Cell Cycle-Dependent Transcription: The Cyclin Dependent Kinase Cdk1 Is a Direct Regulator of Basal Transcription Machineries
Abstract
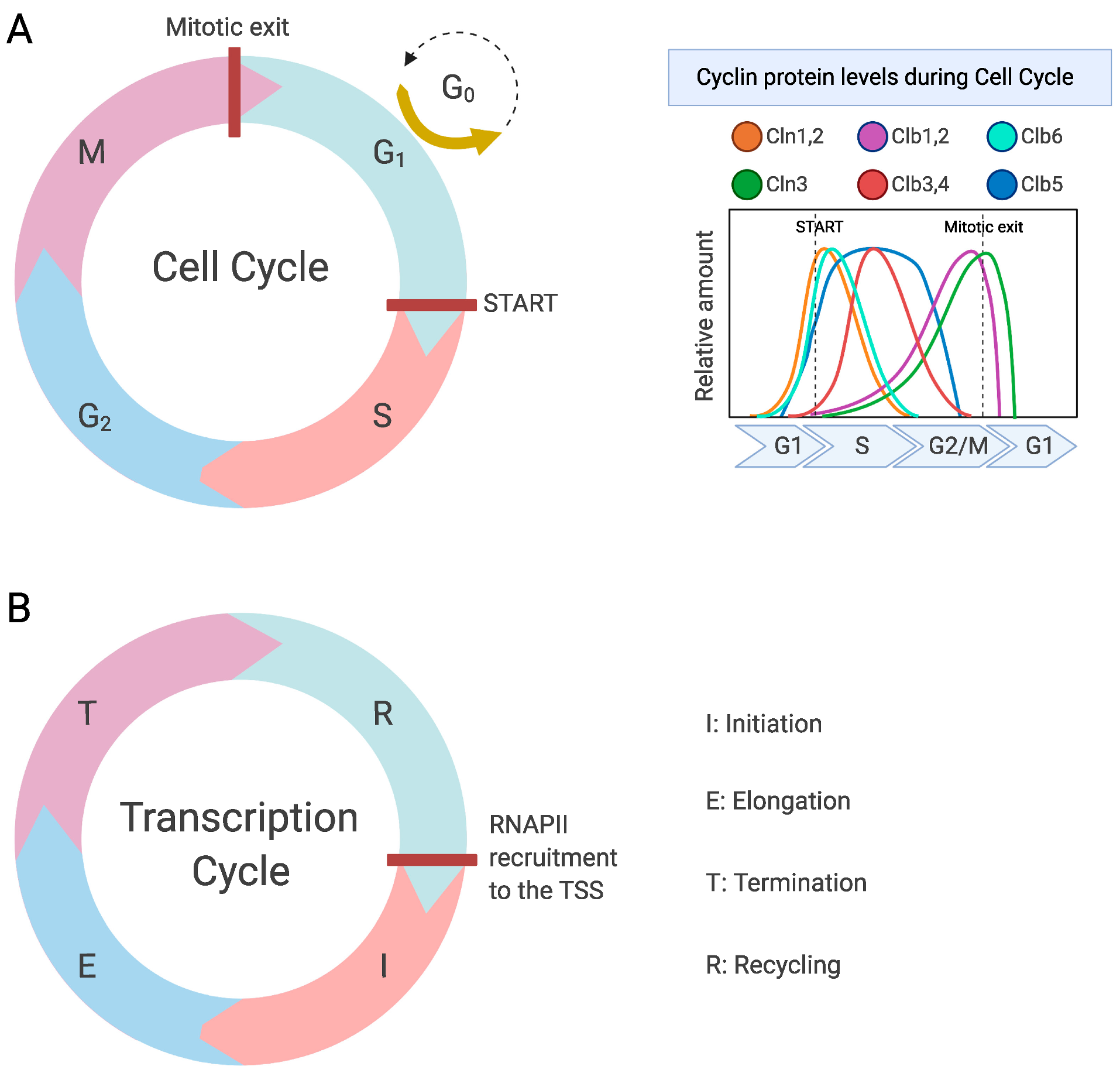
1. Introduction
2. Regulation of Transcription by RNA Polymerase II
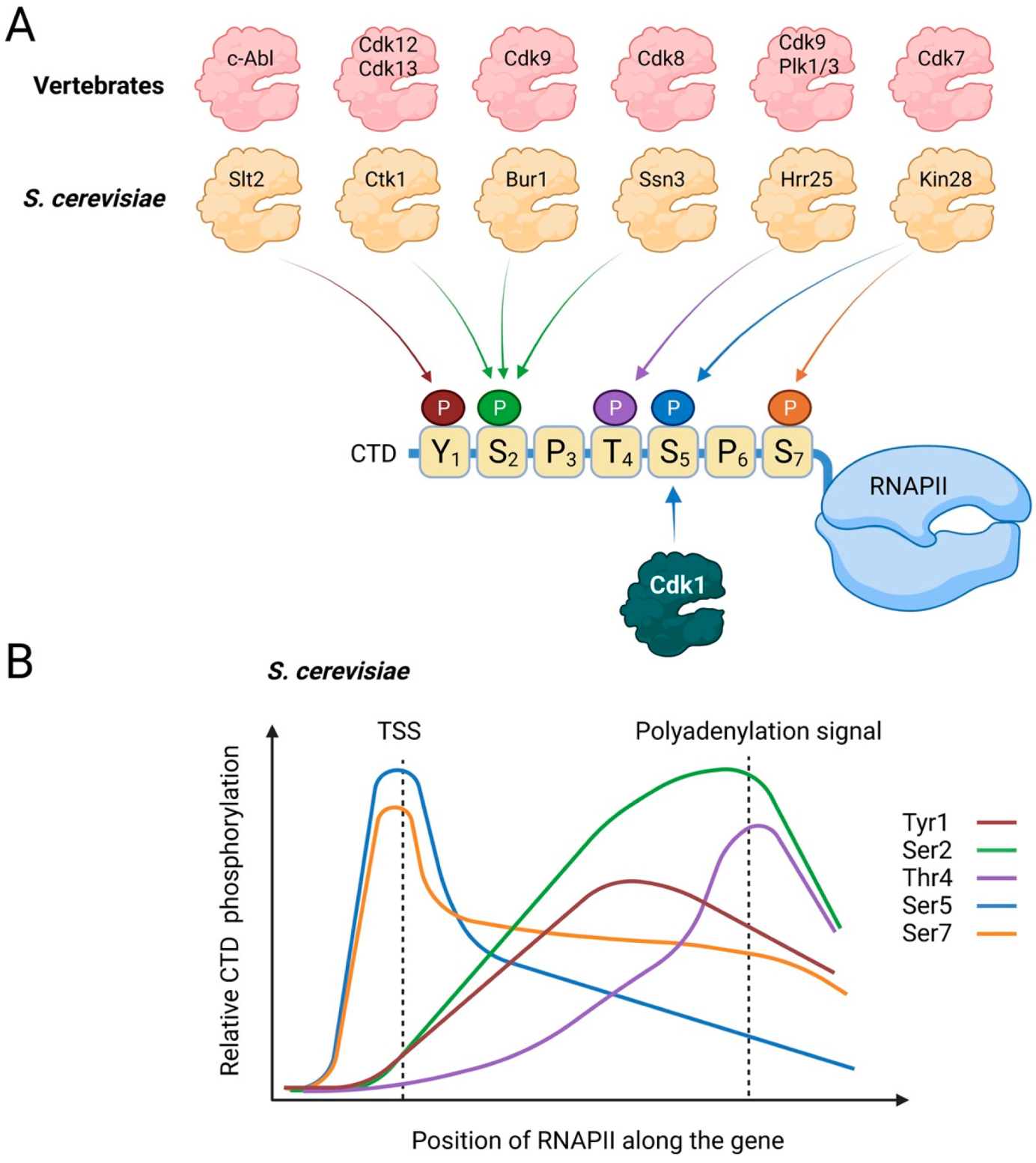
2.1. Regulation of the Transcription Cycle
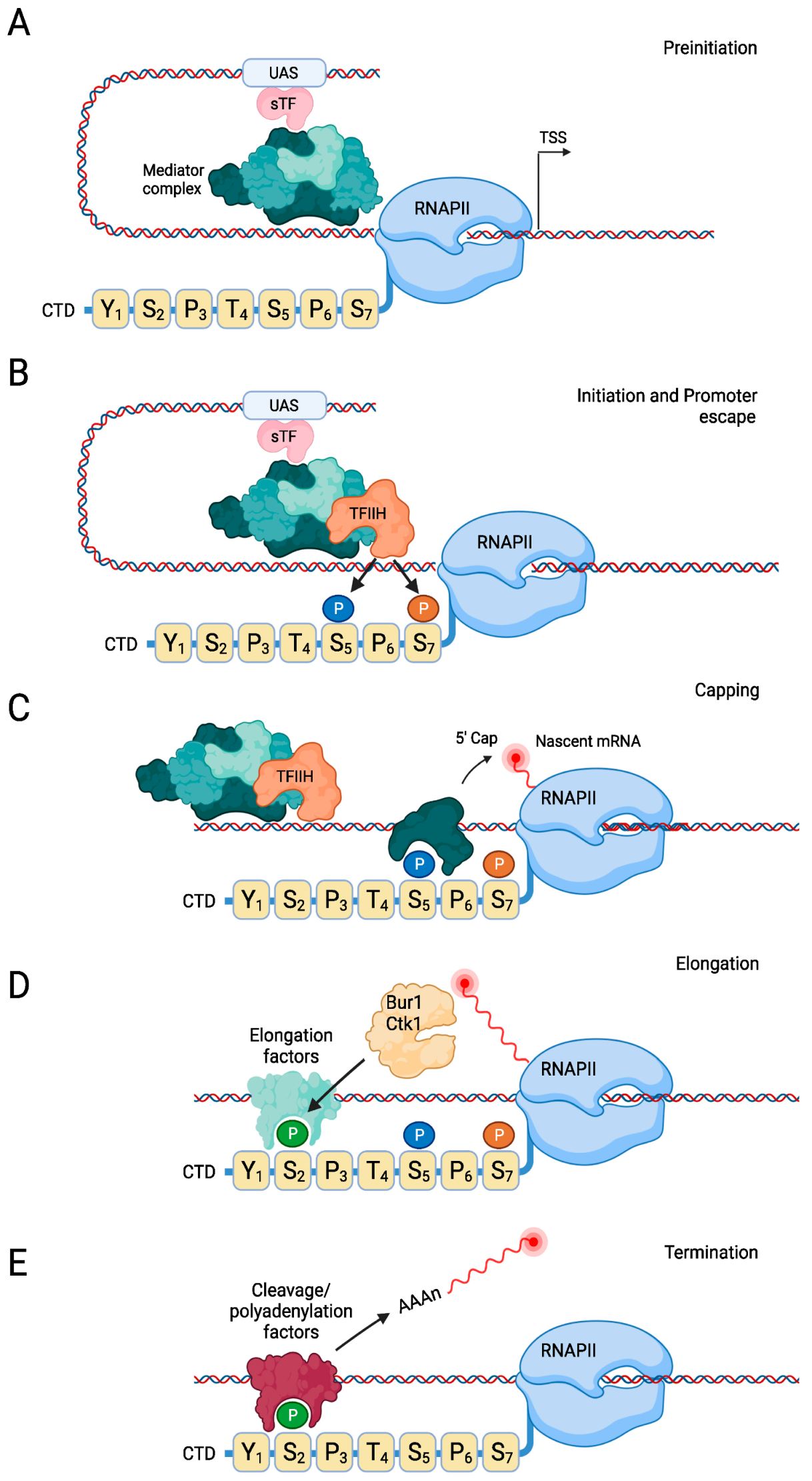
2.2. Transcription Initiation
2.3. Transcription Elongation
2.4. Transcription Termination
2.5. RNAPII Recycling
3. Regulation of RNAPII by Cdk1 during the Cell Cycle
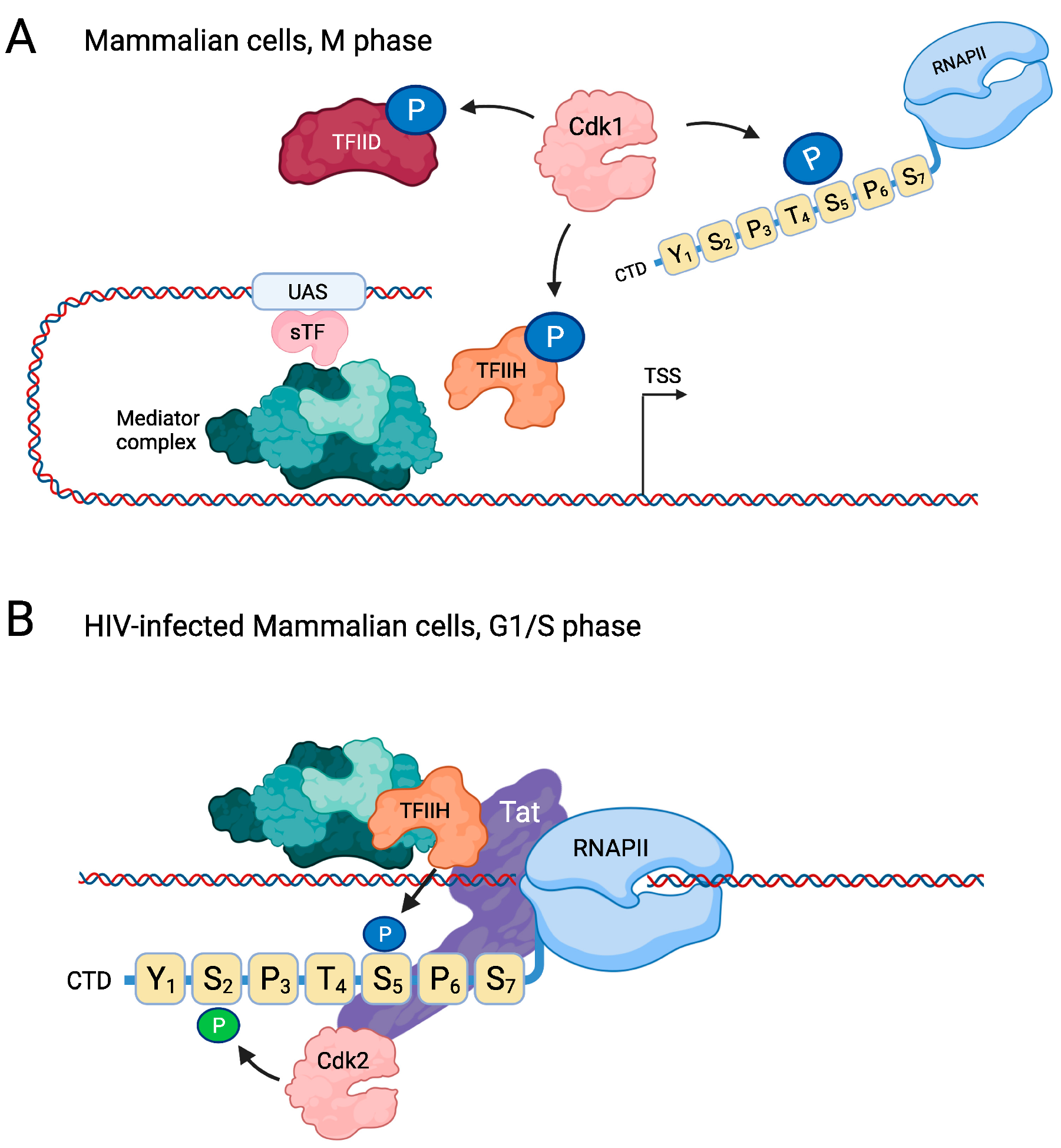
3.1. Downregulation of the Basal Transcription Machinery during M Phase in Vertebrate Cells
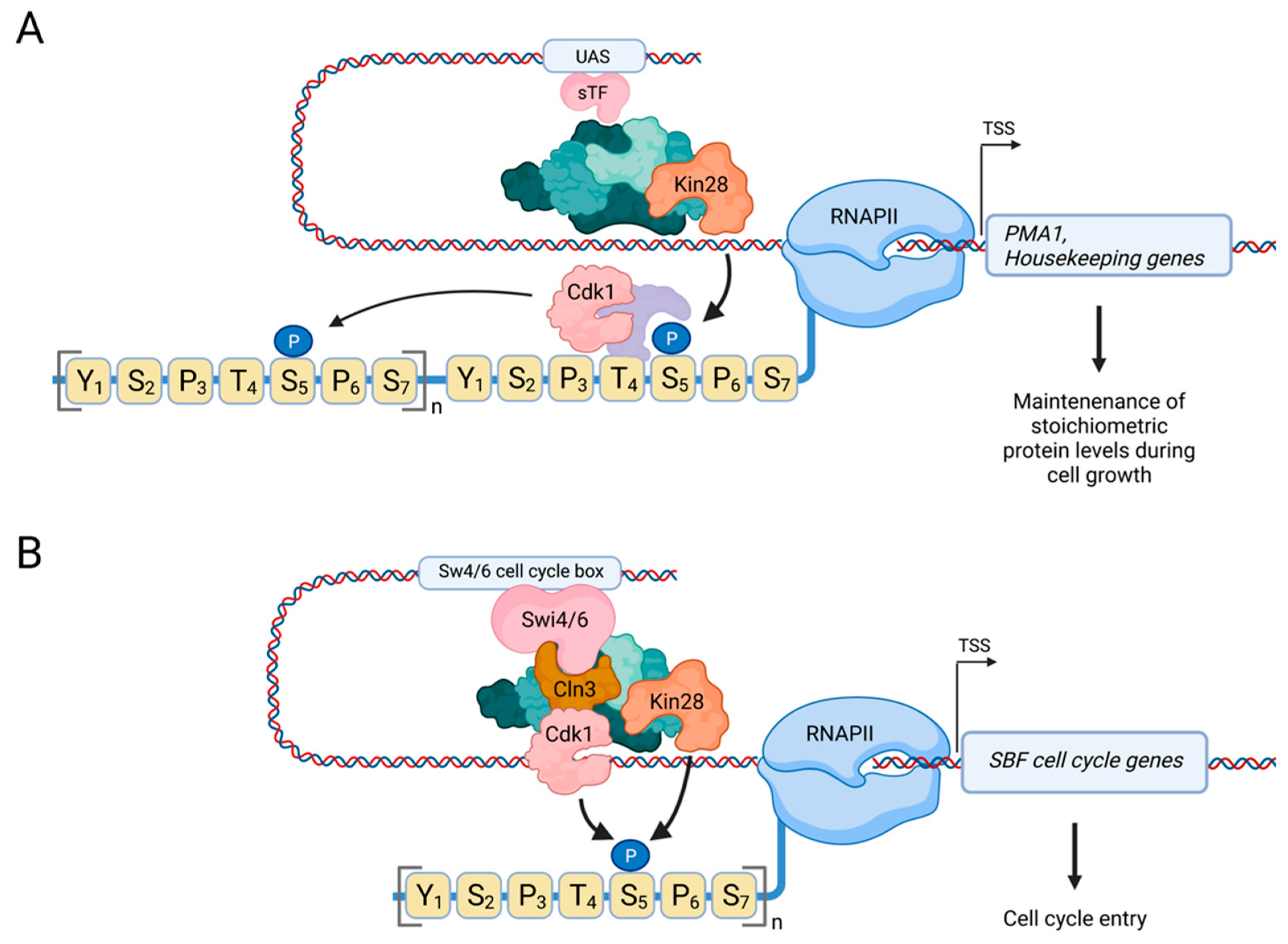
3.2. Activation of the Basal Transcription Machinery by Cdk1 in Budding Yeast
3.3. Direct Regulation of RNAPII by Cdk1 Promotes Cell Cycle Entry
3.4. Indirect Regulation of Basal Transcription by Cdk1
3.5. Is Phosphorylation of RNAPII-CTD the Original Function of Cdk1?
4. Regulation of Transcription by RNA Polymerase I
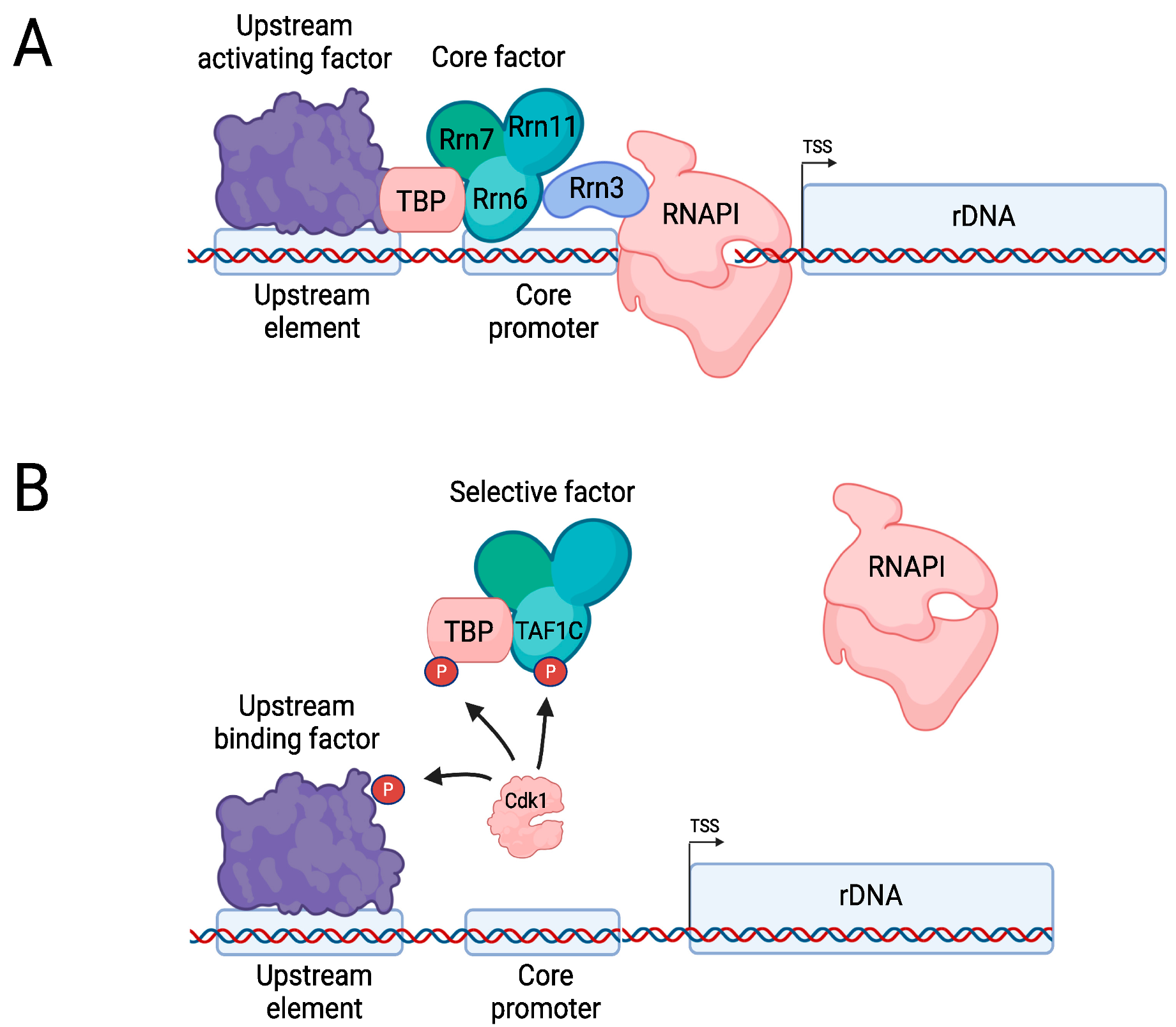
4.1. The RNAPI Transcription Cycle
4.2. Regulation of RNAPI during the Cell Cycle
5. RNA Polymerase III
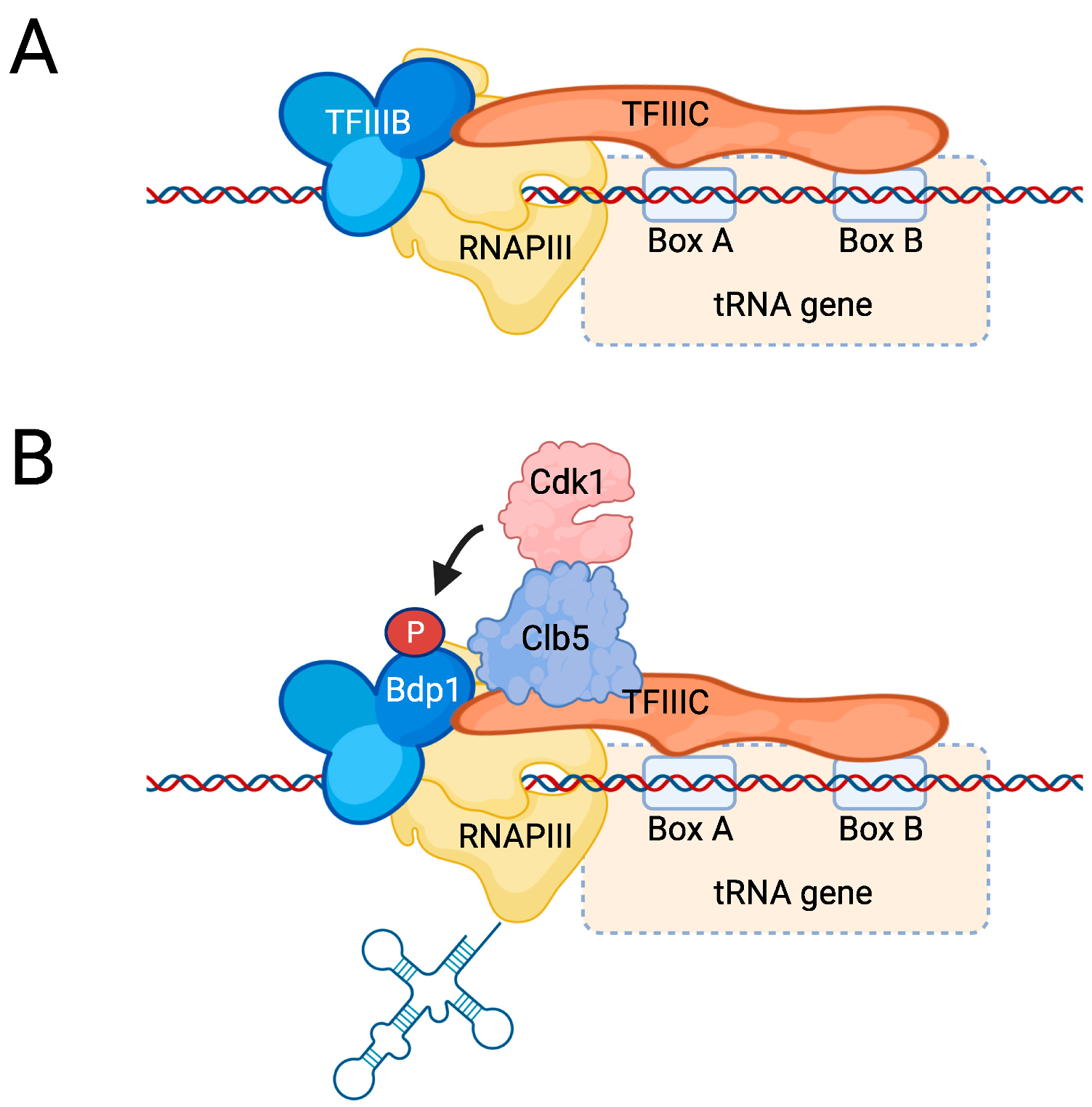
5.1. The RNAPIII Transcription Cycle
5.2. Regulation of RNAPIII by Environmental Cues
5.3. Regulation of RNAPIII Activity by Cdk1
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Enserink, J.M.; Kolodner, R.D. An overview of Cdk1-controlled targets and processes. Cell Div. 2010, 5, 11. [Google Scholar] [CrossRef] [PubMed]
- Morgan, D.O. Cyclin-dependent kinases: Engines, Clocks, and Microprocessors. Annu. Rev. Cell Dev. Biol. 1997, 13, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H. Saccharomyces cerevisiae cell cycle. Bacteriol. Rev. 1974, 38, 164–198. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Culotti, J.; Reid, B. Genetic control of the cell-division cycle in yeast. I. Detection of mutants. Proc. Natl. Acad. Sci. USA 1970, 66, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Hartwell, L.H.; Mortimer, R.K.; Culotti, J.; Culotti, M. Genetic control of the cell division cycle in yeast: V. genetic analysis of cdc mutants. Genetics 1973, 74, 267–286. [Google Scholar] [CrossRef] [PubMed]
- Lorincz, A.T.; Reed, S.I. Primary structure homology between the product of yeast cell division control gene CDC28 and vertebrate oncogenes. Nature 1984, 307, 183–185. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K.A.; Reed, S.I. Isolation of genes by complementation in yeast: Molecular cloning of a cell-cycle gene. Proc. Natl. Acad. Sci. USA 1980, 77, 2119–2123. [Google Scholar] [CrossRef]
- Nurse, P. Cyclin dependent kinases and cell cycle control (nobel lecture). Chembiochem 2002, 3, 596–603. [Google Scholar] [CrossRef]
- Enserink, J.M.; Hombauer, H.; Huang, M.E.; Kolodner, R.D. Cdc28/Cdk1 positively and negatively affects genome stability in S. cerevisiae. J. Cell Biol. 2009, 185, 423–437. [Google Scholar] [CrossRef]
- Nugroho, T.T.; Mendenhall, M.D. An inhibitor of yeast cyclin-dependent protein kinase plays an important role in ensuring the genomic integrity of daughter cells. Mol. Cell. Biol. 1994, 14, 3320–3328. [Google Scholar]
- Lengronne, A.; Schwob, E. The yeast CDK inhibitor Sic1 prevents genomic instability by promoting replication origin licensing in late G(1). Mol. Cell 2002, 9, 1067–1078. [Google Scholar] [CrossRef]
- Kitazono, A.A.; Kron, S.J. An essential function of yeast cyclin-dependent kinase Cdc28 maintains chromosome stability. J. Biol. Chem. 2002, 277, 48627–48634. [Google Scholar] [CrossRef] [PubMed]
- Matthews, H.K.; Bertoli, C.; de Bruin, R.A.M. Cell cycle control in cancer. Nat. Rev. Mol. Cell Biol. 2022, 23, 74–88. [Google Scholar] [CrossRef]
- Malumbres, M. Cyclin-dependent kinases. Genome Biol. 2014, 15, 122. [Google Scholar] [CrossRef] [PubMed]
- Malumbres, M. Revisiting the “Cdk-centric” view of the mammalian cell cycle. Cell Cycle 2005, 4, 206–210. [Google Scholar] [CrossRef]
- Malumbres, M.; Barbacid, M. Mammalian cyclin-dependent kinases. Trends Biochem. Sci. 2005, 30, 630–641. [Google Scholar] [CrossRef]
- Huang, D.; Friesen, H.; Andrews, B. Pho85, a multifunctional cyclin-dependent protein kinase in budding yeast. Mol. Microbiol. 2007, 66, 303–314. [Google Scholar] [CrossRef]
- Jiménez, J.; Ricco, N.; Grijota-Martínez, C.; Fadó, R.; Clotet, J. Redundancy or specificity? The role of the CDK Pho85 in cell cycle control. Int. J. Biochem. Mol. Biol. 2013, 4, 140–149. [Google Scholar]
- Wittenberg, C.; Reed, S.I. Cell cycle-dependent transcription in yeast: Promoters, transcription factors, and transcriptomes. Oncogene 2005, 24, 2746–2755. [Google Scholar] [CrossRef]
- Bertoli, C.; Skotheim, J.M.; de Bruin, R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013, 14, 518–528. [Google Scholar] [CrossRef]
- Lim, S.; Kaldis, P. Cdks, cyclins and CKIs: Roles beyond cell cycle regulation. Development 2013, 140, 3079–3093. [Google Scholar] [CrossRef] [PubMed]
- Haase, S.B.; Wittenberg, C. Topology and control of the cell-cycle-regulated transcriptional circuitry. Genetics 2014, 196, 65–90. [Google Scholar] [CrossRef] [PubMed]
- McInerny, C.J. 2—Cell cycle regulated gene expression in yeasts. In Advances in Genetics; Friedmann, T., Dunlap, J.C., Goodwin, S.F., Eds.; Academic Press: Cambridge, MA, USA, 2011; Volume 73, pp. 51–85. [Google Scholar]
