Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops
Abstract
1. Introduction
1.1. Abiotic Stress Adaptations
1.2. Physiological and Biochemical Responses
1.3. Gene Regulation and Signal Transduction
1.4. Effective Microbes
2. Molecular Tools
2.1. Classic Techniques
2.1.1. Plant Breeding
2.1.2. Grafting and Rootstocks
2.1.3. Random Mutations
2.1.4. Plant Transformation: Biolistics and Agrobacterium
2.2. New Plant Breeding Techniques (NPBT)
2.2.1. Genotyping-by-Sequencing (GBS) and “Omics”
2.2.2. Zinc Finger Nucleases (ZFNs) and Transcription Activator-Like Effector Nucleases (TALENs)
2.2.3. Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)
2.2.4. Oligonucleotide-Directed Mutagenesis (ODM)
3. Crops
3.1. Model and Non-Model Plants
3.2. Rice (Oryza sativa L.)
3.3. Wheat (Triticum aestivum L.)
3.4. Corn (Zea mays L.)
4. Conclusions and Future Perspectives
- Genome/Epigenome (nuclear and organellar) editing and manipulation of key multi-stress-responsive genes or transcription factors have been shown to confer increased tolerance to multiple stressors;
- Altering expression of organellar DNA damage repair system involved genes can lead to more efficient mutagenesis, genetic diversity enhancement, and tolerance improvement to ROS/oxidative stress;
- Emphasis must be considered on post-transcriptional and post-translational regulators (including the huge diversity of types of lncRNAs and recently discovered glycoRNAs) through the use of multiple omics (PlantOmics) integrating genome-wide associations studies and pan-genomic/pan-transcriptomic strategies;
- Plant phenomics will accelerate plant breeding targeted and successful stress-resilient cultivars and their wild relatives under real field conditions;
- It should be taken advantage of multiple cross-talk signaling among diverse challenging atmospheric and soil abiotic (and biotic) factors such as drought, salinity, nutrient deficiency, soil properties, pollution, metal, submergence, anoxia, heat, low/high temperature, wind, light, UV, CO2, methane, N2O, O3, osmotic, oxidative stress, in energy-(sugars), organ-(aerial, roots), tissue-, and phenology-dependent manner;
- CRISPR/Cas9 multiple gene editing for simultaneous expression of structural and regulatory genes represents a promising strategy in order to develop multi-stress-resilient crops;
- Given the evident role of sugar sensing and signaling in abiotic stress responses (sugar-insensitive Arabidopsis mutants are tolerant to abiotic and salt stress), we believe that sugar signaling pathways are key targets to reducing sugar’s negative feedback effect on photosynthesis, which could lead to abiotic stress tolerant phenotypes and increased yields in crops;
- Undoubtedly, much remains to be discovered and learned from the study of resurrection plants and their associated microbiomes, particularly those tolerant to extreme abiotic stress, i.e., Bryum argenteum, Craterostigma plantagineum, Pseudocrossidium replicatum, Selaginella lepidophylla, Syntrichia (Tortula) ruralis, the Arctic and Antarctic moss Sanionia uncinata, desert moss Syntrichia caninervis;
- Sustainable management of agricultural water and soil resources;
- Diversification of food supply (nutritional diversity) with local plant species;
- Multi-stress experimentation in the laboratory considering variable intensity and timing and recovery capacities related to photosynthesis and growth parameters;
- The enrichment of the seed and soil microbiomes through the use of microbe-effective-based inoculants undoubtedly contributes to the integrated management of crops to mitigate the effects of the multiple stressors that challenge them.
- The integration of all available molecular tools to develop smart climate crops without yield penalty and with no increase in cultivated land area is absolutely necessary.
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Springmann, M.; Clark, M.; Mason-D’Croz, D.; Wiebe, K.; Bodirsky, B.L.; Lassaletta, L.; de Vries, W.; Vermeulen, S.J.; Herrero, M.; Carlson, K.M.; et al. Options for keeping the food system within environmental limits. Nature 2018, 562, 519–525. [Google Scholar] [CrossRef] [PubMed]
- Malhi, Y.; Franklin, J.; Seddon, N.; Solan, M.; Turner, M.G.; Field, C.B.; Knowlton, N. Climate change and ecosystems: Threats, opportunities and solutions. Philos. Trans. R. Soc. B 2020, 375, 20190104. [Google Scholar] [CrossRef]
- Feeley, K.J.; Bravo-Avila, C.; Fadrique, B.; Perez, T.M.; Zuleta, D. Climate-driven changes in the composition of New World plant communities. Nat. Clim. Chang. 2020, 10, 965–970. [Google Scholar] [CrossRef]
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.M.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically rethinking agriculture for the 21st century. Science 2010, 327, 833–834. [Google Scholar] [CrossRef]
- Tilman, D.; Balzee, C.; Hill, J.; Befort, B.L. Global food demand and the sustainable intensification of agriculture. Proc. Natl. Acad. Sci. USA 2011, 108, 20260–20264. [Google Scholar] [CrossRef] [PubMed]
- FAO. The Future of Food and Agriculture-Trends and Challenges; Food and Agriculture Organization of the United Nations: Rome, Italy, 2017; pp. 1–163. Available online: https://www.fao.org/3/i6583e/i6583e.pdf (accessed on 14 May 2022).
- Anderson, R.; Bayer, P.E.; Edwards, D. Climate change and the need for agricultural adaptation. Curr. Opin. Plant Biol. 2020, 56, 197–202. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.-K. Abiotic stress signaling and responses in plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef]
- He, M.; He, C.-Q.; Ding, N.-Z. Abiotic stresses: General defenses of land plants and chances for engineering multistress tolerance. Front. Plant Sci. 2018, 9, 1771. [Google Scholar] [CrossRef]
- D’Odorico, P.; Chiarelli, D.D.; Rosa, L.; Bini, A.; Zilberman, D.; Rulli, M.C. The global value of water in agriculture. Proc. Natl. Acad. Sci. USA 2020, 117, 21985–21993. [Google Scholar] [CrossRef] [PubMed]
- Mukarram, M.; Choudhary, S.; Kurjak, D.; Petek, A.; Masroor, M.; Khan, A. Drought: Sensing, signalling, effects and tolerance in higher plants. Physiol. Plant. 2021, 172, 1291–1300. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, Y.; Zhu, J.-K. Developing naturally stress-resistant crops for a sustainable agriculture. Nat. Plants 2018, 4, 989–996. [Google Scholar] [CrossRef]
- Bartels, D.; Hussain, S.S. Resurrection plants: Physiology and molecular biology. In Plant Desiccation Tolerance. Ecological Studies 215; Lüttge, U., Beck, E., Bartels, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2011; pp. 339–364. [Google Scholar]
- Leprince, O.; Pellizzaro, A.; Berriri, S.; Buitink, J. Late seed maturation: Drying without dying. J. Exp. Bot. 2017, 68, 827–841. [Google Scholar] [CrossRef]
- Farrant, J.M.; Moore, J.P. Programming desiccation-tolerance: From plants to seeds to resurrection plants. Curr. Opin. Plant Biol. 2011, 14, 340–345. [Google Scholar] [CrossRef]
- Sachdev, S.; Ansari, S.A.; Ansari, M.I.; Fujita, M.; Hasanuzzaman, M. Abiotic stress and reactive oxygen species: Generation, signaling, and defense mechanisms. Antioxidants 2021, 10, 277. [Google Scholar] [CrossRef]
- Ilyas, M.; Nisar, M.; Khan, N.; Hazrat, A.; Khan, A.H.; Hayat, K.; Fahad, S.; Khan, A.; Ullah, A. Drought tolerance strategies in plants: A mechanistic approach. J. Plant Growth Regul. 2021, 40, 926–944. [Google Scholar] [CrossRef]
- Simkin, A.J.; López-Calcagno, P.E.; Raines, C.A. Feeding the world: Improving photosynthetic efficiency for sustainable crop production. J. Exp. Bot. 2019, 70, 1119–1140. [Google Scholar] [CrossRef]
- López-Calcagno, P.E.; Brown, K.L.; Simkin, A.J.; Fisk, S.J.; Vialet-Chabrand, S.; Lawson, T.; Raines, C.A. Stimulating photosynthetic processes increases productivity and water-use efficiency in the field. Nat. Plants 2020, 6, 1054–1063. [Google Scholar] [CrossRef]
- Condon, A.G. Drying times: Plant traits to improve crop water use efficiency and yield. J. Exp. Bot. 2020, 71, 2239–2252. [Google Scholar] [CrossRef]
- Seo, P.J.; Park, C. Auxin homeostasis during lateral root development under drought condition. Plant Signal Behav. 2009, 4, 1002–1004. [Google Scholar] [CrossRef]
- Chen, H.; Li, Z.; Xiong, L. A plant microRNA regulates the adaptation of roots to drought stress. FEBS Lett. 2012, 586, 1742–1747. [Google Scholar] [CrossRef]
- Abid, M.; Ali, S.; Qi, L.K.; Zahoor, R.; Tian, Z.; Jiang, D.; Snider, J.L.; Dai, T. Physiological and biochemical changes during drought and recovery periods at tillering and jointing stages in wheat (Triticum aestivum L.). Sci. Rep. 2018, 8, 4615. [Google Scholar] [CrossRef] [PubMed]
- Blum, A. Osmotic adjustment is a prime drought stress adaptive engine in support of plant production. Plant Cell Environ. 2017, 40, 4–10. [Google Scholar] [CrossRef] [PubMed]
- Zulfiqar, F.; Akram, N.A.; Ashraf, M. Osmoprotection in plants under abiotic stresses: New insights into a classical phenomenon. Planta 2020, 251, 3. [Google Scholar] [CrossRef] [PubMed]
- Singh, M.; Kumar, J.; Singh, S.; Singh, V.P.; Prasad, S.M. Roles of osmoprotectants in improving salinity and drought tolerance in plants: A review. Rev. Environ. Sci. Biotechnol. 2015, 14, 407–426. [Google Scholar] [CrossRef]
- Giri, J. Glycinebetaine and abiotic stress tolerance in plants. Plant Signal Behav. 2011, 6, 1746–1751. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Conde, A.; Regalado, A.; Rodrigues, D.; Costa, J.M.; Blumwald, E.; Chaves, M.M.; Gerós, H. Polyols in grape berry: Transport and metabolic adjustments as a physiological strategy for water-deficit stress tolerance in grapevine. J. Exp. Bot. 2014, 66, 889–906. [Google Scholar] [CrossRef]
- Iturriaga, G.; Suárez, R.; Nova-Franco, B. Trehalose metabolism: From osmoprotection to signaling. Int. J. Mol. Sci. 2009, 10, 3793–3810. [Google Scholar] [CrossRef]
- Avonce, N.; Mendoza-Vargas, A.; Morett, E.; Iturriaga, G. Insights on the evolution of trehalose biosynthesis. BMC Evol. Biol. 2006, 6, 109. [Google Scholar] [CrossRef]
- Fichtner, F.; Lunn, J.E. The role of Trehalose 6-phosphate (Tre6P) in plant metabolism and development. Annu. Rev. Plant Biol. 2021, 72, 737–760. [Google Scholar] [CrossRef]
- Nuccio, M.; Wu, J.; Mowers, R.; Zhou, H.-P.; Meghji, M.; Primavesi, L.F.; Paul, M.J.; Chen, X.; Gao, Y.; Emdadul Haque, E.; et al. Expression of trehalose-6-phosphate phosphatase in maize ears improves yield in well-watered and drought conditions. Nat. Biotechnol. 2015, 33, 862–869. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, A.; Chakraborty, M.; Saha, J.; Gupta, B.; Gupta, K. Polyamines: Osmoprotectants in plant abiotic stress adaptation. In Osmolytes and Plants Acclimation to Changing Environment: Emerging Omics Technologies; Iqbal, N., Nazar, R., Khan, N.A., Eds.; Springer: New Delhi, India, 2016; pp. 97–127. [Google Scholar]
- Khajuria, A.; Sharma, N.; Bhardwaj, R.; Ohri, P. Emerging role of polyamines in plant stress tolerance. Curr. Protein Pept. Sci. 2018, 19, 1114–1123. [Google Scholar] [CrossRef]
- Van den Ende, W. Multifunctional fructans and raffinose family oligosaccharides. Front. Plant Sci. 2013, 4, 247. [Google Scholar] [PubMed]
- Nagao, M.; Oku, K.; Minami, A.; Mizuno, K.; Sakurai, M.; Arakawa, K.; Fujikawa, S.; Takezawa, D. Accumulation of theanderose in association with development of freezing tolerance in the moss Physcomitrella patens. Phytochemistry 2006, 67, 702–709. [Google Scholar] [CrossRef] [PubMed]
