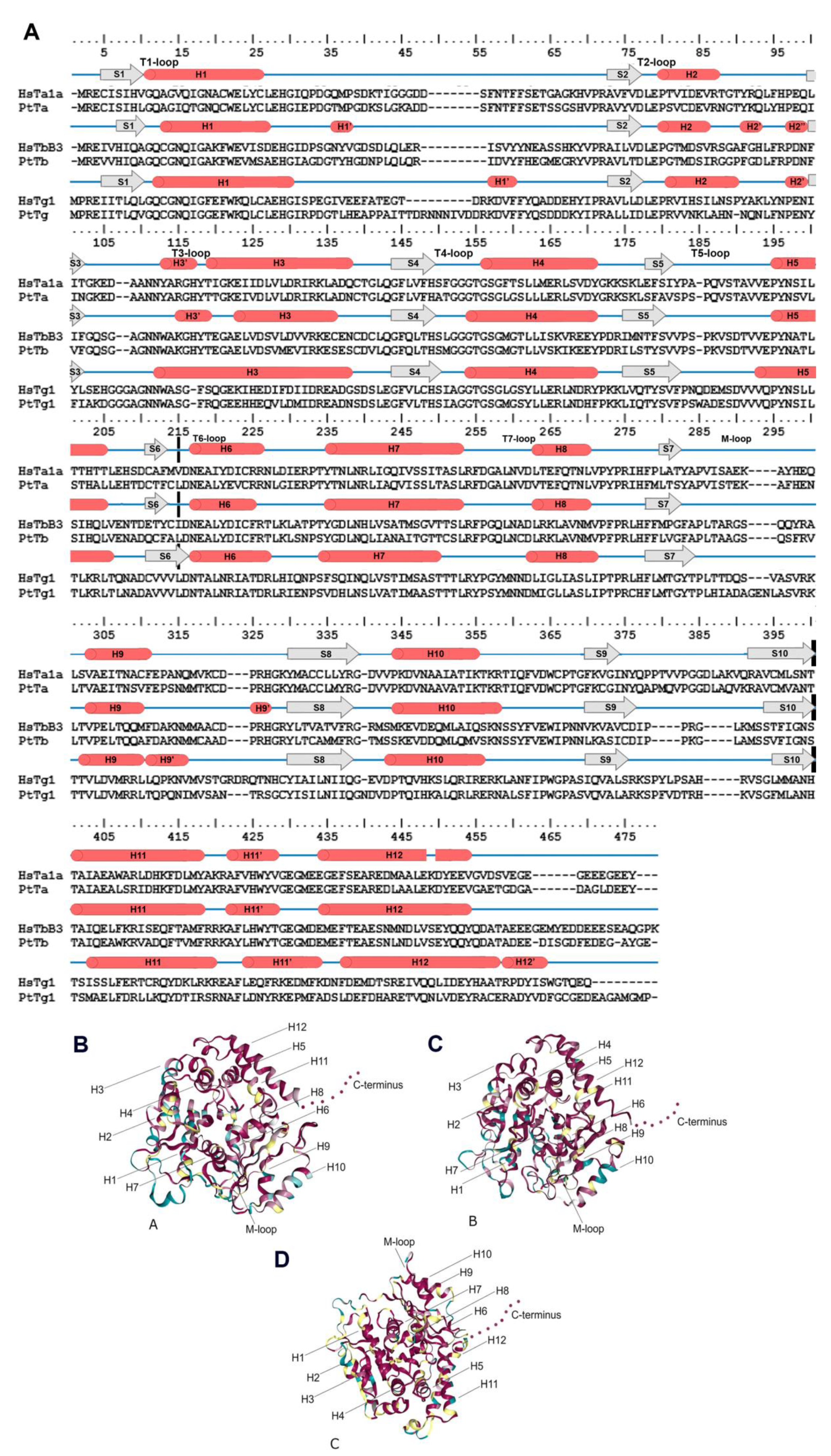
3.1. Features of Diatom Tubulin a.a. Sequences
3.2. Diatom Tubulin Structure and Evolution
4. Materials and Methods
4.1. Identification of Diatom Tubulin Sequences
4.2. Alignment and Comparative Sequence Analysis
4.3. Phylogenetic Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Hashimoto, T. Dissecting the cellular functions of plant microtubules using mutant tubulins. Cytoskeleton 2013, 70, 191–200. [Google Scholar] [CrossRef]
- Janke, C.; Magiera, M.M. The tubulin code and its role in controlling microtubule properties and functions. Nat. Rev. Mol. Cell Biol. 2020, 21, 307–326. [Google Scholar] [CrossRef]
- Cuenca-Zamora, E.J.; Ferrer-Marín, F.; Rivera, J.; Teruel-Montoya, R. Tubulin in platelets: When the shape matters. Int. J. Mol. Sci. 2019, 20, 3484. [Google Scholar] [CrossRef] [PubMed]
- Dutcher, S.K. The tubulin fraternity: Alpha to eta. Curr. Opin. Cell Biol. 2001, 13, 49–54. [Google Scholar] [CrossRef]
- Balaguer, F.D.A.; Mühlethaler, T.; Estévez-Gallego, J.; Calvo, E.; Giménez-Abián, J.F.; Risinger, A.L.; Sorensen, E.J.; Vanderwal, C.D.; Altmann, K.H.; Mooberry, S.L.; et al. Crystal structure of the cyclostreptin-tubulin adduct: Implications for tubulin activation by taxane-site ligands. Int. J. Mol. Sci. 2019, 20, 1392. [Google Scholar] [CrossRef] [PubMed]
- Fong, A.; Durkin, A.; Lee, H. The potential of combining tubulin-targeting anticancer therapeutics and immune therapy. Int. J. Mol. Sci. 2019, 20, 586. [Google Scholar] [CrossRef]
- Soleilhac, E.; Brilletgu, L.; Roussel, V.; Prudent, R.; Touquet, B.; Dass, S.; Acis, S.; Kasam, V.; Barette, C.; Imberty, A.; et al. Specific targeting of plant and Apicomplexa parasite tubulin through differential screening using in silico and assay-based approaches. Int. J. Mol. Sci. 2018, 19, 3085. [Google Scholar] [CrossRef]
- Findeisen, P.; Mühlhausen, S.; Dempewolf, S.; Hertzog, J.; Zietlow, A.; Carlomagno, T.; Kollmar, M. Six subgroups and extensive recent duplications characterize the evolution of the eukaryotic tubulin protein family. Genome Biol. Evol. 2014, 6, 2274–2288. [Google Scholar] [CrossRef] [PubMed]
- Dyer, N. Tubulin and its prokaryotic homologue FtsZ: A structural and functional comparison. Sci. Prog. 2009, 92, 113–137. [Google Scholar] [CrossRef]
- Löwe, J.; Amos, L.A. Crystal structure of the bacterial cell-division protein FtsZ. Nature 1998, 391, 203–206. [Google Scholar] [CrossRef]
- Ludueña, R.F.; Shooter, E.M.; Wilson, L. Structure of the tubulin dimer. J. Biol. Chem. 1977, 252, 7006–7014. [Google Scholar] [CrossRef]
- L’Hernault, S.W.; Rosenbaum, J.L. Chlamydomonas alpha-tubulin is posttranslationally modified by acetylation on the epsilon-amino group of a lysine. Biochemistry 1985, 24, 473–478. [Google Scholar] [CrossRef]
- Ludueña, R.F. Are tubulin isotypes functionally significant. Mol. Biol. Cell. 1993, 4, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Michie, K.A.; Monahan, L.G.; Beech, P.L.; Harry, E.J. Trapping of a spiral-like intermediate of the bacterial cytokinetic protein FtsZ. J. Bacteriol. 2006, 188, 1680–1690. [Google Scholar] [CrossRef]