- Chapman, R.D.; Heidemann, M.; Hintermair, C.; Eick, D. Molecular evolution of the RNA polymerase II CTD. Trends Genet. 2008, 24, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Zaborowska, J.; Egloff, S.; Murphy, S. The pol II CTD: New twists in the tail. Nat. Struct. Mol. Biol. 2016, 23, 771–777. [Google Scholar] [CrossRef] [PubMed]
- Sharma, P.; Lioutas, A.; Fernandez-Fuentes, N.; Quilez, J.; Carbonell-Caballero, J.; Wright, R.H.G.; Di Vona, C.; Le Dily, F.; Schüller, R.; Eick, D.; et al. Arginine citrullination at the c-terminal domain controls RNA polymerase II transcription. Mol. Cell 2019, 73, 84–96.e7. [Google Scholar] [CrossRef] [PubMed]
- Harlen, K.M.; Churchman, L.S. The code and beyond: Transcription regulation by the RNA polymerase II carboxy-terminal domain. Nat. Rev. Mol. Cell Biol. 2017, 18, 263–273. [Google Scholar] [CrossRef]
- Li, X.Y.; Bhaumik, S.R.; Green, M.R. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 2000, 288, 1242–1244. [Google Scholar] [CrossRef]
- Kuras, L.; Kosa, P.; Mencia, M.; Struhl, K. TAF-Containing and TAF-independent forms of transcriptionally active TBP in vivo. Science 2000, 288, 1244–1248. [Google Scholar] [CrossRef] [PubMed]
- Baptista, T.; Grünberg, S.; Minoungou, N.; Koster, M.J.E.; Timmers, H.T.M.; Hahn, S.; Devys, D.; Tora, L. SAGA is a general cofactor for RNA polymerase II transcription. Mol. Cell 2017, 68, 130–143.e5. [Google Scholar] [CrossRef]
- Huisinga, K.L.; Pugh, B.F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol. Cell 2004, 13, 573–585. [Google Scholar] [CrossRef]
- Shen, W.C.; Bhaumik, S.R.; Causton, H.C.; Simon, I.; Zhu, X.; Jennings, E.G.; Wang, T.H.; Young, R.A.; Green, M.R. Systematic analysis of essential yeast TAFs in genome-wide transcription and preinitiation complex assembly. EMBO J. 2003, 22, 3395–3402. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qi, Y.; Wu, Z.; Wang, X.; Li, J.; Zhao, D.; Hou, H.; Li, Y.; Yu, Z.; Liu, W.; et al. Structural insights into preinitiation complex assembly on core promoters. Science 2021, 372, 6541. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, J. Transcription regulation by the Mediator complex. Nat. Rev. Mol. Cell Biol. 2018, 19, 262–274. [Google Scholar] [CrossRef]
- Liao, S.M.; Zhang, J.; Jeffery, D.A.; Koleske, A.J.; Thompson, C.M.; Chao, D.M.; Viljoen, M.; van Vuuren, H.J.; Young, R.A. A kinase-cyclin pair in the RNA polymerase II holoenzyme. Nature 1995, 374, 193–196. [Google Scholar] [CrossRef] [PubMed]
- Kuchin, S.; Yeghiayan, P.; Carlson, M. Cyclin-dependent protein kinase and cyclin homologs SSN3 and SSN8 contribute to transcriptional control in yeast. Proc. Natl. Acad. Sci. USA 1995, 92, 4006–4010. [Google Scholar] [CrossRef]
- Balciunas, D.; Ronne, H. Three subunits of the RNA polymerase II mediator complex are involved in glucose repression. Nucleic Acids Res. 1995, 23, 4421–4425. [Google Scholar] [CrossRef]
- Hengartner, C.J.; Myer, V.E.; Liao, S.M.; Wilson, C.J.; Koh, S.S.; Young, R.A. Temporal regulation of RNA polymerase II by Srb10 and Kin28 cyclin-dependent kinases. Mol. Cell 1998, 2, 43–53. [Google Scholar] [CrossRef]
- van de Peppel, J.; Kettelarij, N.; van Bakel, H.; Kockelkorn, T.T.; van Leenen, D.; Holstege, F.C. Mediator expression profiling epistasis reveals a signal transduction pathway with antagonistic submodules and highly specific downstream targets. Mol. Cell 2005, 19, 511–522. [Google Scholar] [CrossRef]
- Wang, W.; Carey, M.; Gralla, J.D. Polymerase II promoter activation: Closed complex formation and ATP-driven start site opening. Science 1992, 255, 450–453. [Google Scholar] [CrossRef]
- Holstege, F.C.; Fiedler, U.; Timmers, H.T. Three transitions in the RNA polymerase II transcription complex during initiation. EMBO J. 1997, 16, 7468–7480. [Google Scholar] [CrossRef]
- Fishburn, J.; Tomko, E.; Galburt, E.; Hahn, S. Double-stranded DNA translocase activity of transcription factor TFIIH and the mechanism of RNA polymerase II open complex formation. Proc. Natl. Acad. Sci. USA 2015, 112, 3961–3966. [Google Scholar] [CrossRef]
- Chen, X.; Yin, X.; Li, J.; Wu, Z.; Qi, Y.; Wang, X.; Liu, W.; Xu, Y. Structures of the human Mediator and Mediator-bound preinitiation complex. Science 2021, 372, 6546. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, M.S.; Heidemann, M.; Tietjen, J.R.; Zhang, D.W.; Chapman, R.D.; Eick, D.; Ansari, A.Z. TFIIH kinase places bivalent marks on the carboxy-terminal domain of RNA polymerase II. Mol. Cell 2009, 34, 387–393. [Google Scholar] [CrossRef] [PubMed]
- Jeronimo, C.; Robert, F. Kin28 regulates the transient association of Mediator with core promoters. Nat. Struct. Mol. Biol. 2014, 21, 449–455. [Google Scholar] [CrossRef] [PubMed]
- Cho, W.K.; Spille, J.H.; Hecht, M.; Lee, C.; Li, C.; Grube, V.; Cisse, I.I. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 2018, 361, 412–415. [Google Scholar] [CrossRef] [PubMed]
- Boehning, M.; Dugast-Darzacq, C.; Rankovic, M.; Hansen, A.S.; Yu, T.; Marie-Nelly, H.; McSwiggen, D.T.; Kokic, G.; Dailey, G.M.; Cramer, P.; et al. RNA polymerase II clustering through carboxy-terminal domain phase separation. Nat. Struct. Mol. Biol. 2018, 25, 833–840. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; He, X.; Reyes-Reyes, M.; Moore, C.; Hampsey, M. Ssu72 is an RNA polymerase II CTD phosphatase. Mol. Cell 2004, 14, 387–394. [Google Scholar] [CrossRef]
- Qiu, H.; Hu, C.; Hinnebusch, A.G. Phosphorylation of the Pol II CTD by KIN28 enhances BUR1/BUR2 recruitment and Ser2 CTD phosphorylation near promoters. Mol. Cell 2009, 33, 752–762. [Google Scholar] [CrossRef]