- Omari Alzahrani, F. Metabolic engineering of osmoprotectants to elucidate the mechanism(s) of salt stress tolerance in crop plants. Planta 2021, 253, 24. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Bhuyan, M.H.M.B.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef]
- Wang, X.; Liu, H.; Yu, F.; Hu, B.; Jia, Y.; Sha, H.; Zhao, H. Differential activity of the antioxidant defense system and alterations in the accumulation of osmolyte and reactive oxygen species under drought stress and recovery in rice (Oryza sativa L.) tillering. Sci. Rep. 2019, 9, 8543. [Google Scholar] [CrossRef]
- Pommerrenig, B.; Ludewig, F.; Cvetkovic, J.; Trentmann, O.; Klemens, P.A.W.; Neuhaus, H.E. In concert: Orchestrated changes in carbohydrate homeostasis are critical for plant abiotic stress tolerance. Plant Cell Physiol. 2018, 59, 1290–1299. [Google Scholar] [CrossRef]
- Broad, R.C.; Bonneau, J.P.; Hellens, R.P.; Johnson, A.A.T. Manipulation of ascorbate biosynthetic, recycling, and regulatory pathways for improved abiotic stress tolerance in plants. Int. J. Mol. Sci. 2020, 21, 1790. [Google Scholar] [CrossRef]
- Jee, H. Size dependent classification of heat shock proteins: A mini-review. J. Exerc. Rehabil. 2016, 12, 255–259. [Google Scholar] [CrossRef]
- Jacob, P.; Hirt, H.; Bendahmane, A. The heat-shock protein/chaperone network and multiple stress resistance. Plant Biotechnol. J. 2017, 15, 405–414. [Google Scholar] [CrossRef] [PubMed]
- Hong-Bo, S.; Zong-Suo, L.; Ming-An, S. LEA proteins in higher plants: Structure, function, gene expression and regulation. Colloids Surf. B Biointerfaces 2005, 45, 131–135. [Google Scholar] [CrossRef] [PubMed]
- Hibshman, J.D.; Goldstein, B. LEA motifs promote desiccation tolerance in vivo. BMC Biol. 2021, 19, 263. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Lim, C.W.; Lim, S.; Baek, W.; Lee, S.C. The pepper late embryogenesis abundant protein CaLEA1 acts in regulating abscisic acid signaling, drought and salt stress response. Physiol. Plant. 2015, 154, 526–542. [Google Scholar] [CrossRef]
- Afzal, Z.; Howton, T.C.; Sun, Y.; Mukhtar, M.S. The roles of aquaporins in plant stress responses. J. Dev. Biol. 2016, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Benga, G. On the definition, nomenclature and classification of water channel proteins (aquaporins and relatives). Mol. Asp. Med. 2012, 33, 514–517. [Google Scholar] [CrossRef] [PubMed]
- Yepes-Molina, L.; Bárzana, G.; Carvajal, M. Controversial regulation of gene expression and protein transduction of aquaporins under drought and salinity stress. Plants 2020, 9, 1662. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, A.; Swarnam, P.; Subramani, T.; Meena, B.; Kaledhonkar, M.J. Water demand and salinity. In Desalination-Challenges and Opportunities; Farahani, M.H.D.A., Vatanpour, V., Taheri, A., Eds.; IntechOpen Limited: London, UK, 2020; pp. 1–11. [Google Scholar]
- Arif, Y.; Singh, P.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salinity induced physiological and biochemical changes in plants: An omic approach towards salt stress tolerance. Plant Physiol. Biochem. 2020, 156, 64–77. [Google Scholar] [CrossRef]
- van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef]
- Joshi, S.; Nath, J.; Singh, A.K.; Pareek, A.; Joshi, R. Ion transporters and their regulatory signal transduction mechanisms for salinity tolerance in plants. Physiol. Plant. 2022, 174, e13702. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- Skalak, J.; Nicolas, K.L.; Vankova, R.; Hejatko, J. Signal integration in plant abiotic stress responses via multistep phosphorelay signaling. Front. Plant Sci. 2021, 12, 644823. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Verma, V.; Ravindran, P.; Kumar, P.P. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016, 16, 86. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.-A.; Li, M.-Z.; Wang, S.-M.; Yin, H.-J. Revisiting the role of plant transcription factors in the battle against abiotic stress. Int. J. Mol. Sci. 2018, 19, 1634. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Xue, L.; Batelli, G.; Lee, S.; Hou, Y.J.; Van Oosten, M.J.; Zhang, H.; Tao, W.A.; Zhu, J.-K. Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. USA 2013, 110, 11205–11210. [Google Scholar] [CrossRef]
- Ali, M.-S.; Baek, K.-H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Li, W.; Nguyen, K.H.; Fujita, M.; Tran, L.P. Strigolactones in plant adaptation to abiotic stresses: An emerging avenue of plant research. Plant Cell Environ. 2018, 41, 2227–2243. [Google Scholar] [CrossRef] [PubMed]
- Rhaman, M.S.; Imran, S.; Karim, M.M.; Chakrobortty, J.; Mahamud, M.A.; Sarker, P.; Tahjib-Ul-Arif, M.; Robin, A.H.K.; Ye, W.; Murata, Y.; et al. 5-aminolevulinic acid-mediated plant adaptive responses to abiotic stress. Plant Cell Rep. 2021, 40, 1451–1469. [Google Scholar] [CrossRef]
- Tahjib-Ul-Arif, M.; Zahan, M.I.; Karim, M.M.; Imran, S.; Hunter, C.T.; Islam, M.S.; Mia, M.A.; Hannan, M.A.; Rhaman, M.S.; Hossain, M.A.; et al. Citric Acid-Mediated Abiotic Stress Tolerance in Plants. Int. J. Mol. Sci. 2021, 22, 7235. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Joshi, S.; Khare, T.; Patil, S.; Shang, J.; Kumar, V. Nitric oxide, crosstalk with stress regulators and plant abiotic stress tolerance. Plant Cell Rep. 2021, 40, 1395–1414. [Google Scholar] [CrossRef] [PubMed]
- Arnao, M.B.; Hernández-Ruiz, J. Melatonin: A new plant hormone and/or a plant master regulator? Trends Plant Sci. 2019, 24, 38–48. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, F.; Hanada, K.; Kondo, T.; Shinozaki, K. Hormone-like peptides and small coding genes in plant stress signaling and development. Curr. Opin. Plant Biol. 2019, 51, 88–95. [Google Scholar] [CrossRef]
- Liu, Z.; Hou, S.; Rodrigues, O.; Wang, P.; Luo, D.; Munemasa, S.; Lei, J.; Liu, J.; Ortiz-Morea, F.A.; Wang, X.; et al. Phytocytokine signalling reopens stomata in plant immunity and water loss. Nature 2022, 605, 332–339. [Google Scholar] [CrossRef]
- Anwar, A.; Kim, J.-K. Transgenic breeding approaches for improving abiotic stress tolerance: Recent progress and future perspective. Int. J. Mol. Sci. 2020, 21, 2695. [Google Scholar] [CrossRef]
- Chang, Y.-N.; Zhu, C.; Jiang, J.; Zhang, H.; Jian-Kang Zhu, J.-K.; Duan, C.-G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Jha, U.C.; Nayyar, H.; Jha, R.; Khurshid, M.; Zhou, M.; Mantri, N.; Siddique, K.H.M. Long non-coding RNAs: Emerging players regulating plant abiotic stress response and adaptation. BMC Plant Biol. 2020, 20, 466. [Google Scholar] [CrossRef]
- Kim, J.-M.; To, T.K.; Matsui, A.; Tanoi, K.; Kobayashi, N.I.; Matsuda, F.; Habu, Y.; Ogawa, D.; Sakamoto, T.; Matsunaga, S.; et al. Acetate-mediated novel survival strategy against drought in plants. Nat. Plants 2017, 3, 17097. [Google Scholar] [CrossRef]
- Suárez, R.; Wong, A.; Ramírez, M.; Barraza, A.; Orozco, M.C.; Cevallos, M.A.; Lara, M.; Hernández, G.; Iturriaga, G. Improvement of drought tolerance and grain yield in common bean by overexpressing trehalose-6-phosphate synthase in rhizobia. Mol. Plant-Microbe Interact. 2008, 21, 958–966. [Google Scholar] [CrossRef]
- Ilangumaran, G.; Smith, D.L. Plant Growth Promoting Rhizobacteria in Amelioration of Salinity Stress: A Systems Biology Perspective. Front. Plant Sci. 2017, 8, 1768. [Google Scholar] [CrossRef] [PubMed]
- Boutasknit, A.; Baslam, M.; Ait-El-Mokhtar, M.; Anli, M.; Ben-Laouane, R.; Ait-Rahou, Y.; Mitsui, T.; Douira, A.; El Modafar, C.; Wahbi, S.; et al. Assemblage of indigenous arbuscular mycorrhizal fungi and green waste compost enhance drought stress tolerance in carob (Ceratonia siliqua L.) trees. Sci. Rep. 2021, 11, 22835. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Verma, J.P. Does Plant—Microbe interaction confer stress tolerance in plants: A review? Microbiol. Res. 2018, 207, 41–52. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Rajkumar, M.; Oliveira, R.S.; Zhang, C.; Freitas, H. Potential of plant beneficial bacteria and arbuscular mycorrhizal fungi in phytoremediation of metal-contaminated saline soils. J. Hazard. Mater. 2019, 379, 120813. [Google Scholar] [CrossRef] [PubMed]
- Etesami, H.; Maheshwari, D.K. Use of Plant Growth Promoting Rhizobacteria (PGPRs) with multiple plant growth promoting traits in stress agriculture: Action mechanisms and future prospects. Ecotoxicol. Environ. Saf. 2018, 156, 225–246. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Chauhan, P.K.; Upadhyay, S.K.; Singh, R.K.; Dwivedi, P.; Wang, J.; Jain, D.; Jiang, M. Mechanistic insights and potential use of siderophores producing microbes in rhizosphere for mitigation of stress in plants grown in degraded land. Front. Microbiol. 2022, 13, 898979. [Google Scholar] [CrossRef]
- Bhagat, N.; Raghav, M.; Dubey, S.; Bedi, N. Bacterial exopolysaccharides: Insight into their role in plant abiotic stress tolerance. J. Microbiol. Biotechnol. 2021, 31, 1045–1059. [Google Scholar] [CrossRef]
- Hanaka, A.; Ozimek, E.; Reszczyńska, E.; Jaroszuk-Ściseł, J.; Stolarz, M. Plant tolerance to drought dtress in the presence of supporting bacteria and fungi: An efficient strategy in horticulture. Horticulturae 2021, 7, 390. [Google Scholar] [CrossRef]
- Tang, H.; Hassan, M.U.; Feng, L.; Nawaz, M.; Shah, A.N.; Qari, S.H.; Liu, Y.; Miao, J. The Critical role of arbuscular mycorrhizal fungi to improve drought tolerance and nitrogen use efficiency in crops. Front. Plant Sci. 2022, 13, 919166. [Google Scholar] [CrossRef]
- Guler, N.S.; Pehlivan, N.; Karaoglu, S.A.; Guzel, S.; Bozdeveci, A. Trichoderma atroviride ID20G inoculation ameliorates drought stress-induced damages by improving antioxidant defence in maize seedlings. Acta Physiol. Plant. 2016, 38, 132. [Google Scholar] [CrossRef]
- Singh, D.; Singh, V.; Gupta, V.; Shukla, R.; Prabha, R.; Sarma, B.; Patel, J. Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Sci. Rep. 2020, 10, 4818. [Google Scholar] [CrossRef] [PubMed]
- Cañada-Coyote, E.; Ramírez-Pimentel, J.G.; Aguirre-Mancilla, C.L.; Raya-Pérez, J.C.; Acosta-García, G.; Iturriaga, G. Trichoderma harzianum mutants enhance antagonism against phytopathogenic fungi, phosphorus assimilation and drought tolerance in Jalapeño pepper plants. Chil. J. Agric. Res. 2021, 81, 270–280. [Google Scholar] [CrossRef]
- Lee, J.; Chin, J.H.; Ahn, S.N.; Koh, H.-J. Brief history and perspectives on plant breeding. In Current Technologies in Plant Molecular Breeding: A Guide Book of Plant Molecular Breeding for Researchers; Koh, H.-J., Kwon, S.-Y., Thomson, M., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 1–14. ISBN 978-94-017-9996-6. [Google Scholar]
- Liu, J.; Fernie, A.R.; Yan, J. Crop breeding—From experience-based selection to precision design. J. Plant Physiol. 2021, 256, 153313. [Google Scholar] [CrossRef] [PubMed]
- Ahmar, S.; Gill, R.A.; Jung, K.-H.; Faheem, A.; Qasim, M.U.; Mubeen, M.; Zhou, W. Conventional and molecular techniques from simple breeding to speed breeding in crop plants: Recent advances and future outlook. Int. J. Mol. Sci. 2020, 21, 2590. [Google Scholar] [CrossRef]
- Sattar, M.N.; Iqbal, Z.; Al-Khayri, J.M.; Jain, S.M. Induced genetic variations in fruit trees using new breeding tools: Food security and climate resilience. Plants 2021, 10, 1347. [Google Scholar] [CrossRef]
- Wieczorek, A.; Wright, M. History of agricultural biotechnology: How crop development has evolved. Nat. Educ. Knowl. 2012, 3, 9. [Google Scholar]
- Colasuonno, P.; Marcotuli, I.; Gadaleta, A.; Soriano, J.M. From genetic maps to QTL Cloning: An overview for durum wheat. Plants 2021, 10, 315. [Google Scholar] [CrossRef]