- McKean, P.G.; Vaughan, S.; Gull, K. The extended tubulin superfamily. J. Cell Sci. 2001, 114, 2723–2733. [Google Scholar] [CrossRef]
- Oakley, B.R. An abundance of tubulins. Trends Cell Biol. 2000, 10, 537–542. [Google Scholar] [CrossRef]
- Zhao, Z.; Liu, H.; Luo, Y.; Zhou, S.; An, L.; Wang, C.; Jin, Q.; Zhou, M.; Xu, J.-R. Molecular evolution and functional divergence of tubulin superfamily in the fungal tree of life. Sci. Rep. 2014, 4, 6746. [Google Scholar] [CrossRef]
- Breviario, D.; Gianì, S.; Morello, L. Multiple tubulins: Evolutionary aspects and biological implications. Plant J. 2013, 75, 202–218. [Google Scholar] [CrossRef] [PubMed]
- Elliott, E.M.; Henderson, G.; Sarangi, F.; Ling, V. Complete sequence of three alpha-tubulin cDNAs in Chinese hamster ovary cells: Each encodes a distinct alpha-tubulin isoprotein. Mol. Cell Biol. 1986, 6, 906–913. [Google Scholar]
- Good, P.J.; Richter, K.; Dawid, I.B. The sequence of a nervous system-specific, class II beta-tubulin gene from Xenopus laevis. Nucleic Acids Res. 1989, 17, 8000. [Google Scholar] [CrossRef]
- Ludueña, R.F. Multiple forms of tubulin: Different gene products and covalent modifications. Int. Rev. Cytol. 1997, 178, 207–275. [Google Scholar]
- Ludueña, R.F. A hypothesis on the origin and evolution of tubulin. Int. Rev. Cell. Mol. Biol. 2013, 302, 41–185. [Google Scholar]
- Prassanawar, S.S.; Panda, D. Tubulin heterogeneity regulates functions and dynamics of microtubules and plays a role in the development of drug resistance in cancer. Biochem. J. 2019, 476, 1359–1376. [Google Scholar] [CrossRef] [PubMed]
- Vemu, A.; Atherton, J.; Spector, J.O.; Moores, C.A.; Roll-Mecak, A. Tubulin isoform composition tunes microtubule dynamics. Mol. Biol. Cell. 2017, 28, 3564–3572. [Google Scholar] [CrossRef]
- Dutcher, S.K. Long-lost relatives reappear: Identification of new members of the tubulin superfamily. Curr. Opin. Cell Biol. 2003, 6, 634–640. [Google Scholar] [CrossRef] [PubMed]
- Horio, T.; Uzawa, S.; Jung, M.K.; Oakley, B.R.; Tanaka, K.; Yanagida, M. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J. Cell Sci. 1991, 99, 693–700. [Google Scholar] [CrossRef] [PubMed]
- Gigant, B.; Curmi, P.A.; Martin-Barbey, C.; Charbaut, E.; Lachkar, S.; Lebeau, L.; Siavoshian, S.; Sobel, A.; Knossow, M. The 4 Å X-ray structure of a tubulin: Stathmin-like domain complex. Cell 2000, 102, 809–816. [Google Scholar] [CrossRef]
- Li, H.; DeRosier, D.J.; Nicholson, W.V.; Nogales, E.; Downing, K.H. Microtubule structure at 8 Å resolution. Structure 2002, 10, 1317–1328. [Google Scholar] [CrossRef]
- Löwe, J.; Li, H.; Downing, K.H.; Nogales, E. Refined structure of αβ-tubulin at 3.5 Å resolution. J. Mol. Biol. 2001, 313, 1045–1057. [Google Scholar] [CrossRef] [PubMed]