- Liu, Y.; Warfield, L.; Zhang, C.; Luo, J.; Allen, J.; Lang, W.H.; Ranish, J.; Shokat, K.M.; Hahn, S. Phosphorylation of the transcription elongation factor Spt5 by yeast Bur1 kinase stimulates recruitment of the PAF complex. Mol. Cell. Biol. 2009, 29, 4852–4863. [Google Scholar] [CrossRef]
- Chen, Z.; Hankey, W.; Zhao, Y.; Groth, J.; Huang, F.; Wang, H.; Campos, A.R.; Huang, J.; Roeder, R.G.; Wang, Q. Transcription recycling assays identify PAF1 as a driver for RNA Pol II recycling. Nat. Commun. 2021, 12, 6318. [Google Scholar] [CrossRef]
- Francette, A.M.; Tripplehorn, S.A.; Arndt, K.M. The Paf1 complex: A keystone of nuclear regulation operating at the interface of transcription and chromatin. J. Mol. Biol. 2021, 433, 166979. [Google Scholar] [CrossRef]
- Dronamraju, R.; Strahl, B.D. A feed forward circuit comprising Spt6, Ctk1 and PAF regulates Pol II CTD phosphorylation and transcription elongation. Nucleic Acids Res. 2014, 42, 870–881. [Google Scholar] [CrossRef] [PubMed]
- Ardehali, M.B.; Yao, J.; Adelman, K.; Fuda, N.J.; Petesch, S.J.; Webb, W.W.; Lis, J.T. Spt6 enhances the elongation rate of RNA polymerase II in vivo. EMBO J. 2009, 28, 1067–1077. [Google Scholar] [CrossRef] [PubMed]
- Bortvin, A.; Winston, F. Evidence that Spt6p controls chromatin structure by a direct interaction with histones. Science 1996, 272, 1473–1476. [Google Scholar] [CrossRef] [PubMed]
- Youdell, M.L.; Kizer, K.O.; Kisseleva-Romanova, E.; Fuchs, S.M.; Duro, E.; Strahl, B.D.; Mellor, J. Roles for Ctk1 and Spt6 in regulating the different methylation states of histone H3 lysine 36. Mol. Cell. Biol. 2008, 28, 4915–4926. [Google Scholar] [CrossRef]
- Mayer, A.; Heidemann, M.; Lidschreiber, M.; Schreieck, A.; Sun, M.; Hintermair, C.; Kremmer, E.; Eick, D.; Cramer, P. CTD tyrosine phosphorylation impairs termination factor recruitment to RNA polymerase II. Science 2012, 336, 1723–1725. [Google Scholar] [CrossRef] [PubMed]
- Yurko, N.M.; Manley, J.L. The RNA polymerase II CTD “orphan” residues: Emerging insights into the functions of Tyr-1, Thr-4, and Ser-7. Transcription 2018, 9, 30–40. [Google Scholar] [CrossRef]
- Yurko, N.; Liu, X.; Yamazaki, T.; Hoque, M.; Tian, B.; Manley, J.L. MPK1/SLT2 Links Multiple Stress Responses with Gene Expression in Budding Yeast by Phosphorylating Tyr1 of the RNAP II CTD. Mol. Cell 2017, 68, 913–925 e3. [Google Scholar] [CrossRef]
- St Amour, C.V.; Sansó, M.; Bösken, C.A.; Lee, K.M.; Larochelle, S.; Zhang, C.; Shokat, K.M.; Geyer, M.; Fisher, R.P. Separate domains of fission yeast Cdk9 (P-TEFb) are required for capping enzyme recruitment and primed (Ser7-phosphorylated) Rpb1 carboxyl-terminal domain substrate recognition. Mol. Cell. Biol. 2012, 32, 2372–2383. [Google Scholar] [CrossRef]
- Egloff, S.; O’Reilly, D.; Chapman, R.D.; Taylor, A.; Tanzhaus, K.; Pitts, L.; Eick, D.; Murphy, S. Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 2007, 318, 1777–1779. [Google Scholar] [CrossRef]
- Baillat, D.; Hakimi, M.A.; Näär, A.M.; Shilatifard, A.; Cooch, N.; Shiekhattar, R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 2005, 123, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Nemec, C.M.; Singh, A.K.; Ali, A.; Tseng, S.C.; Syal, K.; Ringelberg, K.J.; Ho, Y.H.; Hintermair, C.; Ahmad, M.F.; Kar, R.K.; et al. Noncanonical CTD kinases regulate RNA polymerase II in a gene-class-specific manner. Nat. Chem. Biol. 2019, 15, 123–131. [Google Scholar] [CrossRef] [PubMed]
- Rosonina, E.; Yurko, N.; Li, W.; Hoque, M.; Tian, B.; Manley, J.L. Threonine-4 of the budding yeast RNAP II CTD couples transcription with Htz1-mediated chromatin remodeling. Proc. Natl. Acad. Sci. USA 2014, 111, 11924–11931. [Google Scholar] [CrossRef] [PubMed]
- Nemec, C.M.; Yang, F.; Gilmore, J.M.; Hintermair, C.; Ho, Y.H.; Tseng, S.C.; Heidemann, M.; Zhang, Y.; Florens, L.; Gasch, A.P.; et al. Different phosphoisoforms of RNA polymerase II engage the Rtt103 termination factor in a structurally analogous manner. Proc. Natl. Acad. Sci. USA 2017, 114, E3944–E3953. [Google Scholar] [CrossRef] [PubMed]
- Harlen, K.M.; Trotta, K.L.; Smith, E.E.; Mosaheb, M.M.; Fuchs, S.M.; Churchman, L.S. Comprehensive RNA polymerase II interactomes reveal distinct and varied roles for each phospho-CTD residue. Cell Rep. 2016, 15, 2147–2158. [Google Scholar] [CrossRef] [PubMed]
- Porrua, O.; Libri, D. Transcription termination and the control of the transcriptome: Why, where and how to stop. Nat. Rev. Mol. Cell Biol. 2015, 16, 190–202. [Google Scholar] [CrossRef]
- Hill, C.H.; Boreikaite, V.; Kumar, A.; Casanal, A.; Kubik, P.; Degliesposti, G.; Maslen, S.; Mariani, A.; von Loeffelholz, O.; Girbig, M.; et al. Activation of the endonuclease that defines mRNA 3′ ends requires incorporation into an 8-subunit core cleavage and polyadenylation factor complex. Mol. Cell 2019, 73, 1217–1231 e11. [Google Scholar] [CrossRef]
- Schreieck, A.; Easter, A.D.; Etzold, S.; Wiederhold, K.; Lidschreiber, M.; Cramer, P.; Passmore, L.A. RNA polymerase II termination involves C-terminal-domain tyrosine dephosphorylation by CPF subunit Glc7. Nat. Struct. Mol. Biol. 2014, 21, 175–179. [Google Scholar] [CrossRef]
- Logan, J.; Falck-Pedersen, E.; Darnell, J.E.; Shenk, T. A poly(A) addition site and a downstream termination region are required for efficient cessation of transcription by RNA polymerase II in the mouse beta maj-globin gene. Proc. Natl. Acad. Sci. USA 1987, 84, 8306. [Google Scholar] [CrossRef]
- Connelly, S.; Manley, J.L. A functional mRNA polyadenylation signal is required for transcription termination by RNA polymerase II. Genes Dev. 1988, 2, 440–452. [Google Scholar] [CrossRef]