- Yang, H.-B.; Kang, W.-H.; Nahm, S.-H.; Kang, B.-C.; Yang, H.-B.; Kang, W.-H.; Kang, B.-C.; Nahm, S.-H. Methods for developing molecular markers. In Current Technologies in Plant Molecular Breeding; Springer: Berlin/Heidelberg, Germany, 2015. [Google Scholar]
- Oladosu, Y.; Rafii, M.Y.; Samuel, C.; Fatai, A.; Magaji, U.; Kareem, I.; Kamarudin, Z.S.; Muhammad, I.; Kolapo, K. Drought resistance in rice from conventional to molecular breeding: A Review. Int. J. Mol. Sci. 2019, 20, 3519. [Google Scholar] [CrossRef]
- Yoon, J.B.; Kwon, S.-W.; Ham, T.-H.; Kim, S.; Thomson, M.; Hechanova, S.L.; Jena, K.K.; Park, Y. Marker-Assisted Breeding. In Current Technologies in Plant Molecular Breeding: A Guide Book of Plant Molecular Breeding for Researchers; Koh, H.-J., Kwon, S.-Y., Thomson, M., Eds.; Springer: Dordrecht, The Netherlands, 2015; pp. 95–144. ISBN 978-94-017-9996-6. [Google Scholar]
- Tshikunde, N.M.; Mashilo, J.; Shimelis, H.; Odindo, A. Agronomic and physiological traits, and associated Quantitative Trait Loci (QTL) affecting yield response in wheat (Triticum aestivum L.): A Review. Front. Plant Sci. 2019, 10, 1428. [Google Scholar] [CrossRef]
- Dheer, P.; Rautela, I.; Sharma, V.; Dhiman, M.; Sharma, A.; Sharma, N.; Sharma, M.D. Evolution in crop improvement approaches and future prospects of molecular markers to CRISPR/Cas9 system. Gene 2020, 753, 144795. [Google Scholar] [CrossRef]
- Singh, R.K.; Kota, S.; Flowers, T.J. Salt tolerance in rice: Seedling and reproductive stage QTL mapping come of age. Theor. Appl. Genet. 2021, 1, 3. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Y.; Du, J.; Guo, X.; Wen, W.; Gu, S.; Wang, J.; Fan, J. Crop Phenomics: Current status and perspectives. Front. Plant Sci. 2019, 10, 714. [Google Scholar] [CrossRef] [PubMed]
- Nordey, T.; Schwarz, D.; Kenyon, L.; Manickam, R.; Huat, J. Tapping the potential of grafting to improve the performance of vegetable cropping systems in sub-Saharan Africa. A review. Agron. Sustain. Dev. 2020, 40, 23. [Google Scholar] [CrossRef]
- Singh, H.; Sethi, S.; Kaushik, P.; Fulford, A. Grafting vegetables for mitigating environmental stresses under climate change: A review. J. Water Clim. Chang. 2020, 11, 1784–1797. [Google Scholar] [CrossRef]
- Spanò, R.; Ferrara, M.; Gallitelli, D.; Mascia, T. The role of grafting in the resistance of tomato to viruses. Plants 2020, 9, 1042. [Google Scholar] [CrossRef]
- Tsaballa, A.; Xanthopoulou, A.; Madesis, P.; Tsaftaris, A.; Nianiou-Obeidat, I. Vegetable grafting from a molecular point of view: The involvement of epigenetics in rootstock-scion interactions. Front. Plant Sci. 2021, 11, 621999. [Google Scholar] [CrossRef]
- Mozafarian Meimandi, M.; Kappel, N. Grafting plants to improve abiotic stress tolerance. In Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives II: Mechanisms of Adaptation and Stress Amelioration; Hasanuzzaman, M., Ed.; Springer: Singapore, 2020; pp. 477–490. ISBN 978-981-15-2172-0. [Google Scholar]
- Aloni, R. Vascular regeneration and grafting. In Vascular Differentiation and Plant Hormones; Aloni, R., Ed.; Springer International Publishing: Cham, Switzerland, 2021; pp. 185–198. [Google Scholar]
- Baron, D.; Esteves Amaro, A.C.; Pina, A.; Ferreira, G. An overview of grafting re-establishment in woody fruit species. Sci. Hortic. 2019, 243, 84–91. [Google Scholar] [CrossRef]
- Pérez-Luna, A.; Wehenkel, C.; Prieto-Ruíz, J.Á.; López-Upton, J.; Solís-González, S.; Chávez-Simental, J.A.; Hernández-Díaz, J.C. Grafting in conifers: A review. Pak. J. Bot. 2020, 52, 1369–1378. [Google Scholar] [CrossRef]
- Singh, H.; Kumar, P.; Chaudhari, S.; Edelstein, M. Tomato Grafting: A global perspective. HortScience 2017, 52, 1328–1336. [Google Scholar] [CrossRef]
- Gaion, L.A.; Trevisan Braz, L.; Carvalho, R.F. Grafting in vegetable crops: A great technique for agriculture. Int. J. Veg. Sci. 2018, 24, 85–102. [Google Scholar] [CrossRef]
- Chilukamarri, L.; Ashrafzadeh, S.; Leung, D.W.M. In-vitro grafting–Current applications and future prospects. Sci. Hortic. 2021, 280, 109899. [Google Scholar] [CrossRef]
- Kumar, A.; Rathore, J.P.; Iqbal, U.; Sharma, A.; Nagar, P.K.; Mir, M.M. Rootstocks of stone fruit crops. In Production Technology of Stone Fruits; Mir, M.M., Iqbal, U., Mir, S.A., Eds.; Springer: Singapore, 2021; pp. 131–169. ISBN 978-981-15-8920-1. [Google Scholar]
- Limera, C.; Sabbadini, S.; Sweet, J.B.; Mezzetti, B. New biotechnological tools for the genetic improvement of major woody fruit species. Front. Plant Sci. 2017, 8, 1418. [Google Scholar] [CrossRef] [PubMed]
- Sidorova, T.; Miroshnichenko, D.; Kirov, I.; Pushin, A.; Dolgov, S. Effect of grafting on viral resistance of non-transgenic plum scion combined with transgenic PPV-Resistant rootstock. Front. Plant Sci. 2021, 12, 621954. [Google Scholar] [CrossRef] [PubMed]
- Bartusch, K.; Melnyk, C.W. Insights Into Plant Surgery: An overview of the multiple grafting techniques for Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 613442. [Google Scholar] [CrossRef] [PubMed]
- Vidoy-Mercado, I.; Narváez, I.; Palomo-Ríos, E.; Litz, R.E.; Barceló-Muñoz, A.; Pliego-Alfaro, F. Reinvigoration/Rejuvenation induced through micrografting of tree species: Signaling through graft union. Plants 2021, 10, 1197. [Google Scholar] [CrossRef] [PubMed]
- Tsutsui, H.; Notaguchi, M. The use of grafting to study systemic signaling in plants. Plant Cell Physiol. 2017, 58, 1291–1301. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jiang, L.; Wu, R. Plant grafting: How genetic exchange promotes vascular reconnection. N. Phytol. 2017, 214, 56–65. [Google Scholar] [CrossRef]
- Thomas, H.R.; Frank, M.H. Connecting the pieces: Uncovering the molecular basis for long-distance communication through plant grafting. N. Phytol. 2019, 223, 582–589. [Google Scholar] [CrossRef]
- Rasool, A.; Mansoor, S.; Bhat, K.M.; Hassan, G.I.; Baba, T.R.; Alyemeni, M.N.; Alsahli, A.A.; El-Serehy, H.A.; Paray, B.A.; Ahmad, P. Mechanisms underlying graft union formation and rootstock scion interaction in horticultural plants. Front. Plant Sci. 2020, 11, 590847. [Google Scholar] [CrossRef]
- Kapazoglou, A.; Tani, E.; Avramidou, E.V.; Abraham, E.M.; Gerakari, M.; Megariti, S.; Doupis, G.; Doulis, A.G. Epigenetic changes and transcriptional reprogramming upon woody plant grafting for crop sustainability in a changing environment. Front. Plant Sci. 2021, 11, 613004. [Google Scholar] [CrossRef]
- Oladosu, Y.; Rafii, M.Y.; Abdullah, N.; Hussin, G.; Ramli, A.; Rahim, H.A.; Miah, G.; Usman, M. Principle and application of plant mutagenesis in crop improvement: A review. Biotechnol. Biotechnol. Equip. 2016, 30, 1–16. [Google Scholar] [CrossRef]
- Beyaz, R.; Yildiz, M. The Use of Gamma irradiation in plant mutation breeding. In Plant Engineering; IntechOpen: London, UK, 2017. [Google Scholar]
- Chaudhary, J.; Alisha, A.; Bhatt, V.; Chandanshive, S.; Kumar, N.; Mir, Z.; Kumar, A.; Yadav, S.K.; Shivaraj, S.M.; Sonah, H.; et al. Mutation Breeding in Tomato: Advances, Applicability and Challenges. Plants 2019, 8, 128. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, J.; Deshmukh, R.; Sonah, H. Mutagenesis approaches and their role in crop improvement. Plants 2019, 8, 467. [Google Scholar] [CrossRef]
- Jo, Y.D.; Kim, J.-B. Frequency and spectrum of radiation-induced mutations revealed by whole-genome sequencing analyses of plants. Quantum Beam Sci. 2019, 3, 7. [Google Scholar] [CrossRef]
- Hernández-Muñoz, S.; Pedraza-Santos, M.E.; López, P.A.; Gómez-Sanabria, J.M.; Morales-García, J.L. Mutagenesis in the improvement of ornamental plants. Rev. Chapingo Ser. Hortic. 2019, 25, 151–167. [Google Scholar] [CrossRef]
- Anne, S.; Lim, J.H. Mutation breeding using gamma irradiation in the development of ornamental plants: A review. Flower Res. J. 2020, 28, 102–115. [Google Scholar] [CrossRef]
- Mba, C.; Afza, R.; Bado, S.; Jain, S.M.; Anthony, P. Induced mutagenesis in plants using physical and chemical agents. In Plant cell culture: Essential methods; Wiley-Blackwell: Hoboken, NJ, USA, 2010; ISBN 9780470686485. [Google Scholar]
- Ramkumar, M.K.; Senthil Kumar, S.; Gaikwad, K.; Pandey, R.; Chinnusamy, V.; Singh, N.K.; Singh, A.K.; Mohapatra, T.; Sevanthi, A.M. A Novel Stay-Green Mutant of rice with delayed leaf senescence and better harvest index confers drought tolerance. Plants 2019, 8, 375. [Google Scholar] [CrossRef] [PubMed]
- Kazama, Y.; Hirano, T.; Saito, H.; Liu, Y.; Ohbu, S.; Hayashi, Y.; Abe, T. Characterization of highly efficient heavy-ion mutagenesis in Arabidopsis thaliana. BMC Plant Biol. 2011, 11, 161. [Google Scholar] [CrossRef] [PubMed]
- Shirasawa, K.; Hirakawa, H.; Nunome, T.; Tabata, S.; Isobe, S. Genome-wide survey of artificial mutations induced by ethyl methanesulfonate and gamma rays in tomato. Plant Biotechnol. J. 2016, 14, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Wanga, M.A.; Shimelis, H.; Horn, L.N.; Sarsu, F. The effect of single and combined use of gamma radiation and ethylmethane sulfonate on early growth parameters in sorghum. Plants 2020, 9, 827. [Google Scholar] [CrossRef] [PubMed]
- Shu, Q.Y.; Forster, B.P.; Nakagawa, H. Plant Mutation Breeding and Biotechnology, 1st ed.; FAO: Rome, Italy, 2011; ISBN 978-92-5-105000-0. [Google Scholar]
- Mullins, E.; Bresson, J.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Moreno, F.J.; Naegeli, H.; et al. In vivo and in vitro random mutagenesis techniques in plants. EFSA J. 2021, 19, e06611. [Google Scholar]
- Kumar, K.; Gambhir, G.; Dass, A.; Tripathi, A.K.; Singh, A.; Jha, A.K.; Yadava, P.; Choudhary, M.; Rakshit, S. Genetically modified crops: Current status and future prospects. Planta 2020, 251, 91. [Google Scholar] [CrossRef] [PubMed]
- Ramkumar, T.R.; Lenka, S.K.; Arya, S.S.; Bansal, K.C.; Arya, S.S.; Bansal, K.C. Arya, S.S.; Bansal, K.C.; Arya, S.S.; Bansal, K.C. A short history and perspectives on plant genetic transformation. In Biolistic DNA Delivery in Plants: Methods and Protocols; Rustgi, S., Luo, H., Eds.; Springer Nature: Berlin/Heidelberg, Germany, 2020; Arya, S.S.; Volume 2124, pp. 39–68. ISBN 9781071603567. [Google Scholar]
- Gosal, S.S.; Wani, S.H. Plant genetic transformation and transgenic crops: Methods and applications. In Biotechnologies of Crop Improvement, Volume 2: Transgenic Approaches; Kumar, S., Barone, P., Smith, M., Eds.; Springer Nature: Berlin/Heidelberg, Germany, 2018; Volume 2, pp. 1–23. ISBN 9783319906508. [Google Scholar]
- Soman, J.; Hema, J.; Subramanian, S. Plant Tissue Culture and DNA Delivery Methods. In Advances in Plant Transgenics: Methods and Applications; Springer: Singapore, 2019; ISBN 978-981-13-9623-6. [Google Scholar]
- Kausch, A.P.; Wang, K.; Kaeppler, H.F.; Gordon-Kamm, W. Maize transformation: History, progress, and perspectives. Mol. Breed. 2021, 41, 38. [Google Scholar] [CrossRef]
- Reed, K.M.; Bargmann, B.O.R. Protoplast regeneration and its use in new plant breeding technologies. Front. Genome Ed. 2021, 3, 734951. [Google Scholar] [CrossRef]
- Krenek, P.; Samajova, O.; Luptovciak, I.; Doskocilova, A.; Komis, G.; Samaj, J. Transient plant transformation mediated by Agrobacterium tumefaciens: Principles, methods and applications. Biotechnol. Adv. 2015, 33, 1024–1042. [Google Scholar] [CrossRef]
- Van Eck, J. Genome editing and plant transformation of solanaceous food crops. Curr. Opin. Biotechnol. 2018, 49, 35–41. [Google Scholar] [CrossRef]