- Sept, D.; Baker, N.A.; McCammon, J.A. The physical basis of microtubule structure and stability. Protein Sci. 2003, 12, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- Howes, S.C.; Geyer, E.A.; LaFrance, B.; Zhang, R.; Kellogg, E.H.; Westermann, S.; Rice, L.M.; Nogales, E. Structural and functional differences between porcine brain and budding yeast microtubules. Cell Cycle 2018, 17, 278–287. [Google Scholar] [CrossRef] [PubMed]
- Mghlethaler, T.; Gioia, D.; Prota, A.E.; Sharpe, M.E.; Cavalli, A.; Steinmetz, M.O. Comprehensive analysis of binding sites in tubulin. Angew. Chem. Int. Ed. 2021, 60, 13331–13342. [Google Scholar] [CrossRef] [PubMed]
- Freedman, H.; Luchko, T.; Luduena, R.F.; Tuszynski, J.A. Molecular dynamics modeling of tubulin C-terminal tail interactions with the microtubule surface. Proteins 2011, 79, 2968–2982. [Google Scholar] [CrossRef]
- Keskin, O.; Durell, S.R.; Bahar, I.; Jernigan, R.L.; Covell, D.G. Relating molecular flexibility to function: A case study of tubulin. Biophys J. 2002, 83, 663–680. [Google Scholar] [CrossRef]
- Nogales, E.; Wolf, S.G.; Downing, K.H. Structure of the αβ-tubulin dimer by electron crystallography (Correction). Nature 1998, 393, 191. [Google Scholar] [CrossRef]
- de Pereda, J.M.; Leynadier, D.; Evangelio, J.A.; Chacon, P.; Andreu, J.M. Tubulin secondary structure analysis, limited proteolysis sites, and homology to FtsZ. Biochemistry 1996, 35, 14203–14215. [Google Scholar] [CrossRef]
- Chumová, J.; Kourová, H.; Trögelová, L.; Daniel, G.; Binarov, P. γ-tubulin complexes and fibrillar arrays: Two conserved high molecular forms with many cellular functions. Cells 2021, 10, 776. [Google Scholar] [CrossRef]
- Bigman, L.S.; Levy, Y. Tubulin tails and their modifications regulate protein diffusion on microtubules. Proc. Natl. Acad. Sci. USA 2020, 117, 8876–8883. [Google Scholar] [CrossRef]
- Parrotta, L.; Cresti, M.; Cai, G. Accumulation and post-translational modifications of plant tubulins. Plant Biol. 2014, 16, 521–527. [Google Scholar] [CrossRef]
- Borys, F.; Joachimiak, E.; Krawczyk, H.; Fabczak, H. Intrinsic and extrinsic factors affecting microtubule dynamics in normal and cancer cells. Molecules 2020, 25, 3705. [Google Scholar] [CrossRef]
- Nogales, E.; Whittaker, M.; Milligan, R.A.; Downing, K.H. High-resolution model of the microtubule. Cell 1999, 96, 79–88. [Google Scholar] [CrossRef]
- Cavalier-Smith, T. Kingdom Chromista and its eight phyla: A new synthesis emphasizing periplastid protein targeting, cytoskeletal and periplastid evolution, and ancient divergences. Protoplasma 2018, 255, 297–357. [Google Scholar] [CrossRef]
- Guiry, M.D.; Guiry, G.M. AlgaeBase World-Wide Electronic Publication; National University of Ireland: Galway, Ireland, 2021. [Google Scholar]
- Medlin, L.K.; Desvidevises, Y. Phylogeny of ‘araphid’ diatoms inferred from SSU and LSU rDNA, rbcl and psbA sequences. Vie Milieu. 2016, 66, 129–154. [Google Scholar]
- Sato, S. Phylogeny of Araphid Diatoms Inferred from Morphological and Molecular Data. Ph.D. Thesis, Univiversity Bremen, Bremen, Germany, 2008. [Google Scholar]