- Luo, W.; Johnson, A.W.; Bentley, D.L. The role of Rat1 in coupling mRNA 3′-end processing to transcription termination: Implications for a unified allosteric-torpedo model. Genes Dev. 2006, 20, 954–965. [Google Scholar] [CrossRef] [PubMed]
- Medler, S.; Al Husini, N.; Raghunayakula, S.; Mukundan, B.; Aldea, A.; Ansari, A. Evidence for a complex of transcription factor IIB with poly(A) polymerase and cleavage factor 1 subunits required for gene looping. J. Biol. Chem. 2011, 286, 33709–33718. [Google Scholar] [CrossRef] [PubMed]
- El Kaderi, B.; Medler, S.; Raghunayakula, S.; Ansari, A. Gene looping is conferred by activator-dependent interaction of transcription initiation and termination machineries. J. Biol. Chem. 2009, 284, 25015–25025. [Google Scholar] [CrossRef] [PubMed]
- Al-Husini, N.; Medler, S.; Ansari, A. Crosstalk of promoter and terminator during RNA polymerase II transcription cycle. Biochim. Biophys. Acta Gene Regul. Mech. 2020, 1863, 194657. [Google Scholar] [CrossRef]
- Dichtl, B.; Blank, D.; Ohnacker, M.; Friedlein, A.; Roeder, D.; Langen, H.; Keller, W. A role for SSU72 in balancing RNA polymerase II transcription elongation and termination. Mol. Cell 2002, 10, 1139–1150. [Google Scholar] [CrossRef]
- Sun, Z.W.; Hampsey, M. Synthetic enhancement of a TFIIB defect by a mutation in SSU72, an essential yeast gene encoding a novel protein that affects transcription start site selection in vivo. Mol. Cell. Biol. 1996, 16, 1557–1566. [Google Scholar] [CrossRef]
- Allepuz-Fuster, P.; O’Brien, M.J.; González-Polo, N.; Pereira, B.; Dhoondia, Z.; Ansari, A.; Calvo, O. RNA polymerase II plays an active role in the formation of gene loops through the Rpb4 subunit. Nucleic Acids Res. 2019, 47, 8975–8987. [Google Scholar] [CrossRef]
- Mayfield, J.E.; Burkholder, N.T.; Zhang, Y.J. Dephosphorylating eukaryotic RNA polymerase II. Biochim. Biophys. Acta 2016, 1864, 372–387. [Google Scholar] [CrossRef]
- Kobor, M.S.; Archambault, J.; Lester, W.; Holstege, F.C.; Gileadi, O.; Jansma, D.B.; Jennings, E.G.; Kouyoumdjian, F.; Davidson, A.R.; Young, R.A.; et al. An unusual eukaryotic protein phosphatase required for transcription by RNA polymerase II and CTD dephosphorylation in S. cerevisiae. Mol. Cell 1999, 4, 55–62. [Google Scholar] [CrossRef]
- Archambault, J.; Chambers, R.S.; Kobor, M.S.; Ho, Y.; Cartier, M.; Bolotin, D.; Andrews, B.; Kane, C.M.; Greenblatt, J. An essential component of a C-terminal domain phosphatase that interacts with transcription factor IIF in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1997, 94, 14300–14305. [Google Scholar] [CrossRef]
- Cho, E.J.; Kobor, M.S.; Kim, M.; Greenblatt, J.; Buratowski, S. Opposing effects of Ctk1 kinase and Fcp1 phosphatase at Ser 2 of the RNA polymerase II C-terminal domain. Genes Dev. 2001, 15, 3319–3329. [Google Scholar] [CrossRef] [PubMed]
- Taylor, J.H. Nucleic acid synthesis in relation to the cell division cycle. Ann. N. Y. Acad. Sci. 1960, 90, 409–421. [Google Scholar] [CrossRef] [PubMed]
- Gottesfeld, J.M.; Forbes, D.J. Mitotic repression of the transcriptional machinery. Trends Biochem. Sci. 1997, 22, 197–202. [Google Scholar] [CrossRef]
- Palozola, K.C.; Donahue, G.; Liu, H.; Grant, G.R.; Becker, J.S.; Cote, A.; Yu, H.; Raj, A.; Zaret, K.S. Mitotic transcription and waves of gene reactivation during mitotic exit. Science 2017, 358, 119–122. [Google Scholar] [CrossRef] [PubMed]
- Timmers, H.T.M.; Verrijzer, C.P. Mitotic chromosomes: Not so silent after all. Dev. Cell 2017, 43, 119–121. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.C.; Holland, J.J. Ribonucleic acid and protein synthesis in mitotic HeLa cells. J. Cell Biol. 1965, 27, 565–574. [Google Scholar] [CrossRef] [PubMed]
- Segil, N.; Guermah, M.; Hoffmann, A.; Roeder, R.G.; Heintz, N. Mitotic regulation of TFIID: Inhibition of activator-dependent transcription and changes in subcellular localization. Genes Dev. 1996, 10, 2389–2400. [Google Scholar] [CrossRef]
- Long, J.J.; Leresche, A.; Kriwacki, R.W.; Gottesfeld, J.M. Repression of TFIIH transcriptional activity and TFIIH-associated cdk7 kinase activity at mitosis. Mol. Cell. Biol. 1998, 18, 1467–1476. [Google Scholar] [CrossRef][Green Version]
- Akoulitchev, S.; Reinberg, D. The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 1998, 12, 3541–3550. [Google Scholar] [CrossRef][Green Version]
- Cisek, L.J.; Corden, J.L. Phosphorylation of RNA polymerase by the murine homologue of the cell-cycle control protein cdc2. Nature 1989, 339, 679–684. [Google Scholar] [CrossRef]
- Zhang, J.; Corden, J.L. Identification of phosphorylation sites in the repetitive carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J. Biol. Chem. 1991, 266, 2290–2296. [Google Scholar] [CrossRef]
- Zhang, J.; Corden, J.L. Phosphorylation causes a conformational change in the carboxyl-terminal domain of the mouse RNA polymerase II largest subunit. J. Biol. Chem. 1991, 266, 2297–2302. [Google Scholar] [CrossRef]
- Xu, Y.X.; Hirose, Y.; Zhou, X.Z.; Lu, K.P.; Manley, J.L. Pin1 modulates the structure and function of human RNA polymerase II. Genes Dev. 2003, 17, 2765–2776. [Google Scholar] [CrossRef] [PubMed]
- Gebara, M.M.; Sayre, M.H.; Corden, J.L. Phosphorylation of the carboxy-terminal repeat domain in RNA polymerase II by cyclin-dependent kinases is sufficient to inhibit transcription. J. Cell. Biochem. 1997, 64, 390–402. [Google Scholar] [CrossRef]