- Guo, M.; Ye, J.; Gao, D.; Xu, N.; Yang, J. Agrobacterium-mediated horizontal gene transfer: Mechanism, biotechnological application, potential risk and forestalling strategy. Biotechnol. Adv. 2019, 37, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Que, Q.; Chilton, M.M.; Elumalai, S.; Zhong, H. Repurposing macromolecule delivery tools for plant engineering. In Transgenic Plants: Methods and Protocols; Kumar, S., Barone, P., Smith, M., Eds.; Springer Nature: Berlin/Heidelberg, Germany, 2019; Volume 1864, pp. 3–18. ISBN 9781493987788. [Google Scholar]
- Basso, M.F.; Arraes, F.B.M.; Grossi-de-Sa, M.; Moreira, V.J.V.; Alves-Ferreira, M.; Grossi-de-Sa, M.F. Insights into genetic and molecular elements for transgenic crop development. Front. Plant Sci. 2020, 11, 509. [Google Scholar] [CrossRef]
- Lacroix, B.; Citovsky, V. Biolistic approach for transient gene expression studies in plants. In Biolistic DNA Delivery in Plants: Methods and Protocols; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 125–139. [Google Scholar]
- Anjanappa, R.B.; Gruissem, W. Current progress and challenges in crop genetic transformation. J. Plant Physiol. 2021, 261, 153411. [Google Scholar] [CrossRef] [PubMed]
- Klein, T.M.; Sanford, J.C.; Wolf, E.D.; Wu, R. High-velocity microprojectiles for delivering nucleic acids into living cells. Nature 1987, 327, 70–73. [Google Scholar] [CrossRef]
- Rustgi, S.; Luo, H. Biolistic DNA Delivery in Plants. In Methods in Molecular Biology, 1st ed.; Rustgi, S., Luo, H., Eds.; Springer: New York, NY, USA, 2020; Volume 2124, ISBN 978-1-0716-0355-0. [Google Scholar]
- Dong, O.X.; Ronald, P.C. Targeted DNA insertion in plants. Proc. Natl. Acad. Sci. USA 2021, 118, e2004834117. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, P.; Armarego-Marriott, T.; Bock, R. Plastid transformation and its application in metabolic engineering. Curr. Opin. Biotechnol. 2018, 49, 10–15. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Sharma, L.; Bohra, N.; Anandhan, S.; Ruiz-May, E.; Quiroz-Figueroa, F.R. Recent developments in generation of marker-free transgenic plants. In Advances in Plant Transgenics: Methods and Applications; Springer: Singapore, 2019; pp. 127–142. ISBN 9789811396243. [Google Scholar]
- Yu, Y.; Yu, P.-C.; Chang, W.-J.; Yu, K.; Lin, C.-S. Plastid Transformation: How does it work? Can it Be Applied to Crops? What Can it Offer? Int. J. Mol. Sci. 2020, 21, 4854. [Google Scholar] [CrossRef] [PubMed]
- Bock, R. Engineering chloroplasts for high-level constitutive or inducible transgene expression. Methods Mol. Biol. 2021, 2317, 77–94. [Google Scholar]
- Ozyigit, I.I.; Yucebilgili Kurtoglu, K. Particle bombardment technology and its applications in plants. Mol. Biol. Rep. 2020, 47, 9831–9847. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Nannas, N.J.; Fu, F.; Shi, J.; Aspinwall, B.; Parrott, W.A.; Dawe, R.K. Genome-Scale Sequence Disruption Following Biolistic Transformation in Rice and Maize. Plant Cell 2019, 31, 368–383. [Google Scholar] [CrossRef]
- Jacob, S.S.; Veluthambi, K. Generation of selection marker-free transgenic plants by cotransformation of a cointegrate vector T-DNA and a binary vector T-DNA in one Agrobacterium tumefaciens strain. Plant Sci. 2002, 163, 801–806. [Google Scholar] [CrossRef]
- Permingeat, H.R.; Alvarez, M.L.; Cervigni, G.D.L.; Ravizzini, R.A.; Vallejos, R.H. Stable wheat transformation obtained without selectable markers. Plant Mol. Biol. 2003, 52, 415–419. [Google Scholar] [CrossRef]
- Kyndt, T.; Quispe, D.; Zhai, H.; Jarret, R.; Ghislain, M.; Liu, Q.; Gheysen, G.; Kreuze, J.F. The genome of cultivated sweet potato contains Agrobacterium T-DNAs with expressed genes: An example of a naturally transgenic food crop. Proc. Natl. Acad. Sci. USA 2015, 112, 5844–5849. [Google Scholar] [CrossRef]
- Matveeva, T.V.; Otten, L. Widespread occurrence of natural genetic transformation of plants by Agrobacterium. Plant Mol. Biol. 2019, 101, 415–437. [Google Scholar] [CrossRef]
- Nogué, F.; Mara, K.; Collonnier, C.; Casacuberta, J.M. Genome engineering and plant breeding: Impact on trait discovery and development. Plant Cell Rep. 2016, 35, 1475–1486. [Google Scholar] [CrossRef] [PubMed]
- Naegeli, H.; Birch, A.N.; Casacuberta, J.; De Schrijver, A.; Gralak, M.A.; Guerche, P.; Jones, H.; Manachini, B.; Messéan, A.; Nielsen, E.E.; et al. Guidance on allergenicity assessment of genetically modified plants. EFSA J. 2017, 15, e04862. [Google Scholar] [PubMed]
- Roberts, R.J. The Nobel Laureates’ Campaign Supporting GMOs. J. Innov. Knowl. 2018, 3, 61–65. [Google Scholar] [CrossRef]
- Naegeli, H.; Bresson, J.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Moreno, F.J.; Nogue, F.; et al. Evaluation of existing guidelines for their adequacy for the molecular characterisation and environmental risk assessment of genetically modified plants obtained through synthetic biology. EFSA J. 2021, 19, e06301. [Google Scholar] [PubMed]
- Naegeli, H.; Bresson, J.; Dalmay, T.; Dewhurst, I.C.; Epstein, M.M.; Firbank, L.G.; Guerche, P.; Hejatko, J.; Moreno, F.J.; Mullins, E.; et al. Statement on in vitro protein digestibility tests in allergenicity and protein safety assessment of genetically modified plants. EFSA J. 2021, 19, e06350. [Google Scholar] [PubMed]
- Holme, I.B.; Wendt, T.; Holm, P.B. Intragenesis and Cisgenesis as Alternatives to Transgenic Crop Development. Plant Biotechnol. J. 2013, 11, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Gibson, D.G.; Young, L.; Chuang, R.-Y.; Venter, J.C.; Hutchison, C.A.; Smith, H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 2009, 6, 343–345. [Google Scholar] [CrossRef] [PubMed]
- Engler, C.; Kandzia, R.; Marillonnet, S. A One Pot, One Step, Precision Cloning Method with High Throughput Capability. PLoS ONE 2008, 3, e3647. [Google Scholar] [CrossRef] [PubMed]
- Sarrion-Perdigones, A.; Falconi, E.E.; Zandalinas, S.I.; Juárez, P.; Fernández-del-Carmen, A.; Granell, A.; Orzaez, D. GoldenBraid: An Iterative Cloning System for Standardized Assembly of Reusable Genetic Modules. PLoS ONE 2011, 6, e21622. [Google Scholar]
- Sarrion-Perdigones, A.; Vazquez-Vilar, M.; Palaci, J.; Castelijns, B.; Forment, J.; Ziarsolo, P.; Blanca, J.; Granell, A.; Orzaez, D. GoldenBraid 2.0: A Comprehensive DNA Assembly Framework for Plant Synthetic Biology. Plant Physiol. 2013, 162, 1618–1631. [Google Scholar] [CrossRef] [PubMed]
- Gelvin, S.B. Agrobacterium-Mediated plant transformation: The Biology behind the “Gene-Jockeying” tool. Microbiol. Mol. Biol. Rev. 2003, 67, 16–37. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.-H.; Yu, M.; Lai, E.-M. Agrobacterium-Mediated plant transformation: Biology and applications. Arab. Book 2017, 15, e0186. [Google Scholar] [CrossRef] [PubMed]
- Bevan, M.W.; Flavell, R.B.; Chilton, M.-D. A chimaeric antibiotic resistance gene as a selectable marker for plant cell transformation. Nature 1983, 304, 184–187. [Google Scholar] [CrossRef]
- Fraley, R.T.; Rogers, S.G.; Horsch, R.B.; Sanders, P.R.; Flick, J.S.; Adams, S.P.; Bittner, M.L.; Brand, L.A.; Fink, C.L.; Fry, J.S.; et al. Expression of bacterial genes in plant cells. Proc. Natl. Acad. Sci. USA 1983, 80, 4803–4807. [Google Scholar] [CrossRef] [PubMed]
- Herrera-Estrella, L.; Depicker, A.; Van Montagu, M.; Schell, J. Expression of chimaeric genes transferred into plant cells using a Ti-plasmid-derived vector. Nature 1983, 303, 209–213. [Google Scholar] [CrossRef]
- Hoekema, A.; Hirsch, P.R.; Hooykaas, P.J.J.; Schilperoort, R.A. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 1983, 303, 179–180. [Google Scholar] [CrossRef]
- Bevan, M. Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 1984, 12, 8711–8721. [Google Scholar] [CrossRef]
- Wang, X.; Chang, S.; Lu, J.; Fray, R.; Grierson, D.; Han, Y. Plant genetic engineering and genetically modified crop breeding: History and current status. Front. Agric. Sci. Eng. 2017, 4, 5–27. [Google Scholar] [CrossRef]
- Anami, S.; Njuguna, E.; Coussens, G.; Aesaert, S.; Lijsebettens, M. Van Higher plant transformation: Principles and molecular tools. Int. J. Dev. Biol. 2013, 57, 483–494. [Google Scholar] [CrossRef]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple Genotyping-by-Sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Yu, J.-K. Advanced Breeding Technologies for Accelerating Genetic Gain. Plant Breed. Biotechnol. 2020, 8, 203–210. [Google Scholar] [CrossRef]
- He, J.; Zhao, X.; Laroche, A.; Lu, Z.-X.; Liu, H.; Li, Z. Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front. Plant Sci. 2014, 5, 484. [Google Scholar] [CrossRef] [PubMed]
- Peterson, G.; Dong, Y.; Horbach, C.; Fu, Y.-B. Genotyping-By-Sequencing for plant genetic diversity analysis: A lab guide for SNP Genotyping. Diversity 2014, 6, 665–680. [Google Scholar] [CrossRef]
- Sonah, H.; O’Donoughue, L.; Cober, E.; Rajcan, I.; Belzile, F. Identification of loci governing eight agronomic traits using a GBS-GWAS approach and validation by QTL mapping in soya bean. Plant Biotechnol. J. 2015, 13, 211–221. [Google Scholar] [CrossRef] [PubMed]
- Andrews, K.R.; Good, J.M.; Miller, M.R.; Luikart, G.; Hohenlohe, P.A. Harnessing the power of RADseq for ecological and evolutionary genomics. Nat. Rev. Genet. 2016, 17, 81–92. [Google Scholar] [CrossRef]
- Wu, X.; Blair, M.W. Diversity in grain amaranths and relatives distinguished by Genotyping by Sequencing (GBS). Front. Plant Sci. 2017, 8, 1960. [Google Scholar] [CrossRef]
- Carbonell, P.; Alonso, A.; Grau, A.; Salinas, J.; García-Martínez, S.; Ruiz, J. Twenty years of tomato breeding at EPSO-UMH: Transfer resistance from wild types to local landraces—From the first molecular markers to genotyping by sequencing (GBS). Diversity 2018, 10, 12. [Google Scholar] [CrossRef]
- Challa, S.; Neelapu, N.R.R. Genome-Wide Association Studies (GWAS) for abiotic stress tolerance in plants. In Biochemical, Physiological and Molecular Avenues for Combating Abiotic Stress Tolerance in Plants; Wani, S.H., Ed.; Elsevier: London, UK, 2018; pp. 135–150. ISBN 9780128130667. [Google Scholar]
- Edet, O.U.; Gorafi, Y.S.A.; Nasuda, S.; Tsujimoto, H. DArTseq-based analysis of genomic relationships among species of tribe Triticeae. Sci. Rep. 2018, 8, 16397. [Google Scholar] [CrossRef]
- Nwosisi, S.; Dhakal, K.; Nandwani, D.; Raji, J.I.; Krishnan, S.; Beovides-García, Y. Genetic diversity in vegetable and fruit crops. In Genetic Diversity in Horticultural Plants; Nandwani, D., Ed.; Springer International Publishing: Cham, Switzerland, 2019; pp. 87–125. ISBN 9783319964546. [Google Scholar]
- Allan, V.; Vetriventhan, M.; Senthil, R.; Geetha, S.; Deshpande, S.; Rathore, A.; Kumar, V.; Singh, P.; Reddymalla, S.; Azevedo, V.C.R. Genome-Wide DArTSeq genotyping and phenotypic based assessment of within and among accessions diversity and effective sample size in the diverse sorghum, pearl millet, and pigeonpea landraces. Front. Plant Sci. 2020, 11, 587426. [Google Scholar] [CrossRef]
- Gahlaut, V.; Zinta, G.; Jaiswal, V.; Kumar, S. Quantitative Epigenetics: A new avenue for crop improvement. Epigenomes 2020, 4, 25. [Google Scholar] [CrossRef]
- Kulkarni, K.P.; Vorsa, N.; Natarajan, P.; Elavarthi, S.; Iorizzo, M.; Reddy, U.K.; Melmaiee, K. Admixture analysis using genotyping-by-sequencing reveals genetic relatedness and parental lineage distribution in highbush blueberry genotypes and cross derivatives. Int. J. Mol. Sci. 2020, 22, 163. [Google Scholar] [CrossRef] [PubMed]