- Medlin, L.K. A timescale for diatom evolution based on four molecular markers: Reassessment of ghost lineages and major steps defining diatom evolution. Vie Milieu 2015, 65, 219–238. [Google Scholar]
- Armbrust, E.V. Structural features of nuclear genes in the centric diatom Thalassiosira weissflogii (Bacillariophyceae). J. Phycol. 2000, 36, 942–946. [Google Scholar] [CrossRef]
- Aumeier, C. The Cytoskeleton of Diatoms Structural and Genomic Analysis. Doctoral Thesis, Mathematisch-Naturwissenschaftlichen Fakultät Rheinischen Friedrich-Wilhelms-Universität Bonn, Bonn, Germany, 2012. [Google Scholar]
- De Martino, A.; Amato, A.; Bowler, C. Mitosis in diatoms: Rediscovering an old model for cell division. Bioassays 2009, 31, 874–884. [Google Scholar] [CrossRef] [PubMed]
- Bedoshvili, Y.; Gneusheva, K.; Popova, M.; Morozov, A.; Likhoshway, Y. Anomalies in the valve morphogenesis of the centric diatom alga Aulacoseira islandica caused by microtubule inhibitors. Biol. Open 2018, 7, bio035519. [Google Scholar]
- Blank, G.; Sullivan, C. Diatom mineralization of silicon acid. VI. The effects of microtubule inhibitors on silicic acid metabolism in Navicula saprophila. J. Phycol. 1983, 19, 39–44. [Google Scholar] [CrossRef]
- Blank, G.; Sullivan, C. Diatom mineralization of silicon acid. VII. Influence of microtubule drugs on symmetry and pattern formation in valves of Navicula saprophila during morphogenesis. J. Phycol. 1983, 19, 294–301. [Google Scholar] [CrossRef]
- Cohn, S.; Nash, J.; Pickett-Heaps, J. The effects of drugs on diatom valve morphogenesis. Protoplasma 1989, 149, 130–143. [Google Scholar] [CrossRef]
- Kharitonenko, K.V.; Bedoshvili, Y.D.; Likhoshway, Y.V. Changes in the micro-and nanostructure of siliceous valves in the diatom Synedra acus under the effect of colchicine treatment at different stages of the cell cycle. J. Struct. Biol. 2015, 190, 73–80. [Google Scholar] [CrossRef]
- Oey, J.L.; Schnepf, E. Uber die Auslösung der Valvenbildungbei der Diatomee Cyclotella cryptica. Arch. Mikrobiol. 1970, 71, 199–213. [Google Scholar] [CrossRef]
- Van de Meene, A.; Pickett-Heaps, J. Valve morphogenesis in the centric diatom Proboscia alata Sundström. J. Phycol. 2002, 38, 351–363. [Google Scholar] [CrossRef]
- Van de Meene, A.; Pickett-Heaps, J. Valve morphogenesis in the centric diatom Rhizosolenia setigera (Bacillariophyceae, Centrales) and its taxonomic implications. Eur. J. Phycol. 2004, 39, 93–104. [Google Scholar] [CrossRef]
- Tesson, B.; Hildebrand, M. Extensive and intimate association of the cytoskeleton with forming silica in diatoms: Control over patterning on the meso- and micro-scale. PLoS ONE 2010, 5, e14300. [Google Scholar] [CrossRef] [PubMed]
- Tesson, B.; Hildebrand, M. Dynamics of silica cell wall morphogenesis in the diatom Cyclotella cryptica: Substructure formation and the role of microfilaments. J. Struct. Biol. 2010, 169, 62–74. [Google Scholar] [CrossRef]