- Van Lint, C.; Bouchat, S.; Marcello, A. HIV-1 transcription and latency: An update. Retrovirology 2013, 10, 67. [Google Scholar] [CrossRef]
- Deng, L.; Ammosova, T.; Pumfery, A.; Kashanchi, F.; Nekhai, S. HIV-1 Tat interaction with RNA polymerase II C-terminal domain (CTD) and a dynamic association with CDK2 induce CTD phosphorylation and transcription from HIV-1 promoter. J. Biol. Chem. 2002, 277, 33922–33929. [Google Scholar] [CrossRef]
- Nekhai, S.; Zhou, M.; Fernandez, A.; Lane, W.S.; Lamb, N.J.; Brady, J.; Kumar, A. HIV-1 Tat-associated RNA polymerase C-terminal domain kinase, CDK2, phosphorylates CDK7 and stimulates Tat-mediated transcription. Biochem. J. 2002, 364 Pt 3, 649–657. [Google Scholar] [CrossRef]
- Agbottah, E.; Deng, L.; Dannenberg, L.O.; Pumfery, A.; Kashanchi, F. Effect of SWI/SNF chromatin remodeling complex on HIV-1 Tat activated transcription. Retrovirology 2006, 3, 48. [Google Scholar] [CrossRef]
- Granovskaia, M.V.; Jensen, L.J.; Ritchie, M.E.; Toedling, J.; Ning, Y.; Bork, P.; Huber, W.; Steinmetz, L.M. High-resolution transcription atlas of the mitotic cell cycle in budding yeast. Genome Biol. 2010, 11, R24. [Google Scholar] [CrossRef]
- Elliott, S.G.; McLaughlin, C.S. Regulation of RNA synthesis in yeast. III. Synthesis during the cell cycle. Mol. Gen. Genet. 1979, 169, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Chymkowitch, P.; Eldholm, V.; Lorenz, S.; Zimmermann, C.; Lindvall, J.M.; Bjoras, M.; Meza-Zepeda, L.A.; Enserink, J.M. Cdc28 kinase activity regulates the basal transcription machinery at a subset of genes. Proc. Natl. Acad. Sci. USA 2012, 109, 10450–10455. [Google Scholar] [CrossRef]
- Serrano, R.; Kielland-Brandt, M.C.; Fink, G.R. Yeast plasma membrane ATPase is essential for growth and has homology with (Na+ + K+), K+- and Ca2+-ATPases. Nature 1986, 319, 689–693. [Google Scholar] [CrossRef]
- Ahn, S.H.; Kim, M.; Buratowski, S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell 2004, 13, 67–76. [Google Scholar] [CrossRef]
- Thayer, N.H.; Leverich, C.K.; Fitzgibbon, M.P.; Nelson, Z.W.; Henderson, K.A.; Gafken, P.R.; Hsu, J.J.; Gottschling, D.E. Identification of long-lived proteins retained in cells undergoing repeated asymmetric divisions. Proc. Natl. Acad. Sci. USA 2014, 111, 14019–14026. [Google Scholar] [CrossRef]
- Henderson, K.A.; Hughes, A.L.; Gottschling, D.E. Mother-daughter asymmetry of pH underlies aging and rejuvenation in yeast. Elife 2014, 3, e03504. [Google Scholar] [CrossRef] [PubMed]
- Spellman, P.T.; Sherlock, G.; Zhang, M.Q.; Iyer, V.R.; Anders, K.; Eisen, M.B.; Brown, P.O.; Botstein, D.; Futcher, B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 1998, 9, 3273–3297. [Google Scholar] [CrossRef] [PubMed]
- Chymkowitch, P.; Enserink, J.M. The cell cycle rallies the transcription cycle: Cdc28/Cdk1 is a cell cycle-regulated transcriptional CDK. Transcription 2013, 4, 3–6. [Google Scholar] [CrossRef] [PubMed]
- Koivomagi, M.; Valk, E.; Venta, R.; Iofik, A.; Lepiku, M.; Balog, E.R.; Rubin, S.M.; Morgan, D.O.; Loog, M. Cascades of multisite phosphorylation control Sic1 destruction at the onset of S phase. Nature 2011, 480, 128–131. [Google Scholar] [CrossRef]
- Cosma, M.P.; Panizza, S.; Nasmyth, K. Cdk1 triggers association of RNA polymerase to cell cycle promoters only after recruitment of the mediator by SBF. Mol. Cell 2001, 7, 1213–1220. [Google Scholar] [CrossRef]
- Koivomagi, M.; Swaffer, M.P.; Turner, J.J.; Marinov, G.; Skotheim, J.M. G1 cyclin-Cdk promotes cell cycle entry through localized phosphorylation of RNA polymerase II. Science 2021, 374, 347–351. [Google Scholar] [CrossRef]
- de Bruin, R.A.; McDonald, W.H.; Kalashnikova, T.I.; Yates, J., 3rd; Wittenberg, C. Cln3 activates G1-specific transcription via phosphorylation of the SBF bound repressor Whi5. Cell 2004, 117, 887–898. [Google Scholar] [CrossRef] [PubMed]
- Costanzo, M.; Nishikawa, J.L.; Tang, X.; Millman, J.S.; Schub, O.; Breitkreuz, K.; Dewar, D.; Rupes, I.; Andrews, B.; Tyers, M. CDK activity antagonizes Whi5, an inhibitor of G1/S transcription in yeast. Cell 2004, 117, 899–913. [Google Scholar] [CrossRef] [PubMed]
- Hortobagyi, G.N.; Stemmer, S.M.; Burris, H.A.; Yap, Y.S.; Sonke, G.S.; Paluch-Shimon, S.; Campone, M.; Blackwell, K.L.; André, F.; Winer, E.P.; et al. Ribociclib as First-Line Therapy for HR-Positive, Advanced Breast Cancer. N. Engl. J. Med. 2016, 375, 1738–1748. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. The retinoblastoma protein and cell cycle control. Cell 1995, 81, 323–330. [Google Scholar] [CrossRef]
- Goel, S.; DeCristo, M.J.; McAllister, S.S.; Zhao, J.J. CDK4/6 Inhibition in Cancer: Beyond Cell Cycle Arrest. Trends Cell Biol. 2018, 28, 911–925. [Google Scholar] [CrossRef]
- Brickner, D.G.; Brickner, J.H. Cdk phosphorylation of a nucleoporin controls localization of active genes through the cell cycle. Mol. Biol. Cell 2010, 21, 3421–3432. [Google Scholar] [CrossRef]
- Taddei, A.; Van Houwe, G.; Hediger, F.; Kalck, V.; Cubizolles, F.; Schober, H.; Gasser, S.M. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature 2006, 441, 774–778. [Google Scholar] [CrossRef]
- Casolari, J.M.; Brown, C.R.; Komili, S.; West, J.; Hieronymus, H.; Silver, P.A. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell 2004, 117, 427–439. [Google Scholar] [CrossRef]
- Menon, B.B.; Sarma, N.J.; Pasula, S.; Deminoff, S.J.; Willis, K.A.; Barbara, K.E.; Andrews, B.; Santangelo, G.M. Reverse recruitment: The Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc. Natl. Acad. Sci. USA 2005, 102, 5749–5754. [Google Scholar] [CrossRef]