- Brandt, K.M.; Chen, X.; Tabima, J.F.; See, D.R.; Vining, K.J.; Zemetra, R.S. QTL analysis of adult plant resistance to stripe rust in a winter wheat recombinant inbred population. Plants 2021, 10, 572. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Soto, R.; Salvatierra, A.; Maldonado, C.; Mashilo, J. The genetic diversity and population structure of different geographical populations of bottle gourd (Lagenaria siceraria) accessions based on genotyping-by-sequencing. Agronomy 2021, 11, 1677. [Google Scholar] [CrossRef]
- Hyun, D.Y.; Sebastin, R.; Lee, G.-A.; Lee, K.J.; Kim, S.-H.; Yoo, E.; Lee, S.; Kang, M.-J.; Lee, S.B.; Jang, I.; et al. Genome-Wide SNP markers for genotypic and phenotypic differentiation of melon (Cucumis melo L.) varieties using genotyping-by-sequencing. Int. J. Mol. Sci. 2021, 22, 6722. [Google Scholar] [CrossRef]
- Ryu, J.; Lyu, J.I.; Kim, D.-G.; Koo, K.M.; Yang, B.; Jo, Y.D.; Kim, S.H.; Kwon, S.-J.; Ha, B.-K.; Kang, S.-Y.; et al. Single Nucleotide Polymorphism (SNP) discovery and association study of flowering times, crude fat and fatty acid composition in rapeseed (Brassica napus L.) mutant lines using Genotyping-by-Sequencing (GBS). Agronomy 2021, 11, 508. [Google Scholar] [CrossRef]
- Nadeem, M.A.; Guizado, S.J.V.; Shahid, M.Q.; Nawaz, M.A.; Habyarimana, E.; Ercişli, S.; Ali, F.; Karaköy, T.; Aasim, M.; Hatipoğlu, R.; et al. In-Depth genetic diversity and population structure of endangered peruvian amazon rosewood germplasm using Genotyping by Sequencing (GBS) technology. Forests 2021, 12, 197. [Google Scholar] [CrossRef]
- Naeem, M.; Demirel, U.; Yousaf, M.F.; Caliskan, S.; Caliskan, M.E. Overview on domestication, breeding, genetic gain and improvement of tuber quality traits of potato using fast forwarding technique (GWAS): A review. Plant Breed. 2021, 140, 519–542. [Google Scholar] [CrossRef]
- Nankar, A.N.; Pratt, R.C. Genotyping by sequencing reveals genetic relatedness of Southwestern U.S. Blue Maize Landraces. Int. J. Mol. Sci. 2021, 22, 3436. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Dong, A.; Duan, H.; Kumar, J.; Rai, M.; Verma, P.C. Advances in cereal crop genomics for resilience under climate change. Life 2021, 11, 502. [Google Scholar] [CrossRef]
- Zuluaga, D.L.; Lioi, L.; Delvento, C.; Pavan, S.; Sonnante, G. Genotyping-by-Sequencing in Vigna unguiculata Landraces and its utility for assessing taxonomic relationships. Plants 2021, 10, 509. [Google Scholar] [CrossRef]
- De Donato, M.; Peters, S.O.; Mitchell, S.E.; Hussain, T.; Imumorin, I.G. Genotyping-by-Sequencing (GBS): A novel, efficient and cost-effective genotyping method for cattle using Next-Generation Sequencing. PLoS ONE 2013, 8, e62137. [Google Scholar] [CrossRef] [PubMed]
- Sahu, P.K.; Sao, R.; Mondal, S.; Vishwakarma, G.; Gupta, S.K.; Kumar, V.; Singh, S.; Sharma, D.; Das, B.K. Next Generation Sequencing based forward genetic approaches for identification and mapping of causal mutations in crop plants: A comprehensive review. Plants 2020, 9, 1355. [Google Scholar] [CrossRef] [PubMed]
- Danilevicz, M.F.; Tay Fernandez, C.G.; Marsh, J.I.; Bayer, P.E.; Edwards, D. Plant pangenomics: Approaches, applications and advancements. Curr. Opin. Plant Biol. 2020, 54, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Sherman, R.M.; Salzberg, S.L. Pan-genomics in the human genome era. Nat. Rev. Genet. 2020, 21, 243–254. [Google Scholar] [CrossRef]
- Lei, L.; Goltsman, E.; Goodstein, D.; Wu, G.A.; Rokhsar, D.S.; Vogel, J.P. Plant Pan-Genomics comes of age. Annu. Rev. Plant Biol. 2021, 72, 411–435. [Google Scholar] [CrossRef]
- Petereit, J.; Bayer, P.E.; Thomas, W.J.W.; Tay Fernandez, C.G.; Amas, J.; Zhang, Y.; Batley, J.; Edwards, D. Pangenomics and Crop Genome Adaptation in a Changing Climate. Plants 2022, 11, 1949. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.W.; Garg, V.; Roorkiwal, M.; Golicz, A.A.; Edwards, D.; Varshney, R.K. Super-Pangenome by Integrating the Wild Side of a Species for Accelerated Crop Improvement. Trends Plant Sci. 2020, 25, 148–158. [Google Scholar] [CrossRef]
- Shah, T.; Xu, J.; Zou, X.; Cheng, Y.; Nasir, M.; Zhang, X. Omics approaches for engineering wheat production under abiotic stresses. Int. J. Mol. Sci. 2018, 19, 2390. [Google Scholar] [CrossRef]
- Della Coletta, R.; Qiu, Y.; Ou, S.; Hufford, M.B.; Hirsch, C.N. How the pan-genome is changing crop genomics and improvement. Genome Biol. 2021, 22, 3. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Chen, Z.; Liang, C. Oryza pan-genomics: A new foundation for future rice research and improvement. Crop J. 2021, 9, 622–632. [Google Scholar] [CrossRef]
- Razzaq, M.K.; Aleem, M.; Mansoor, S.; Khan, M.A.; Rauf, S.; Iqbal, S.; Siddique, K.H.M. Omics and crispr-cas9 approaches for molecular insight, functional gene analysis, and stress tolerance development in crops. Int. J. Mol. Sci. 2021, 22, 1292. [Google Scholar] [CrossRef] [PubMed]
- Ruperao, P.; Thirunavukkarasu, N.; Gandham, P.; Selvanayagam, S.; Govindaraj, M.; Nebie, B.; Manyasa, E.; Gupta, R.; Das, R.R.; Odeny, D.A.; et al. Sorghum pan-genome explores the functional utility for genomic-assisted breeding to accelerate the genetic gain. Front. Plant Sci. 2021, 12, 666342. [Google Scholar] [CrossRef]
- Li, W.; Liu, J.; Zhang, H.; Liu, Z.; Wang, Y.; Xing, L.; He, Q.; Du, H. Plant pan-genomics: Recent advances, new challenges, and roads ahead. J. Genet. Genom. 2022, 49, 833–846. [Google Scholar] [CrossRef] [PubMed]
- Kong, W.; Jiang, M.; Wang, Y.; Chen, S.; Zhang, S.; Lei, W.; Chai, K.; Wang, P.; Liu, R.; Zhang, X. Pan-transcriptome assembly combined with multiple association analysis provides new insights into the regulatory network of specialized metabolites in the tea plant Camellia sinensis. Hortic. Res. 2022, 9, uhac100. [Google Scholar] [CrossRef] [PubMed]
- Woldegiorgis, S.T.; Wu, T.; Gao, L.; Huang, Y.; Zheng, Y.; Qiu, F.; Xu, S.; Tao, H.; Harrison, A.; Liu, W.; et al. Identification of heat-tolerant genes in non-reference sequences in rice by integrating pan-genome, transcriptomics, and QTLs. Genes 2022, 13, 1353. [Google Scholar] [CrossRef]
- Arbona, V.; Manzi, M.; de Ollas, C.; Gómez-Cadenas, A. Metabolomics as a tool to investigate abiotic stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 4885–4911. [Google Scholar] [CrossRef]
- Bashir, K.; Matsui, A.; Rasheed, S.; Seki, M. Recent advances in the characterization of plant transcriptomes in response to drought, salinity, heat, and cold stress. F1000Research 2019, 8, 658. [Google Scholar] [CrossRef]
- Charoensawan, V.; Cortijo, S.; Domijan, M.; Negrão, S. Editorial: Multi-Disciplinary Approaches to Plant Responses to Climate Change. Front. Plant Sci. 2022, 13, 544–563. [Google Scholar] [CrossRef]
- Danilevicz, M.F.; Bayer, P.E.; Nestor, B.J.; Bennamoun, M.; Edwards, D. Resources for image-based high-throughput phenotyping in crops and data sharing challenges. Plant Physiol. 2021, 187, 699–715. [Google Scholar] [CrossRef] [PubMed]
- Marsh, J.I.; Hu, H.; Gill, M.; Batley, J.; Edwards, D. Crop breeding for a changing climate: Integrating phenomics and genomics with bioinformatics. Theor. Appl. Genet. 2021, 134, 1677–1690. [Google Scholar] [CrossRef]
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Hussain, M.A.; Wei, S.; He, H.; Zaman, Q.U.; Xuekun, Z.; Hasanuzzaman, M. Omics: The way forward to enhance abiotic stress tolerance in Brassica napus L. GM Crop. Food 2021, 12, 251–281. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Sood, P.; Prasad, A.; Prasad, M. Advances in omics technology for improving crop yield and stress resilience. Plant Breed. 2021, 140, 719–731. [Google Scholar] [CrossRef]
- Yang, Y.; Saand, M.A.; Huang, L.; Abdelaal, W.B.; Zhang, J.; Wu, Y.; Li, J.; Sirohi, M.H.; Wang, F. Applications of Multi-Omics Technologies for Crop Improvement. Front. Plant Sci. 2021, 12, 563953. [Google Scholar] [CrossRef]
- Zenda, T.; Liu, S.; Dong, A.; Li, J.; Wang, Y.; Liu, X.; Wang, N.; Duan, H. Omics-Facilitated Crop Improvement for Climate Resilience and Superior Nutritive Value. Front. Plant Sci. 2021, 12, 774994. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, M.F.; Hou, D.; Hussain, Q.; Imran, M.; Pei, J.; Ali, M.; Shehzad, A.; Anwar, M.; Noman, A.; Waseem, M.; et al. Entailing the Next-Generation Sequencing and Metabolome for Sustainable Agriculture by Improving Plant Tolerance. Int. J. Mol. Sci. 2022, 23, 651. [Google Scholar] [CrossRef] [PubMed]
- Javaid, M.H.; Khan, A.R.; Salam, A.; Neelam, A.; Azhar, W.; Ulhassan, Z.; Gan, Y. Exploring the Adaptive Responses of Plants to Abiotic Stresses Using Transcriptome Data. Agriculture 2022, 12, 211. [Google Scholar] [CrossRef]
- Ninomiya, S. High-throughput field crop phenotyping: Current status and challenges. Breed. Sci. 2022, 72, 21069. [Google Scholar] [CrossRef]
- Rahman, S.U.; Nawaz, M.F.; Gul, S.; Yasin, G.; Hussain, B.; Li, Y.; Cheng, H. State-of-the-art OMICS strategies against toxic effects of heavy metals in plants: A review. Ecotoxicol. Environ. Saf. 2022, 242, 113952. [Google Scholar] [CrossRef]
- Shen, Y.; Zhou, G.; Liang, C.; Tian, Z. Omics-based interdisciplinarity is accelerating plant breeding. Curr. Opin. Plant Biol. 2022, 66, 102167. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.; Gantait, S.; Azizi, P.; Mazumdar, P. Drought tolerance improvement in Solanum lycopersicum: An insight into “OMICS” approaches and genome editing. 3 Biotech 2022, 12, 63. [Google Scholar] [CrossRef]
- Zhou, R.; Jiang, F.; Niu, L.; Song, X.; Yu, L.; Yang, Y.; Wu, Z. Increase Crop Resilience to Heat Stress Using Omic Strategies. Front. Plant Sci. 2022, 13, 891861. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liu, Z. Unlocking plant metabolic diversity: A (pan)-genomic view. Plant Commun. 2022, 3, 100300. [Google Scholar] [CrossRef]
- Pazhamala, L.T.; Kudapa, H.; Weckwerth, W.; Millar, A.H.; Varshney, R.K. Systems biology for crop improvement. Plant Genome 2021, 14, e20098. [Google Scholar] [CrossRef]
- Anonymous. Method of the Year 2011. Nat. Methods 2012, 9, 1. [Google Scholar] [CrossRef] [PubMed]
- Anonymous. The Runners-Up. Science 2012, 338, 1525–1532. [Google Scholar] [CrossRef] [PubMed]
- Porteus, M.H. Zinc fingers on target. Nature 2009, 459, 337–338. [Google Scholar] [CrossRef] [PubMed]
- Shukla, V.K.; Doyon, Y.; Miller, J.C.; DeKelver, R.C.; Moehle, E.A.; Worden, S.E.; Mitchell, J.C.; Arnold, N.L.; Gopalan, S.; Meng, X.; et al. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 2009, 459, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Townsend, J.A.; Wright, D.A.; Winfrey, R.J.; Fu, F.; Maeder, M.L.; Joung, J.K.; Voytas, D.F. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 2009, 459, 442–445. [Google Scholar] [CrossRef] [PubMed]
- Davies, J.P.; Kumar, S.; Sastry-Dent, L. Use of Zinc-Finger Nucleases for crop improvement. In Progress in Molecular Biology and Translational Science; Elsevier Inc.: Amsterdam, The Netherlands, 2017; Volume 149, pp. 47–63. [Google Scholar]
- Beerli, R.R.; Segal, D.J.; Dreier, B.; Barbas, C.F. Toward controlling gene expression at will: Specific regulation of the erbB-2/HER-2 promoter by using polydactyl zinc finger proteins constructed from modular building blocks. Proc. Natl. Acad. Sci. USA 1998, 95, 14628–14633. [Google Scholar] [CrossRef] [PubMed]
- Beerli, R.R.; Dreier, B.; Barbas, C.F. Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl. Acad. Sci. USA 2000, 97, 1495–1500. [Google Scholar] [CrossRef]