- Murphy, S.M.; Urbani, L.; Stearns, T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J. Cell Biol. 1998, 141, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Murphy, S.M.; Preble, A.M.; Patel, U.K.; O’Connell, K.L.; Dias, D.P.; Moritz, M.; Agard, D.; Stults, J.T.; Stearns, T. GCP5 and GCP6: Two new members of the human gamma-tubulin complex. Mol. Biol. Cell. 2001, 12, 3340–3352. [Google Scholar] [CrossRef] [PubMed]
- Bedoshvili, Y.D.; Gneusheva, K.V.; Popova, M.S.; Avezova, T.N.; Arsentyev, K.Y.; Likhoshway, Y.V. Frustule morphogenesis of raphid pennate diatom Encyonema ventricosum (Agardh) Grunow. Protoplasma 2018, 255, 911–921. [Google Scholar] [CrossRef] [PubMed]
- Parkinson, J.; Brechet, Y.; Gordon, R. Centric diatom morphogenesis: A model based on a DLA algorithm investigating the potential role of microtubules. Biochim. Biophys. Acta 1999, 1452, 89–102. [Google Scholar] [CrossRef][Green Version]
- Grachev, M.A.; Annenkov, V.V.; Likhoshway, Y.V. Silicon nanotechnologies of pigmented heterokonts. BioEssays 2008, 30, 328–337. [Google Scholar] [CrossRef]
- Fukushima, N.; Furuta, D.; Hidaka, Y.; Moriyama, R.; Tsujiuchi, T. Post-translational modifications of tubulin in the nervous system. J. Neurochem. 2009, 109, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Heale, K.A.; Alisaraie, L. C-terminal tail of β-tubulin and its role in the alterations of dynein binding mode. Cell Biochem. Biophys. 2020, 78, 331–345. [Google Scholar] [CrossRef]
- Stearns, T.; Evans, L.; Kirschner, M. γ-tubulin is a highly conserved component of the centrosome. Cell 1991, 65, 825–836. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Giannakakou, P.; Gussio, R.; Nogales, E.; Downing, K.H.; Zaharevitz, D.; Bollbuck, B.; Poy, G.; Sackett, D.; Nicolaou, K.C.; Fojo, T. A common pharmacophore for epothilone and taxanes: Molecular basis for drug resistance conferred by tubulin mutations in human cancer cells. Proc. Natl. Acad. Sci. USA 2000, 97, 2904–2909. [Google Scholar] [CrossRef]
- Maccioni, R.B.; Rivas, C.I.; Vera., J.C. Differential interaction of synthetic peptides from the carboxyl-terminal regulatory domain of tubulin with microtubule-associated proteins. EMBO J. 1988, 7, 1957–1963. [Google Scholar] [CrossRef] [PubMed]
- Paschal, B.M.; Obar, R.A.; Vallee, R.B. Interaction of brain cytoplasmic dynein and MAP2 with a common sequence at the C terminus of tubulin. Nature 1989, 342, 569–572. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Li, C.; Ganesan, L.; Oak, J.; Tsai, S.; Sept, D.; Morrissette, N.S. Mutations in α-tubulin confer dinitroaniline resistance at a cost to microtubule function. Mol. Biol. Cell. 2007, 18, 4711–4720. [Google Scholar] [CrossRef][Green Version]
- Hargreaves, J.; Wandosell, F.; Avila, J. Phosphorylation of tubulin enhances its interaction with membranes. Nature 1986, 323, 827–828. [Google Scholar] [CrossRef]