- Morris, M.C.; Kaiser, P.; Rudyak, S.; Baskerville, C.; Watson, M.H.; Reed, S.I. Cks1-dependent proteasome recruitment and activation of CDC20 transcription in budding yeast. Nature 2003, 423, 1009–1013. [Google Scholar] [CrossRef]
- Chaves, S.; Baskerville, C.; Yu, V.; Reed, S.I. Cks1, Cdk1, and the 19S proteasome collaborate to regulate gene induction-dependent nucleosome eviction in yeast. Mol. Cell. Biol. 2010, 30, 5284–5294. [Google Scholar] [CrossRef] [PubMed]
- Yu, V.P.; Baskerville, C.; Grunenfelder, B.; Reed, S.I. A kinase-independent function of Cks1 and Cdk1 in regulation of transcription. Mol. Cell 2005, 17, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.; Stiller, J.W. Comparative genomics of cyclin-dependent kinases suggest co-evolution of the RNAP II C-terminal domain and CTD-directed CDKs. BMC Genom. 2004, 5, 69. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Stiller, J.W. Evolutionary diversity and taxon-specific modifications of the RNA polymerase II C-terminal domain. Proc. Natl. Acad. Sci. USA 2014, 111, 5920–5925. [Google Scholar] [CrossRef] [PubMed]
- Burton, Z.F. The old and new testaments of gene regulation. Evolution of multi-subunit RNA polymerases and co-evolution of eukaryote complexity with the RNAP II CTD. Transcription 2014, 5, e28674. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Kipreos, E.T. Evolution of cyclin-dependent kinases (CDKs) and CDK-activating kinases (CAKs): Differential conservation of CAKs in yeast and metazoa. Mol. Biol. Evol. 2000, 17, 1061–1074. [Google Scholar] [CrossRef] [PubMed]
- Warner, J.R. The economics of ribosome biosynthesis in yeast. Trends Biochem. Sci. 1999, 24, 437–440. [Google Scholar] [CrossRef]
- Birch, J.L.; Zomerdijk, J.C. Structure and function of ribosomal RNA gene chromatin. Biochem. Soc. Trans. 2008, 36 Pt 4, 619–624. [Google Scholar] [CrossRef]
- Jackobel, A.J.; Han, Y.; He, Y.; Knutson, B.A. Breaking the mold: Structures of the RNA polymerase I transcription complex reveal a new path for initiation. Transcription 2018, 9, 255–261. [Google Scholar] [CrossRef]
- Kuhn, C.D.; Geiger, S.R.; Baumli, S.; Gartmann, M.; Gerber, J.; Jennebach, S.; Mielke, T.; Tschochner, H.; Beckmann, R.; Cramer, P. Functional architecture of RNA polymerase I. Cell 2007, 131, 1260–1272. [Google Scholar] [CrossRef]
- Zhang, Y.; Smith, A.D.; Renfrow, M.B.; Schneider, D.A. The RNA polymerase-associated factor 1 complex (Paf1C) directly increases the elongation rate of RNA polymerase I and is required for efficient regulation of rRNA synthesis. J. Biol. Chem. 2010, 285, 14152–14159. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.A.; French, S.L.; Osheim, Y.N.; Bailey, A.O.; Vu, L.; Dodd, J.; Yates, J.R.; Beyer, A.L.; Nomura, M. RNA polymerase II elongation factors Spt4p and Spt5p play roles in transcription elongation by RNA polymerase I and rRNA processing. Proc. Natl. Acad. Sci. USA 2006, 103, 12707–12712. [Google Scholar] [CrossRef] [PubMed]
- Schneider, D.A.; Michel, A.; Sikes, M.L.; Vu, L.; Dodd, J.A.; Salgia, S.; Osheim, Y.N.; Beyer, A.L.; Nomura, M. Transcription elongation by RNA polymerase I is linked to efficient rRNA processing and ribosome assembly. Mol. Cell 2007, 26, 217–229. [Google Scholar] [CrossRef] [PubMed]
- Kos, M.; Tollervey, D. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 2010, 37, 809–820. [Google Scholar] [CrossRef]
- Reiter, A.; Hamperl, S.; Seitz, H.; Merkl, P.; Perez-Fernandez, J.; Williams, L.; Gerber, J.; Németh, A.; Léger, I.; Gadal, O.; et al. The Reb1-homologue Ydr026c/Nsi1 is required for efficient RNA polymerase I termination in yeast. EMBO J. 2012, 31, 3480–3493. [Google Scholar] [CrossRef]
- Lang, W.H.; Reeder, R.H. Transcription termination of RNA polymerase I due to a T-rich element interacting with Reb1p. Proc. Natl. Acad. Sci. USA 1995, 92, 9781–9785. [Google Scholar] [CrossRef]
- Németh, A.; Perez-Fernandez, J.; Merkl, P.; Hamperl, S.; Gerber, J.; Griesenbeck, J.; Tschochner, H. RNA polymerase I termination: Where is the end? Biochim. Biophys. Acta Gene Regul. Mech. 2013, 1829, 306–317. [Google Scholar] [CrossRef]
- Li, H.; Tsang, C.K.; Watkins, M.; Bertram, P.G.; Zheng, X.F. Nutrient regulates Tor1 nuclear localization and association with rDNA promoter. Nature 2006, 442, 1058–1061. [Google Scholar] [CrossRef]
- González-Jiménez, A.; Campos, A.; Navarro, F.; Clemente-Blanco, A.; Calvo, O. Regulation of eukaryotic RNAPs activities by phosphorylation. Front. Mol. Biosci. 2021, 8, 681865. [Google Scholar] [CrossRef]
- Heix, J.; Vente, A.; Voit, R.; Budde, A.; Michaelidis, T.M.; Grummt, I. Mitotic silencing of human rRNA synthesis: Inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998, 17, 7373–7381. [Google Scholar] [CrossRef]
- Gorski, J.J.; Pathak, S.; Panov, K.; Kasciukovic, T.; Panova, T.; Russell, J.; Zomerdijk, J.C. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007, 26, 1560–1568. [Google Scholar] [CrossRef] [PubMed]
- Russell, J.; Zomerdijk, J.C. The RNA polymerase I transcription machinery. Biochem. Soc. Symp. 2006, 73, 203–216. [Google Scholar]
- Kuhn, A.; Vente, A.; Doree, M.; Grummt, I. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J. Mol. Biol. 1998, 284, 1–5. [Google Scholar] [CrossRef]
- Voit, R.; Seiler, J.; Grummt, I. Cooperative action of Cdk1/cyclin B and SIRT1 is required for mitotic repression of rRNA synthesis. PLoS Genet. 2015, 11, e1005246. [Google Scholar] [CrossRef] [PubMed]