- Papworth, M.; Kolasinska, P.; Minczuk, M. Designer zinc-finger proteins and their applications. Gene 2006, 366, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Sprink, T.; Metje, J.; Hartung, F. Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr. Opin. Biotechnol. 2015, 32, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Lee, H.J.; Kim, H.; Cho, S.W.; Kim, J.-S. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009, 19, 1279–1288. [Google Scholar] [CrossRef] [PubMed]
- Saha, S.K.; Saikot, F.K.; Rahman, M.S.; Jamal, M.A.H.M.; Rahman, S.M.K.; Islam, S.M.R.; Kim, K.-H. Programmable molecular scissors: Applications of a new tool for genome editing in biotech. Mol. Ther.-Nucleic Acids 2019, 14, 212–238. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA–Guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Zetsche, B.; Gootenberg, J.S.; Abudayyeh, O.O.; Slaymaker, I.M.; Makarova, K.S.; Essletzbichler, P.; Volz, S.E.; Joung, J.; van der Oost, J.; Regev, A.; et al. Cpf1 is a single rna-guided endonuclease of a Class 2 CRISPR-Cas system. Cell 2015, 163, 759–771. [Google Scholar] [CrossRef]
- Miglani, G.S.; Kaur, A.; Kaur, L. Plant gene expression control using genome- and epigenome-editing technologies. J. Crop Improv. 2020, 34, 1–63. [Google Scholar] [CrossRef]
- Qasim, W.; Zhan, H.; Samarasinghe, S.; Adams, S.; Amrolia, P.; Stafford, S.; Butler, K.; Rivat, C.; Wright, G.; Somana, K.; et al. Molecular remission of infant B-ALL after infusion of universal TALEN gene-edited CAR T cells. Sci. Transl. Med. 2017, 9, eaaj2013. [Google Scholar] [CrossRef]
- Menz, J.; Modrzejewski, D.; Hartung, F.; Wilhelm, R.; Sprink, T. Genome edited crops touch the market: A view on the global development and regulatory environment. Front. Plant Sci. 2020, 11, 1525. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Voytas, D.F. TAL Effectors: Customizable proteins for DNA targeting. Science 2011, 333, 1843–1846. [Google Scholar] [CrossRef]
- Mahfouz, M.M.; Piatek, A.; Stewart, C.N. Genome engineering via TALENs and CRISPR/Cas9 systems: Challenges and perspectives. Plant Biotechnol. J. 2014, 12, 1006–1014. [Google Scholar] [CrossRef] [PubMed]
- Becker, S.; Boch, J. TALE and TALEN genome editing technologies. Gene Genome Ed. 2021, 2, 100007. [Google Scholar] [CrossRef]
- Boch, J.; Scholze, H.; Schornack, S.; Landgraf, A.; Hahn, S.; Kay, S.; Lahaye, T.; Nickstadt, A.; Bonas, U. Breaking the Code of DNA Binding Specificity of TAL-Type III Effectors. Science 2009, 326, 1509–1512. [Google Scholar] [CrossRef]
- Boch, J.; Bonas, U. Xanthomonas AvrBs3 Family-Type III Effectors: Discovery and function. Annu. Rev. Phytopathol. 2010, 48, 419–436. [Google Scholar] [CrossRef] [PubMed]
- Kay, S.; Hahn, S.; Marois, E.; Hause, G.; Bonas, U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 2007, 318, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Schornack, S.; Moscou, M.J.; Ward, E.R.; Horvath, D.M. Engineering plant disease resistance based on TAL effectors. Annu. Rev. Phytopathol. 2013, 51, 383–406. [Google Scholar] [CrossRef] [PubMed]
- Deng, D.; Yan, C.; Pan, X.; Mahfouz, M.; Wang, J.; Zhu, J.-K.; Shi, Y.; Yan, N. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 2012, 335, 720–723. [Google Scholar] [CrossRef]
- Doyle, E.L.; Stoddard, B.L.; Voytas, D.F.; Bogdanove, A.J. TAL effectors: Highly adaptable phytobacterial virulence factors and readily engineered DNA-targeting proteins. Trends Cell Biol. 2013, 23, 390–398. [Google Scholar] [CrossRef][Green Version]
- Shah, T.; Andleeb, T.; Lateef, S.; Noor, M.A. Genome editing in plants: Advancing crop transformation and overview of tools. Plant Physiol. Biochem. 2018, 131, 12–21. [Google Scholar] [CrossRef]
- Sedeek, K.E.M.; Mahas, A.; Mahfouz, M. Plant genome engineering for targeted improvement of crop traits. Front. Plant Sci. 2019, 10, 114. [Google Scholar] [CrossRef]
- Christian, M.; Cermak, T.; Doyle, E.L.; Schmidt, C.; Zhang, F.; Hummel, A.; Bogdanove, A.J.; Voytas, D.F. Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 2010, 186, 756–761. [Google Scholar] [CrossRef] [PubMed]
- Mahfouz, M.M.; Li, L.; Shamimuzzaman, M.; Wibowo, A.; Fang, X.; Zhu, J.K. De novo-engineered transcription activator-like effector (TALE) hybrid nuclease with novel DNA binding specificity creates double-strand breaks. Proc. Natl. Acad. Sci. USA 2011, 108, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Miller, J.C.; Tan, S.; Qiao, G.; Barlow, K.A.; Wang, J.; Xia, D.F.; Meng, X.; Paschon, D.E.; Leung, E.; Hinkley, S.J.; et al. A TALE nuclease architecture for efficient genome editing. Nat. Biotechnol. 2011, 29, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Huang, S.; Jiang, W.Z.; Wright, D.; Spalding, M.H.; Weeks, D.P.; Yang, B. TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011, 39, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Bhardwaj, A.; Nain, V. TALENs—An indispensable tool in the era of CRISPR: A mini review. J. Genet. Eng. Biotechnol. 2021, 19, 125. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Gao, C. TALENs: Customizable molecular DNA scissors for genome engineering of plants. J. Genet. Genom. 2013, 40, 271–279. [Google Scholar] [CrossRef]
- Wang, Y.; Cheng, X.; Shan, Q.; Zhang, Y.; Liu, J.; Gao, C.; Qiu, J.-L. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 2014, 32, 947–951. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Wei, Z.; Rohila, J.S.; Zhao, K. Multiplex Genome-Editing technologies for revolutionizing plant biology and crop improvement. Front. Plant Sci. 2021, 12, 721203. [Google Scholar] [CrossRef]
- Li, D.; Tang, N.; Fang, Z.; Xia, Y.; Cao, M. Co-Transfer of TALENs Construct Targeted for Chloroplast Genome and Chloroplast Transformation Vector Into Rice Using Particle Bombardment. J. Nanosci. Nanotechnol. 2016, 16, 12194–12201. [Google Scholar] [CrossRef]
- Kazama, T.; Okuno, M.; Watari, Y.; Yanase, S.; Koizuka, C.; Tsuruta, Y.; Sugaya, H.; Toyoda, A.; Itoh, T.; Tsutsumi, N.; et al. Curing cytoplasmic male sterility via TALEN-mediated mitochondrial genome editing. Nat. Plants 2019, 5, 722–730. [Google Scholar] [CrossRef]
- Huang, C.-H.; Liu, Y.-C.; Shen, J.-Y.; Lu, F.-I.; Shaw, S.-Y.; Huang, H.-J.; Chang, C.-C. Repairing TALEN-mediated double-strand break by microhomology-mediated recombination in tobacco plastids generates abundant subgenomic DNA. Plant Sci. 2021, 313, 111028. [Google Scholar] [CrossRef] [PubMed]
- Rascón-Cruz, Q.; González-Barriga, C.D.; Iglesias-Figueroa, B.F.; Trejo-Muñoz, J.C.; Siqueiros-Cendón, T.; Sinagawa-García, S.R.; Arévalo-Gallegos, S.; Espinoza-Sánchez, E.A. Plastid transformation: Advances and challenges for its implementation in agricultural crops. Electron. J. Biotechnol. 2021, 51, 95–109. [Google Scholar] [CrossRef]
- Arimura, S. MitoTALENs: A method for targeted gene disruption in plant mitochondrial genomes. In Plant Mitochondria; Van Aken, O., Rasmusson, A.G., Eds.; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2022; Volume 2363, pp. 335–340. ISBN 978-1-0716-1653-6. [Google Scholar]
- Khan, S.H. Genome-Editing Technologies: Concept, Pros, and Cons of various genome-editing techniques and bioethical concerns for clinical application. Mol. Ther.-Nucleic Acids 2019, 16, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Di Blasi, R.; Zouein, A.; Ellis, T.; Ceroni, F. Genetic toolkits to design and build mammalian synthetic systems. Trends Biotechnol. 2021, 39, 1004–1018. [Google Scholar] [PubMed]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef]
- Schmid-Burgk, J.L.; Schmidt, T.; Kaiser, V.; Höning, K.; Hornung, V. A ligation-independent cloning technique for high-throughput assembly of transcription activator–like effector genes. Nat. Biotechnol. 2013, 31, 76–81. [Google Scholar] [CrossRef]
- Bogdanove, A.J.; Booher, N.J. Online Tools for TALEN Design. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2016; Volume 1338, pp. 43–47. [Google Scholar]
- Gogolok, S.; Garcia-Diaz, C.; Pollard, S.M. STAR: A simple TAL effector assembly reaction using isothermal assembly OPEN. Sci. Rep. 2016, 6, 33209. [Google Scholar] [CrossRef]
- Ma, A.C.; Mcnulty, M.S.; Poshusta, T.L.; Campbell, J.M.; Martínez-Gálvez, G.; Argue, D.P.; Lee, H.B.; Urban, M.D.; Bullard, C.E.; Blackburn, P.R.; et al. FusX: A rapid one-step transcription activator-like effector assembly system for genome science. Hum. Gene Ther. 2016, 27, 451–463. [Google Scholar] [CrossRef]
- Khan, Z.; Khan, S.H.; Mubarik, M.S.; Sadia, B.; Ahmad, A. Use of TALEs and TALEN technology for genetic improvement of plants. Plant Mol. Biol. Report. 2017, 35, 1–19. [Google Scholar] [CrossRef]
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. Elife 2013, 2, e00471. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-Guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Teng, F.; Li, T.; Zhou, Q. Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 2013, 31, 684–686. [Google Scholar] [CrossRef]
- Belhaj, K.; Chaparro-Garcia, A.; Kamoun, S.; Nekrasov, V. Plant genome editing made easy: Targeted mutagenesis in model and crop plants using the CRISPR/Cas system. Plant Methods 2013, 9, 39. [Google Scholar] [CrossRef]
- Feng, Z.; Zhang, B.; Ding, W.; Liu, X.; Yang, D.-L.; Wei, P.; Cao, F.; Zhu, S.; Zhang, F.; Mao, Y.; et al. Efficient genome editing in plants using a CRISPR/Cas system. Cell Res. 2013, 23, 1229–1232. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, H.; Bi, H.; Fromm, M.; Yang, B.; Weeks, D.P. Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 2013, 41, e188. [Google Scholar] [CrossRef] [PubMed]
- Li, J.-F.; Norville, J.E.; Aach, J.; McCormack, M.; Zhang, D.; Bush, J.; Church, G.M.; Sheen, J. Multiplex and homologous recombination–mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 2013, 31, 688–691. [Google Scholar] [CrossRef]
- Mao, Y.; Zhang, H.; Xu, N.; Zhang, B.; Gou, F.; Zhu, J.-K. Application of the CRISPR–Cas system for efficient genome engineering in plants. Mol. Plant 2013, 6, 2008–2011. [Google Scholar] [CrossRef]
- Miao, J.; Guo, D.; Zhang, J.; Huang, Q.; Qin, G.; Zhang, X.; Wan, J.; Gu, H.; Qu, L.-J. Targeted mutagenesis in rice using CRISPR-Cas system. Cell Res. 2013, 23, 1233–1236. [Google Scholar] [CrossRef]
- Shan, Q.; Wang, Y.; Li, J.; Zhang, Y.; Chen, K.; Liang, Z.; Zhang, K.; Liu, J.; Xi, J.J.; Qiu, J.-L.; et al. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 686–688. [Google Scholar] [CrossRef] [PubMed]
- Xie, K.; Yang, Y. RNA-Guided genome editing in plants using a CRISPR–Cas system. Mol. Plant 2013, 6, 1975–1983. [Google Scholar] [CrossRef] [PubMed]
- Wright, A.V.; Nuñez, J.K.; Doudna, J.A. Biology and applications of CRISPR systems: Harnessing Nature’s toolbox for genome engineering. Cell 2016, 164, 29–44. [Google Scholar] [CrossRef] [PubMed]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovič, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef] [PubMed]
- Chennakesavulu, K.; Singh, H.; Trivedi, P.K.; Jain, M.; Yadav, S.R. State-of-the-Art in CRISPR technology and engineering drought, salinity, and thermo-tolerant crop plants. Plant Cell Rep. 2021, 1, 3. [Google Scholar] [CrossRef]
- Chylinski, K.; Le Rhun, A.; Charpentier, E. The tracrRNA and Cas9 families of type II CRISPR-Cas immunity systems. RNA Biol. 2013, 10, 726–737. [Google Scholar] [CrossRef]
- Makarova, K.S.; Wolf, Y.I.; Alkhnbashi, O.S.; Costa, F.; Shah, S.A.; Saunders, S.J.; Barrangou, R.; Brouns, S.J.J.; Charpentier, E.; Haft, D.H.; et al. An updated evolutionary classification of CRISPR–Cas systems. Nat. Rev. Microbiol. 2015, 13, 722–736. [Google Scholar] [CrossRef]
- Makarova, K.S.; Koonin, E.V. Annotation and classification of CRISPR-Cas systems. In Methods in Molecular Biology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 1311, pp. 47–75. [Google Scholar]