- Chu, C.W.; Hou, F.; Zhang, J.; Phu, L.; Loktev, A.V.; Kirkpatrick, D.S.; Jackson, P.K.; Zhao, Y.; Zou, H. A novel acetylation of beta-tubulin by San modulates microtubule polymerization via down-regulating tubulin incorporation. Mol. Biol. Cell 2011, 22, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Park, I.Y.; Powell, R.T.; Tripathi, D.N.; Dere, R.; Ho, T.H.; Blasius, T.L.; Chiang, Y.C.; Davis, I.J.; Fahey, C.C.; Hacker, K.E.; et al. Dual chromatin and cytoskeletal remodeling by SETD2. Cell 2016, 166, 950–962. [Google Scholar] [CrossRef]
- Ozols, J.; Caron, J.M. Posttranslational modification of tubulin by palmitoylation: II. Identification of sites of palmitoylation. Mol. Biol. Cell 1997, 8, 637–645. [Google Scholar] [CrossRef] [PubMed]
- Zambito, M.; Wolff, J. Palmitoylation of tubulin. Biochem. Biophys. Res. Commun. 1997, 239, 650–654. [Google Scholar] [CrossRef]
- Wang, Q.; Peng, Z.; Long, H.; Deng, X.; Huang, K. Polyubiquitylation of alpha- tubulin at K304 is required for flagellar disassembly in Chlamydomonas. J. Cell Sci. 2019, 132, jcs229047. [Google Scholar] [CrossRef]
- Song, Y.; Kirkpatrick, L.L.; Schilling, A.B.; Helseth, D.L.; Chabot, N.; Keillor, J.W.; Johnson, G.V.; Brady, S.T. Transglutaminase and polyamination of tubulin: Posttranslational modification for stabilizing axonal microtubules. Neuron 2013, 78, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Eshun-Wilson, L.; Zhang, R.; Portran, D.; Nachury, M.V.; Toso, D.B.; Löhr, T.; Vendruscolo, M.; Bonomi, M.; Fraser, J.S.; Nogales, E. Effects of alpha-tubulin acetylation on microtubule structure and stability. Proc. Natl. Acad. Sci. USA 2019, 116, 10366–10371. [Google Scholar] [CrossRef]
- Nieuwenhuis, J.; Adamopoulos, J.A.; Bleijerveld, O.B.; Mazouzi, A.; Stickel, E.; Celie, P.; Altelaar, M.; Knipscheer, P.; Perrakis, A.; Blomen, V.A.; et al. Vasohibins encode tubulin detyrosinating activity. Science 2017, 358, 1453–1456. [Google Scholar] [CrossRef]
- Eddé, B.; Rossier, J.; Le Caer, J.P.; Desbruyères, E.; Gros, F.; Denoulet, P. Posttranslational glutamylation of alpha-tubulin. Science 1990, 247, 83–85. [Google Scholar] [CrossRef]
- Wloga, D.; Rogowski, K.; Sharma, N.; Van Dijk, J.; Janke, C.; Eddé, B.; Bré, M.H.; Levilliers, N.; Redeker, V.; Duan, J.; et al. Glutamylation on alpha-tubulin is not essential but affects the assembly and functions of a subset of microtubules in Tetrahymena thermophila. Eukaryot Cell. 2008, 7, 1362–1372. [Google Scholar] [CrossRef]
- Ori-McKenney, K.M.; McKenney, R.J.; Huang, H.H.; Li, T.; Meltzer, S.; Jan, L.Y. Phosphorylation of β tubulin by the Down syndrome kinase, Minibrain/DYRK1a, regulates microtubule dynamics and dendrite morphogenesis. Neuron 2016, 90, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Rüdiger, M.; Plessman, U.; Kloppel, K.D.; Wehland, J.; Weber, K. Class II tubulin, the major brain beta tubulin isotype is polyglutamylated on glutamic acid residue 435. FEBS Lett. 1992, 308, 101–105. [Google Scholar] [CrossRef]
- Alexander, J.E.; Hunt, D.F.; Lee, M.K.; Shabanowitz, J.; Michel, H.; Berlin, S.C.; MacDonald, T.L.; Sundberg, R.J.; Rebhun, L.I.; Frankfurter, A. Characterization of posttranslational modifications in neuron- specific class III beta- tubulin by mass spectrometry. Proc. Natl. Acad. Sci. USA 1991, 88, 4685–4689. [Google Scholar] [CrossRef]