- Klein, J.; Grummt, I. Cell cycle-dependent regulation of RNA polymerase I transcription: The nucleolar transcription factor UBF is inactive in mitosis and early G1. Proc. Natl. Acad. Sci. USA 1999, 96, 6096–6101. [Google Scholar] [CrossRef] [PubMed]
- Clemente-Blanco, A.; Mayan-Santos, M.; Schneider, D.A.; Machin, F.; Jarmuz, A.; Tschochner, H.; Aragon, L. Cdc14 inhibits transcription by RNA polymerase I during anaphase. Nature 2009, 458, 219–222. [Google Scholar] [CrossRef]
- Acker, J.; Conesa, C.; Lefebvre, O. Yeast RNA polymerase III transcription factors and effectors. Biochim. Biophys. Acta 2013, 1829, 283–295. [Google Scholar] [CrossRef]
- Wang, C.K.; Weil, P.A. Purification and characterization of Saccharomyces cerevisiae transcription factor IIIA. J. Biol. Chem. 1989, 264, 1092–1099. [Google Scholar] [CrossRef]
- Brun, I.; Sentenac, A.; Werner, M. Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J. 1997, 16, 5730–5741. [Google Scholar] [CrossRef]
- Vorländer, M.K.; Khatter, H.; Wetzel, R.; Hagen, W.J.H.; Müller, C.W. Molecular mechanism of promoter opening by RNA polymerase III. Nature 2018, 553, 295–300. [Google Scholar] [CrossRef]
- Abascal-Palacios, G.; Ramsay, E.P.; Beuron, F.; Morris, E.; Vannini, A. Structural basis of RNA polymerase III transcription initiation. Nature 2018, 553, 301–306. [Google Scholar] [CrossRef] [PubMed]
- Khoo, S.-K.; Wu, C.-C.; Lin, Y.-C.; Chen, H.-T. The TFIIE-related Rpc82 subunit of RNA polymerase III interacts with the TFIIB-related transcription factor Brf1 and the polymerase cleft for transcription initiation. Nucleic Acids Res. 2018, 46, 1157–1166. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Yan, C.; Fishbain, S.; Ivanov, I.; He, Y. Structural visualization of RNA polymerase III transcription machineries. Cell Discov. 2018, 4, 40. [Google Scholar] [CrossRef]
- Turowski, T.W.; Leśniewska, E.; Delan-Forino, C.; Sayou, C.; Boguta, M.; Tollervey, D. Global analysis of transcriptionally engaged yeast RNA polymerase III reveals extended tRNA transcripts. Genome Res. 2016, 26, 933–944. [Google Scholar] [CrossRef] [PubMed]
- Arimbasseri, A.G.; Maraia, R.J. Mechanism of transcription termination by RNA polymerase III utilizes a non-template strand sequence-specific signal element. Mol. Cell 2015, 58, 1124–1132. [Google Scholar] [CrossRef] [PubMed]
- Dieci, G.; Sentenac, A. Facilitated recycling pathway for RNA polymerase III. Cell 1996, 84, 245–252. [Google Scholar] [CrossRef]
- Moir, R.D.; Willis, I.M. Regulation of pol III transcription by nutrient and stress signaling pathways. Biochim. Biophys. Acta. 2013, 1829, 361–375. [Google Scholar] [CrossRef]
- Nguéa, P.A.; Robertson, J.; Herrera, M.C.; Chymkowitch, P.; Enserink, J.M. Desumoylation of RNA polymerase III lies at the core of the Sumo stress response in yeast. J. Biol. Chem. 2019, 294, 18784–18795. [Google Scholar] [CrossRef]
- Pluta, K.; Lefebvre, O.; Martin, N.C.; Smagowicz, W.J.; Stanford, D.R.; Ellis, S.R.; Hopper, A.K.; Sentenac, A.; Boguta, M. Maf1p, a negative effector of RNA polymerase III in Saccharomyces cerevisiae. Mol. Cell. Biol. 2001, 21, 5031–5040. [Google Scholar] [CrossRef]
- Roberts, D.N.; Wilson, B.; Huff, J.T.; Stewart, A.J.; Cairns, B.R. Dephosphorylation and genome-wide association of Maf1 with Pol III-transcribed genes during repression. Mol. Cell 2006, 22, 633–644. [Google Scholar] [CrossRef]
- Towpik, J.; Graczyk, D.; Gajda, A.; Lefebvre, O.; Boguta, M. Derepression of RNA polymerase III transcription by phosphorylation and nuclear export of its negative regulator, Maf1. J. Biol. Chem. 2008, 283, 17168–17174. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Tsang, C.K.; Zheng, X.F. Mechanisms of regulation of RNA polymerase III-dependent transcription by TORC1. EMBO J. 2009, 28, 2220–2230. [Google Scholar] [CrossRef] [PubMed]
- Huber, A.; Bodenmiller, B.; Uotila, A.; Stahl, M.; Wanka, S.; Gerrits, B.; Aebersold, R.; Loewith, R. Characterization of the rapamycin-sensitive phosphoproteome reveals that Sch9 is a central coordinator of protein synthesis. Genes Dev. 2009, 23, 1929–1943. [Google Scholar] [CrossRef]
- Graczyk, D.; Debski, J.; Muszynska, G.; Bretner, M.; Lefebvre, O.; Boguta, M. Casein kinase II-mediated phosphorylation of general repressor Maf1 triggers RNA polymerase III activation. Proc. Natl. Acad. Sci. USA 2011, 108, 4926–4931. [Google Scholar] [CrossRef] [PubMed]
- Moir, R.D.; Lee, J.; Haeusler, R.A.; Desai, N.; Engelke, D.R.; Willis, I.M. Protein kinase A regulates RNA polymerase III transcription through the nuclear localization of Maf1. Proc. Natl. Acad. Sci. USA 2006, 103, 15044–15049. [Google Scholar] [CrossRef]
- Oler, A.J.; Cairns, B.R. PP4 dephosphorylates Maf1 to couple multiple stress conditions to RNA polymerase III repression. EMBO J. 2012, 31, 1440–1452. [Google Scholar] [CrossRef]
- Oficjalska-Pham, D.; Harismendy, O.; Smagowicz, W.J.; de Peredo, A.G.; Boguta, M.; Sentenac, A.; Lefebvre, O. General repression of RNA polymerase III transcription is triggered by protein phosphatase type 2A-mediated dephosphorylation of Maf1. Mol. Cell 2006, 22, 623–632. [Google Scholar] [CrossRef]
- Vannini, A.; Ringel, R.; Kusser, A.G.; Berninghausen, O.; Kassavetis, G.A.; Cramer, P. Molecular basis of RNA polymerase III transcription repression by Maf1. Cell 2010, 143, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Moir, R.D.; McIntosh, K.B.; Willis, I.M. TOR signaling regulates ribosome and tRNA synthesis via LAMMER/Clk and GSK-3 family kinases. Mol. Cell 2012, 45, 836–843. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, C.; Aslanian, A.; Yates, J.R., 3rd; Hunter, T. Defective RNA polymerase III is negatively regulated by the SUMO-Ubiquitin-Cdc48 pathway. Elife 2018, 7, e35447. [Google Scholar] [CrossRef]