- Shmakov, S.; Smargon, A.; Scott, D.; Cox, D.; Pyzocha, N.; Yan, W.; Abudayyeh, O.O.; Gootenberg, J.S.; Makarova, K.S.; Wolf, Y.I.; et al. Diversity and evolution of class 2 CRISPR–Cas systems. Nat. Rev. Microbiol. 2017, 15, 169–182. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Aman, R.; Ali, Z.; Butt, H.; Mahas, A.; Aljedaani, F.; Khan, M.Z.; Ding, S.; Mahfouz, M. RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 2018, 19, 1. [Google Scholar] [CrossRef]
- Ahmad, N.; Rahman, M.-U.; Mukhtar, Z.; Zafar, Y.; Zhang, B. A critical look on CRISPR-based genome editing in plants. J. Cell. Physiol. 2020, 235, 666–682. [Google Scholar] [CrossRef] [PubMed]
- Mushtaq, M.; Dar, A.A.; Basu, U.; Bhat, B.A.; Mir, R.A.; Vats, S.; Dar, M.S.; Tyagi, A.; Ali, S.; Bansal, M.; et al. Integrating CRISPR-Cas and Next Generation Sequencing in plant virology. Front. Genet. 2021, 12, 735489. [Google Scholar] [CrossRef]
- Landsberger, M.; Gandon, S.; Meaden, S.; Rollie, C.; Chevallereau, A.; Chabas, H.; Buckling, A.; Westra, E.R.; van Houte, S. Anti-CRISPR phages cooperate to overcome CRISPR-Cas immunity. Cell 2018, 174, 908–916. [Google Scholar] [CrossRef]
- Li, Y.; Bondy-Denomy, J. Anti-CRISPRs go viral: The infection biology of CRISPR-Cas inhibitors. Cell Host Microbe 2021, 29, 704–714. [Google Scholar] [CrossRef]
- Sternberg, S.H.; LaFrance, B.; Kaplan, M.; Doudna, J.A. Conformational control of DNA target cleavage by CRISPR–Cas9. Nature 2015, 527, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef] [PubMed]
- Collonnier, C.; Epert, A.; Mara, K.; Maclot, F.; Guyon-Debast, A.; Charlot, F.; White, C.; Schaefer, D.G.; Nogué, F. CRISPR-Cas9-mediated efficient directed mutagenesis and RAD51-dependent and RAD51-independent gene targeting in the moss Physcomitrella patens. Plant Biotechnol. J. 2017, 15, 122–131. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Obando, M.; Hoffmann, B.; Géry, C.; Guyon-Debast, A.; Téoulé, E.; Rameau, C.; Bonhomme, S.; Nogué, F. Simple and Efficient Targeting of Multiple Genes Through CRISPR-Cas9 in Physcomitrella patens. G3 Genes Genomes Genet. 2016, 6, 3647–3653. [Google Scholar] [CrossRef]
- Tang, Y.; Fu, Y. Class 2 CRISPR/Cas: An expanding biotechnology toolbox for and beyond genome editing. Cell Biosci. 2018, 8, 59. [Google Scholar] [CrossRef]
- Saurabh, S. Genome Editing: Revolutionizing the Crop Improvement. Plant Mol. Biol. Report. 2021, 39, 752–772. [Google Scholar] [CrossRef]
- Yue, J.-J.; Yuan, J.-L.; Wu, F.-H.; Yuan, Y.-H.; Cheng, Q.-W.; Hsu, C.-T.; Lin, C.-S. Protoplasts: From Isolation to CRISPR/Cas Genome Editing Application. Front. Genome Ed. 2021, 3, 717017. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, S.; Kumar, R.; Kumar, V.; Won, S.Y.; Shukla, P. Engineering disease resistant plants through CRISPR-Cas9 technology. GM Crop. Food 2021, 12, 125–144. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, X.; Liu, H.; Li, Z. Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat. Plants 2020, 6, 773–779. [Google Scholar] [CrossRef] [PubMed]
- Hahn, F.; Korolev, A.; Sanjurjo Loures, L.; Nekrasov, V. A modular cloning toolkit for genome editing in plants. BMC Plant Biol. 2020, 20, 179. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.M.; Zhang, Y.; Yuan, G.; De, K.; Chen, J.-G.; Muchero, W.; Tuskan, G.A.; Qi, Y.; Yang, X. Construct design for CRISPR/Cas-based genome editing in plants. Trends Plant Sci. 2021, 26, 1133–1152. [Google Scholar] [CrossRef]
- Moradpour, M.; Abdulah, S.N.A. CRISPR/dCas9 platforms in plants: Strategies and applications beyond genome editing. Plant Biotechnol. J. 2020, 18, 32–44. [Google Scholar] [CrossRef]
- Moreno-Mateos, M.A.; Fernandez, J.P.; Rouet, R.; Vejnar, C.E.; Lane, M.A.; Mis, E.; Khokha, M.K.; Doudna, J.A.; Giraldez, A.J. CRISPR-Cpf1 mediates efficient homology-directed repair and temperature-controlled genome editing. Nat. Commun. 2017, 8, 2024. [Google Scholar] [CrossRef]
- Bandyopadhyay, A.; Kancharla, N.; Javalkote, V.S.; Dasgupta, S.; Brutnell, T.P. CRISPR-Cas12a (Cpf1): A versatile tool in the plant genome editing tool box for agricultural advancement. Front. Plant Sci. 2020, 11, 584151. [Google Scholar] [CrossRef]
- Komor, A.C.; Kim, Y.B.; Packer, M.S.; Zuris, J.A.; Liu, D.R. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 2016, 533, 420–424. [Google Scholar] [CrossRef]
- Gaudelli, N.M.; Komor, A.C.; Rees, H.A.; Packer, M.S.; Badran, A.H.; Bryson, D.I.; Liu, D.R. Programmable base editing of A•T to G•C in genomic DNA without DNA cleavage. Nature 2017, 551, 464–471. [Google Scholar] [CrossRef] [PubMed]
- Shimatani, Z.; Kashojiya, S.; Takayama, M.; Terada, R.; Arazoe, T.; Ishii, H.; Teramura, H.; Yamamoto, T.; Komatsu, H.; Miura, K.; et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017, 35, 441–443. [Google Scholar] [CrossRef] [PubMed]
- Veillet, F.; Perrot, L.; Guyon-Debast, A.; Kermarrec, M.-P.; Chauvin, L.; Chauvin, J.-E.; Gallois, J.-L.; Mazier, M.; Nogué, F. Expanding the CRISPR Toolbox in P. patens Using SpCas9-NG variant and application for gene and base editing in solanaceae crops. Int. J. Mol. Sci. 2020, 21, 1024. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef]
- Guyon-Debast, A.; Alboresi, A.; Terret, Z.; Charlot, F.; Berthier, F.; Vendrell-Mir, P.; Casacuberta, J.M.; Veillet, F.; Morosinotto, T.; Gallois, J.-L.; et al. A blueprint for gene function analysis through Base Editing in the model plant Physcomitrium (Physcomitrella) patens. New Phytol. 2021, 230, 1258–1272. [Google Scholar] [CrossRef]
- Li, C.; Zong, Y.; Wang, Y.; Jin, S.; Zhang, D.; Song, Q.; Zhang, R.; Gao, C. Expanded base editing in rice and wheat using a Cas9-adenosine deaminase fusion. Genome Biol. 2018, 19, 59. [Google Scholar] [CrossRef]
- Li, C.; Zhang, R.; Meng, X.; Chen, S.; Zong, Y.; Lu, C.; Qiu, J.-L.; Chen, Y.-H.; Li, J.; Gao, C. Targeted, random mutagenesis of plant genes with dual cytosine and adenine base editors. Nat. Biotechnol. 2020, 38, 875–882. [Google Scholar] [CrossRef]
- Cox, D.B.T.; Gootenberg, J.S.; Abudayyeh, O.O.; Franklin, B.; Kellner, M.J.; Joung, J.; Zhang, F. RNA editing with CRISPR-Cas13. Science 2017, 358, 1019–1027. [Google Scholar] [CrossRef]
- Ali, Z.; Mahas, A.; Mahfouz, M. CRISPR/Cas13 as a tool for RNA interference. Trends Plant Sci. 2018, 23, 374–378. [Google Scholar] [CrossRef]
- Balderston, S.; Clouse, G.; Ripoll, J.-J.; Pratt, G.K.; Gasiunas, G.; Bock, J.-O.; Bennett, E.P.; Aran, K. Diversification of the CRISPR Toolbox: Applications of CRISPR-Cas Systems Beyond Genome Editing. Cris. J. 2021, 4, 400–415. [Google Scholar] [CrossRef] [PubMed]
- Molla, K.A.; Sretenovic, S.; Bansal, K.C.; Qi, Y. Precise plant genome editing using base editors and prime editors. Nat. Plants 2021, 7, 1166–1187. [Google Scholar] [CrossRef] [PubMed]
- Abudayyeh, O.O.; Gootenberg, J.S.; Franklin, B.; Koob, J.; Kellner, M.J.; Ladha, A.; Joung, J.; Kirchgatterer, P.; Cox, D.B.T.; Zhang, F. A cytosine deaminase for programmable single-base RNA editing. Science 2019, 365, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Lin, Q.; Zong, Y.; Xue, C.; Wang, S.; Jin, S.; Zhu, Z.; Wang, Y.; Anzalone, A.V.; Raguram, A.; Doman, J.L.; et al. Prime genome editing in rice and wheat. Nat. Biotechnol. 2020, 38, 582–585. [Google Scholar] [CrossRef] [PubMed]
- Perroud, P.-F.; Guyon-Debast, A.; Veillet, F.; Kermarrec, M.-P.; Chauvin, L.; Chauvin, J.-E.; Gallois, J.-L.; Nogué, F. Prime Editing in the model plant Physcomitrium patens and its potential in the tetraploid potato. Plant Sci. 2022, 316, 111162. [Google Scholar] [CrossRef]
- Gallego-Bartolomé, J.; Gardiner, J.; Liu, W.; Papikian, A.; Ghoshal, B.; Kuo, H.Y.; Zhao, J.M.-C.; Segal, D.J.; Jacobsen, S.E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proc. Natl. Acad. Sci. USA 2018, 115, E2125–E2134. [Google Scholar] [CrossRef] [PubMed]
- Papikian, A.; Liu, W.; Gallego-Bartolomé, J.; Jacobsen, S.E. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019, 10, 729. [Google Scholar] [CrossRef]
- Zheng, H.-X.; Sun, X.; Zhang, X.-S.; Sui, N. m6A Editing: New tool to improve crop quality? Trends Plant Sci. 2020, 25, 859–867. [Google Scholar] [CrossRef]
- Maher, M.F.; Nasti, R.A.; Vollbrecht, M.; Starker, C.G.; Clark, M.D.; Voytas, D.F. Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 2020, 38, 84–89. [Google Scholar] [CrossRef]
- Wang, X.; Ye, L.; Lyu, M.; Ursache, R.; Löytynoja, A.; Mähönen, A.P. An inducible genome editing system for plants. Nat. Plants 2020, 6, 766–772. [Google Scholar] [CrossRef]
- Nandy, S.; Pathak, B.; Zhao, S.; Srivastava, V. Heat-shock-inducible CRISPR/Cas9 system generates heritable mutations in rice. Plant Direct 2019, 3, e00145. [Google Scholar] [CrossRef]
- Decaestecker, W.; Buono, R.A.; Pfeiffer, M.L.; Vangheluwe, N.; Jourquin, J.; Karimi, M.; Van Isterdael, G.; Beeckman, T.; Nowack, M.K.; Jacobs, T.B. CRISPR-TSKO: A technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell 2019, 31, 2868–2887. [Google Scholar] [CrossRef] [PubMed]
- Gapinske, M.; Luu, A.; Winter, J.; Woods, W.S.; Kostan, K.A.; Shiva, N.; Song, J.S.; Perez-Pinera, P. CRISPR-SKIP: Programmable gene splicing with single base editors. Genome Biol. 2018, 19, 107. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xie, W.; Liu, Z.; Shan, H.; Chen, M.; Song, Y.; Yu, H.; Lai, L.; Li, Z. CRISPR Start-Loss: A novel and practical alternative for gene silencing through base-editing-induced start codon mutations. Mol. Ther.-Nucleic Acids 2020, 21, 1062–1073. [Google Scholar] [CrossRef]
- Woo, J.W.; Kim, J.; Kwon, S.I.; Corvalán, C.; Cho, S.W.; Kim, H.; Kim, S.-G.; Kim, S.-T.; Choe, S.; Kim, J.-S. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015, 33, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Svitashev, S.; Schwartz, C.; Lenderts, B.; Young, J.K.; Mark Cigan, A. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016, 7, 13274. [Google Scholar] [CrossRef]
- Zhang, Y.; Liang, Z.; Zong, Y.; Wang, Y.; Liu, J.; Chen, K.; Qiu, J.-L.; Gao, C. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016, 7, 12617. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, K.; Li, T.; Zhang, Y.; Wang, Y.; Zhao, Q.; Liu, J.; Zhang, H.; Liu, C.; Ran, Y.; et al. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 2017, 8, 14261. [Google Scholar] [CrossRef]
- Li, S.; Li, J.; He, Y.; Xu, M.; Zhang, J.; Du, W.; Zhao, Y.; Xia, L. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat. Biotechnol. 2019, 37, 445–450. [Google Scholar] [CrossRef] [PubMed]
- Toda, E.; Koiso, N.; Takebayashi, A.; Ichikawa, M.; Kiba, T.; Osakabe, K.; Osakabe, Y.; Sakakibara, H.; Kato, N.; Okamoto, T. An efficient DNA- and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants 2019, 5, 363–368. [Google Scholar] [CrossRef] [PubMed]
- Kuscu, C.; Parlak, M.; Tufan, T.; Yang, J.; Szlachta, K.; Wei, X.; Mammadov, R.; Adli, M. CRISPR-STOP: Gene silencing through base-editing-induced nonsense mutations. Nat. Methods 2017, 14, 710–712. [Google Scholar] [CrossRef]
- Lee, C.; Hyun Jo, D.; Hwang, G.-H.; Yu, J.; Kim, J.H.; Park, S.; Kim, J.-S.; Kim, J.H.; Bae, S. CRISPR-Pass: Gene Rescue of Nonsense Mutations Using Adenine Base Editors. Mol. Ther. 2019, 27, 1364–1371. [Google Scholar] [CrossRef] [PubMed]
- Al-Shayeb, B.; Sachdeva, R.; Chen, L.-X.; Ward, F.; Munk, P.; Devoto, A.; Castelle, C.J.; Olm, M.R.; Bouma-Gregson, K.; Amano, Y.; et al. Clades of huge phages from across Earth’s ecosystems. Nature 2020, 578, 425–431. [Google Scholar] [CrossRef]