- Teixidó-Travesa, N.; Roig, J.; Lüders, J. The where, when and how of microtubule nucleation—one ring to rule them all. J. Cell Sci. 2012, 125 Pt 19, 4445–4456. [Google Scholar] [CrossRef]
- Nielsen, M.G.; Gadagkar, S.R.; Gutzwiller, L. Tubulin evolution in insects: Gene duplication and subfunctionalization provide specialized isoforms in a functionally constrained gene family. BMC Evol. Biol. 2010, 10, 113. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef] [PubMed]
- Keeling, P.J.; Burki, F.; Wilcox, H.M.; Allam, B.; Allen, E.E.; Amaral-Zettler, L.A.; Armbrust, E.V.; Archibald, J.M.; Bharti, A.K.; Bell, C.J.; et al. The marine microbial eukaryote transcriptome sequencing project (MMETSP): Illuminating the functional diversity of eukaryotic life in the oceans through transcriptome sequencing. PLoS Biol. 2014, 12, e1001889. [Google Scholar] [CrossRef] [PubMed]
- Madeira, F.; Park, Y.M.; Lee, J.; Buso, N.; Gur, T.; Madhusoodanan, N.; Basutkar, P.; Tivey, A.R.N.; Potter, S.C.; Finn, R.D.; et al. The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res. 2019, 47, W636–W641. [Google Scholar] [CrossRef] [PubMed]
- Rozewickim, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment (describes web interface for sequence and structural alignments). Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar]
- Ashkenazy, H.; Abadi, S.; Martz, E.; Chay, O.; Mayrose, I.; Pupko, T. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016, 44, W344–W350. [Google Scholar] [CrossRef]
- Capella-Gutiérrez, S.; Silla-Martínez, J.M.; Gabaldón, T. trimAl: A tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 2009, 25, 1972–1973. [Google Scholar] [CrossRef] [PubMed]
- Kalyaanamoorthy, S.; Minh, B.Q.; Wong, T.K.; Von Haeseler, A.; Jermiin, L.S. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat. Methods 2017, 14, 587–589. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource f or gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khabudaev, K.V.; Petrova, D.P.; Bedoshvili, Y.D.; Likhoshway, Y.V.; Grachev, M.A. Molecular Evolution of Tubulins in Diatoms. Int. J. Mol. Sci. 2022, 23, 618. https://doi.org/10.3390/ijms23020618
Khabudaev KV, Petrova DP, Bedoshvili YD, Likhoshway YV, Grachev MA. Molecular Evolution of Tubulins in Diatoms. International Journal of Molecular Sciences. 2022; 23(2):618. https://doi.org/10.3390/ijms23020618
Chicago/Turabian StyleKhabudaev, Kirill V., Darya P. Petrova, Yekaterina D. Bedoshvili, Yelena V. Likhoshway, and Mikhail A. Grachev. 2022. "Molecular Evolution of Tubulins in Diatoms" International Journal of Molecular Sciences 23, no. 2: 618. https://doi.org/10.3390/ijms23020618
APA StyleKhabudaev, K. V., Petrova, D. P., Bedoshvili, Y. D., Likhoshway, Y. V., & Grachev, M. A. (2022). Molecular Evolution of Tubulins in Diatoms. International Journal of Molecular Sciences, 23(2), 618. https://doi.org/10.3390/ijms23020618