- Chymkowitch, P.; Nguea, P.A.; Aanes, H.; Robertson, J.; Klungland, A.; Enserink, J.M. TORC1-dependent sumoylation of Rpc82 promotes RNA polymerase III assembly and activity. Proc. Natl. Acad. Sci. USA 2017, 114, 1039–1044. [Google Scholar] [CrossRef] [PubMed]
- Nwagwu, M.; Nana, M. Ribonucleic acid synthesis in embryonic chick muscle, rates of synthesis and half-lives of transfer and ribosomal RNA species. J. Embryol. Exp. Morphol. 1980, 56, 253–267. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, A.; Chernyakov, I.; Gu, W.; Hiley, S.L.; Hughes, T.R.; Grayhack, E.J.; Phizicky, E.M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell 2006, 21, 87–96. [Google Scholar] [CrossRef] [PubMed]
- Svenningsen, S.L.; Kongstad, M.; Stenum, T.S.; Munoz-Gomez, A.J.; Sorensen, M.A. Transfer RNA is highly unstable during early amino acid starvation in Escherichia coli. Nucleic Acids Res. 2017, 45, 793–804. [Google Scholar] [CrossRef] [PubMed]
- Scott, P.H.; Cairns, C.A.; Sutcliffe, J.E.; Alzuherri, H.M.; McLees, A.; Winter, A.G.; White, R.J. Regulation of RNA polymerase III transcription during cell cycle entry. J. Biol. Chem. 2001, 276, 1005–1014. [Google Scholar] [CrossRef]
- Frenkel-Morgenstern, M.; Danon, T.; Christian, T.; Igarashi, T.; Cohen, L.; Hou, Y.M.; Jensen, L.J. Genes adopt non-optimal codon usage to generate cell cycle-dependent oscillations in protein levels. Mol. Syst. Biol. 2012, 8, 572. [Google Scholar] [CrossRef]
- Chen, M.; Gartenberg, M.R. Coordination of tRNA transcription with export at nuclear pore complexes in budding yeast. Genes Dev. 2014, 28, 959–970. [Google Scholar] [CrossRef]
- Herrera, M.C.; Chymkowitch, P.; Robertson, J.M.; Eriksson, J.; Boe, S.O.; Alseth, I.; Enserink, J.M. Cdk1 gates cell cycle-dependent tRNA synthesis by regulating RNA polymerase III activity. Nucleic Acids Res. 2018, 46, 11698–11711. [Google Scholar] [CrossRef]
- Fairley, J.A.; Scott, P.H.; White, R.J. TFIIIB is phosphorylated, disrupted and selectively released from tRNA promoters during mitosis in vivo. EMBO J. 2003, 22, 5841–5850. [Google Scholar] [CrossRef]
- White, R.J.; Gottlieb, T.M.; Downes, C.S.; Jackson, S.P. Mitotic regulation of a TATA-binding-protein-containing complex. Mol. Cell. Biol. 1995, 15, 1983–1992. [Google Scholar] [CrossRef]
- Kitamura, E.; Tanaka, K.; Kitamura, Y.; Tanaka, T.U. Kinetochore microtubule interaction during S phase in Saccharomyces cerevisiae. Genes Dev. 2007, 21, 3319–3330. [Google Scholar] [CrossRef] [PubMed]
- Johnston, G.C.; Pringle, J.R.; Hartwell, L.H. Coordination of growth with cell division in the yeast Saccharomyces cerevisiae. Exp. Cell Res. 1977, 105, 79–98. [Google Scholar] [CrossRef]
| CDK | Cyclin | Function |
|---|---|---|
| Cdk1 | Essential CDK with similarity to mammalian Cdk1, main cell cycle regulator. | |
| Cln1, Cln2 | G1 cyclins, regulate G1–S transition | |
| Cln3 | G1 cyclin that is transcribed throughout the cell cycle, although protein levels peak in M phase. Regulates initial expression of Cln1 and Cln2 | |
| Clb1,2 | B-type cyclins involved in the transition from G2 to M phase | |
| Clb3,4 | B-type cyclins mainly expressed in S–G2 phase and involved in spindle formation. | |
| Clb5, Clb6 | S phase cyclins involved in DNA replication and transcription | |
| Pho85 | Clg1, Pcl1, Pcl2, Pcl5, Pcl6, Pcl7, Pcl8, Pcl9, Pcl10, Pho80 | Non-essential CDK with similarity to mammalian Cdk5. Associates with ten different cyclins to regulate environmental and nutrient responses and to promote cell cycle progression. |
| Bur1 | Bur2 | CDK with similarity to mammalian Cdk9. Regulates transcriptional elongation through the phosphorylation of Ser2 of the CTD of RNAPII. |
| Ctk1 | Ctk2 | CDK with similarity to mammalian Cdk12 and Cdk13. Regulates transcriptional elongation through phosphorylation of Ser2 of the CTD of RNAPII. |
| Kin28 | Ccl1 | CDK with similarity to mammalian Cdk7. Involved in transcription initiation and mRNA capping through phosphorylation of Ser5 and Ser7 in the CTD of RNAPII. |
| Ssn3 | Ssn8 | CDK with similarity to human Cdk8. Part of the Mediator complex. Mainly involved in suppression of transcription. |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Enserink, J.M.; Chymkowitch, P. Cell Cycle-Dependent Transcription: The Cyclin Dependent Kinase Cdk1 Is a Direct Regulator of Basal Transcription Machineries. Int. J. Mol. Sci. 2022, 23, 1293. https://doi.org/10.3390/ijms23031293
Enserink JM, Chymkowitch P. Cell Cycle-Dependent Transcription: The Cyclin Dependent Kinase Cdk1 Is a Direct Regulator of Basal Transcription Machineries. International Journal of Molecular Sciences. 2022; 23(3):1293. https://doi.org/10.3390/ijms23031293
Chicago/Turabian StyleEnserink, Jorrit M., and Pierre Chymkowitch. 2022. "Cell Cycle-Dependent Transcription: The Cyclin Dependent Kinase Cdk1 Is a Direct Regulator of Basal Transcription Machineries" International Journal of Molecular Sciences 23, no. 3: 1293. https://doi.org/10.3390/ijms23031293
APA StyleEnserink, J. M., & Chymkowitch, P. (2022). Cell Cycle-Dependent Transcription: The Cyclin Dependent Kinase Cdk1 Is a Direct Regulator of Basal Transcription Machineries. International Journal of Molecular Sciences, 23(3), 1293. https://doi.org/10.3390/ijms23031293