- Pausch, P.; Al-Shayeb, B.; Bisom-Rapp, E.; Tsuchida, C.A.; Li, Z.; Cress, B.F.; Knott, G.J.; Jacobsen, S.E.; Banfield, J.F.; Doudna, J.A. CRISPR-Cas from huge phages is a hypercompact genome editor. Science 2020, 369, 333–337. [Google Scholar] [CrossRef]
- Kwak, S.-Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.-H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef]
- Zhu, H.; Li, C.; Gao, C. Applications of CRISPR–Cas in agriculture and plant biotechnology. Nat. Rev. Mol. Cell Biol. 2020, 21, 661–677. [Google Scholar] [CrossRef]
- Sauer, N.J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Walker, K.A.; Beetham, P.R.; Schöpke, C.R.; Gocal, G.F.W. Oligonucleotide-directed mutagenesis for precision gene editing. Plant Biotechnol. J. 2016, 14, 496–502. [Google Scholar] [CrossRef] [PubMed]
- Sauer, N.J.; Narváez-Vásquez, J.; Mozoruk, J.; Miller, R.B.; Warburg, Z.J.; Woodward, M.J.; Mihiret, Y.A.; Lincoln, T.A.; Segami, R.E.; Sanders, S.L.; et al. Oligonucleotide-mediated genome editing provides precision and function to engineered nucleases and antibiotics in plants. Plant Physiol. 2016, 170, 1917–1928. [Google Scholar] [CrossRef]
- Moerschell, R.P.; Tsunasawa, S.; Sherman, F. Transformation of yeast with synthetic oligonucleotides. Proc. Natl. Acad. Sci. USA 1988, 85, 524–528. [Google Scholar] [CrossRef]
- Yoon, K.; Cole-Strauss, A.; Kmiec, E.B. Targeted gene correction of episomal DNA in mammalian cells mediated by a chimeric RNA.DNA oligonucleotide. Proc. Natl. Acad. Sci. USA 1996, 93, 2071–2076. [Google Scholar] [CrossRef]
- Aarts, M.; Dekker, M.; de Vries, S.; van der Wal, A.; te Riele, H. Generation of a mouse mutant by oligonucleotide-mediated gene modification in ES cells. Nucleic Acids Res. 2006, 34, e147. [Google Scholar] [CrossRef]
- Beetham, P.R.; Kipp, P.B.; Sawycky, X.L.; Arntzen, C.J.; May, G.D. A tool for functional plant genomics: Chimeric RNA/DNA oligonucleotides cause in vivo gene-specific mutations. Proc. Natl. Acad. Sci. USA 1999, 96, 8774–8778. [Google Scholar] [CrossRef]
- Hohn, B.; Puchta, H. Gene therapy in plants. Proc. Natl. Acad. Sci. USA 1999, 96, 8321–8323. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Peterson, D.J.; Tagliani, L.; St. Clair, G.; Baszczynski, C.L.; Bowen, B. Targeted manipulation of maize genes in vivo using chimeric RNA/DNA oligonucleotides. Proc. Natl. Acad. Sci. USA 1999, 96, 8768–8773. [Google Scholar] [CrossRef]
- Gamper, H.B.; Parekh, H.; Rice, M.C.; Bruner, M.; Youkey, H.; Kmiec, E.B. The DNA strand of chimeric RNA/DNA oligonucleotides can direct gene repair/conversion activity in mammalian and plant cell-free extracts. Nucleic Acids Res. 2000, 28, 4332–4339. [Google Scholar] [CrossRef] [PubMed]
- Breyer, D.; Herman, P.; Brandenburger, A.; Gheysen, G.; Remaut, E.; Soumillion, P.; Van Doorsselaere, J.; Custers, R.; Pauwels, K.; Sneyers, M.; et al. Commentary: Genetic modification through oligonucleotide-mediated mutagenesis. A GMO regulatory challenge? Environ. Biosaf. Res. 2009, 8, 57–64. [Google Scholar] [CrossRef]
- Songstad, D.D.; Petolino, J.F.; Voytas, D.F.; Reichert, N.A. Genome editing of plants. CRC. Crit. Rev. Plant Sci. 2017, 36, 1–23. [Google Scholar] [CrossRef]
- Holme, I.B.; Gregersen, P.L.; Brinch-Pedersen, H. Induced genetic variation in crop plants by random or targeted mutagenesis: Convergence and differences. Front. Plant Sci. 2019, 10, 1468. [Google Scholar] [CrossRef] [PubMed]
- Cole-Strauss, A.; Yoon, K.; Xiang, Y.; Byrne, B.C.; Rice, M.C.; Gryn, J.; Holloman, W.K.; Kmiec, E.B. Correction of the mutation responsible for sickle cell anemia by an RNA-DNA oligonucleotide. Science 1996, 273, 1386–1389. [Google Scholar] [CrossRef] [PubMed]
- Ruiter, R.; Van Den Brande, I.; Stals, E.; Delauré, S.; Cornelissen, M.; D’Halluin, K. Spontaneous mutation frequency in plants obscures the effect of chimeraplasty. Plant Mol. Biol. 2003, 53, 715–729. [Google Scholar] [CrossRef] [PubMed]
- Zhu, T.; Mettenburg, K.; Peterson, D.J.; Tagliani, L.; Baszczynski, C.L. Engineering herbicide-resistant maize using chimeric RNA/DNA oligonucleotides. Nat. Biotechnol. 2000, 18, 555–558. [Google Scholar] [CrossRef]
- Tan, S.; Evans, R.R.; Dahmer, M.L.; Singh, B.K.; Shaner, D.L. Imidazolinone-tolerant crops: History, current status and future. Pest Manag. Sci. 2005, 61, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Gocal, G. Non-Transgenic trait development in crop plants using oligo-directed mutagenesis: Cibus’ rapid trait development system. In NABC Report 26. New DNA-Editing Approaches: Methods, Applications and Policy for Agriculture; North American Agricultural Biotechnology Council: Ithaca, NY, USA, 2015; pp. 97–106. [Google Scholar]
- Gocal, G.F.W.; Schöpke, C.R.; Beetham, P.R. Advances in New Technology for Targeted Modification of Plant Genomes; Zhang, F., Puchta, H., Thomson, J.G., Eds.; Springer: New York, NY, USA, 2015; ISBN 978-1-4939-2555-1. [Google Scholar]
- Wolt, J.D.; Wang, K.; Yang, B. The regulatory status of genome-edited crops. Plant Biotechnol. J. 2016, 14, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Jones, H.D. Regulatory uncertainty over genome editing. Nat. Plants 2015, 1, 14011. [Google Scholar] [CrossRef] [PubMed]
- Kmiec, E.B.; Johnson, C.; May, G.D. Chloroplast lysates support directed mutagenesis via modified DNA and chimeric RNA/DNA oligonucleotides. Plant J. 2001, 27, 267–274. [Google Scholar] [CrossRef][Green Version]
- Yu, J.; Xu, F.; Wei, Z.; Zhang, X.; Chen, T.; Pu, L. Epigenomic Landscape and Epigenetic Regulation in Maize. Theor. Appl. Genet. 2020, 133, 1467–1489. [Google Scholar] [CrossRef]
- Rensing, S.A.; Goffinet, B.; Meyberg, R.; Wu, S.Z.; Bezanilla, M. The Moss Physcomitrium (Physcomitrella) patens: A Model Organism for Non-Seed Plants. Plant Cell 2020, 32, 1361–1376. [Google Scholar] [CrossRef]
- Oh, D.H.; Barkla, B.J.; Vera-Estrella, R.; Pantoja, O.; Lee, S.Y.; Bohnert, H.J.; Dassanayake, M. Cell type-specific responses to salinity-the epidermal bladder cell transcriptome of Mesembryanthemum crystallinum. New Phytol. 2015, 207, 627–644. [Google Scholar] [CrossRef]
- Himabindu, Y.; Chakradhar, T.; Reddy, M.C.; Kanygin, A.; Redding, K.E.; Chandrasekhar, T. Salt-tolerant genes from halophytes are potential key players of salt tolerance in glycophytes. Environ. Exp. Bot. 2016, 124, 39–63. [Google Scholar] [CrossRef]
- Giarola, V.; Bartels, D. What can we learn from the transcriptome of the resurrection plant Craterostigma plantagineum? Planta 2015, 242, 427–434. [Google Scholar] [CrossRef]
- Pampurova, S.; van Dijck, P. The desiccation tolerant secrets of Selaginella lepidophylla: What we have learned so far? Plant Physiol. Biochem. 2014, 80, 285–290. [Google Scholar] [CrossRef]
- Ríos-Meléndez, S.; Valadez-Hernández, E.; Delgadillo, C.; Luna-Guevara, M.L.; Martínez-Núñez, M.A.; Sánchez-Pérez, M.; Martínez-y-Pérez, J.L.; Arroyo-Becerra, A.; Cárdenas, L.; Bibbins-Martínez, M.; et al. Zander is a fully desiccation-tolerant moss that expresses an inducible molecular mechanism in response to severe abiotic stress. Plant Mol. Biol. 2021, 107, 387–404. [Google Scholar] [CrossRef] [PubMed]
- Bitocchi, E.; Rau, D.; Bellucci, E.; Rodriguez, M.; Murgia, M.L.; Gioia, T.; Santo, D.; Nanni, L.; Attene, G.; Papa, R. Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci. 2017, 8, 722. [Google Scholar] [CrossRef] [PubMed]
- Rothan, C.; Diouf, I.; Causse, M. Trait discovery and editing in tomato. Plant J. 2019, 97, 73–90. [Google Scholar] [CrossRef] [PubMed]
- Mrízová, K.; Holasková, E.; Öz, M.T.; Jiskrová, E.; Frébort, I.; Galuszka, P. Transgenic barley: A prospective tool for biotechnology and agriculture. Biotechnol. Adv. 2014, 32, 137–157. [Google Scholar] [CrossRef] [PubMed]
- Gürel, F.; Öztürk, Z.N.; Uçarlı, C.; Rosellini, D. Barley genes as tools to confer abiotic stress tolerance in crops. Front. Plant Sci. 2016, 7, 1137. [Google Scholar] [CrossRef] [PubMed]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef]
- Mwando, E.; Angessa, T.T.; Han, Y.; Li, C. Salinity tolerance in barley during germination—Homologs and potential genes. J. Zhejiang Univ. Sci. B 2020, 21, 93–121. [Google Scholar] [CrossRef]
- Jeyasri, R.; Muthuramalingam, P.; Satish, L.; Karutha Pandian, S.; Chen, J.-T.; Ahmar, S.; Wang, X.; Mora-Poblete, F.; Ramesh, M.; Labudda, M.; et al. An overview of abiotic stress in cereal crops: Negative impacts, regulation, biotechnology and integrated Omics. Plants 2021, 10, 1472. [Google Scholar] [CrossRef]
- Kikuchi, A.; Huynh, H.D.; Endo, T.; Watanabe, K. Review of recent transgenic studies on abiotic stress tolerance and future molecular breeding in potato. Breed. Sci. 2015, 65, 85–102. [Google Scholar] [CrossRef]
- Dahal, K.; Li, X.Q.; Tai, H.; Creelman, A.; Bizimungu, B. Improving potato stress tolerance and tuber yield under a climate change scenario–a current overview. Front. Plant Sci. 2019, 10, 563. [Google Scholar] [CrossRef]
- Handayani, T.; Gilani, S.A.; Watanabe, K.N. Climatic changes and potatoes: How can we cope with the abiotic stresses? Breed. Sci. 2019, 69, 545–563. [Google Scholar] [CrossRef] [PubMed]
- Ezquer, I.; Salameh, I.; Colombo, L.; Kalaitzis, P. Plant cell walls tackling climate change: Insights into plant cell wall remodeling, its regulation, and biotechnological strategies to improve crop adaptations and photosynthesis in response to global warming. Plants 2020, 9, 212. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Mishra, A. Ectopic expression of C4 photosynthetic pathway genes improves carbon assimilation and alleviate stress tolerance for future climate change. Physiol. Mol. Biol. Plants 2020, 26, 195–209. [Google Scholar] [CrossRef] [PubMed]
- Beebe, S.E.; Rao, I.M.; Blair, M.W.; Acosta-Gallegos, J.A. Phenotyping common beans for adaptation to drought. Front. Physiol. 2013, 4, 35. [Google Scholar] [CrossRef]
- Valliyodan, B.; Ye, H.; Song, L.; Murphy, M.; Grover Shannon, J.; Nguyen, H.T. Genetic diversity and genomic strategies for improving drought and waterlogging tolerance in soybeans. J. Exp. Bot. 2017, 68, 1835–1849. [Google Scholar] [CrossRef]
- Mousavi-Derazmahalleh, M.; Bayer, P.E.; Hane, J.K.; Valliyodan, B.; Nguyen, H.T.; Nelson, M.N.; Erskine, W.; Varshney, R.K.; Papa, R.; Edwards, D. Adapting legume crops to climate change using genomic approaches. Plant Cell Environ. 2019, 42, 6–19. [Google Scholar] [CrossRef]
- Mukankusi, C.; Raatz, B.; Nkalubo, S.; Berhanu, F.; Binagwa, P.; Kilango, M.; Williams, M.; Enid, K.; Chirwa, R.; Beebe, S. Genomics, genetics and breeding of common bean in Africa: A review of tropical legume project. Plant Breed. 2018, 138, 401–414. [Google Scholar] [CrossRef]
- Oladzad, A.; Porch, T.; Rosas, J.C.; Moghaddam, S.M.; Beaver, J.; Beebe, S.E.; Burridge, J.; Jochua, C.N.; Miguel, M.A.; Miklas, P.N.; et al. Single and Multi-Trait GWAS identify genetic factors associated with production traits in common bean under abiotic stress environments. G3 Genes Genomes Genet. 2019, 9, 1881–1892. [Google Scholar]
- Thudi, M.; Palakurthi, R.; Schnable, J.C.; Chitikineni, A.; Dreisigacker, S.; Mace, E.; Srivastava, R.K.; Satyavathi, C.T.; Odeny, D.; Tiwari, V.K.; et al. Genomic resources in plant breeding for sustainable agriculture. J. Plant Physiol. 2021, 257, 153351. [Google Scholar] [CrossRef]
- Parmar, N.; Singh, K.H.; Sharma, D.; Singh, L.; Kumar, P.; Nanjundan, J.; Khan, Y.J.; Chauhan, D.K.; Thakur, A.K. Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: A comprehensive review. 3 Biotech 2017, 7, 239. [Google Scholar] [CrossRef]
- Chaudhary, J.; Khatri, P.; Singla, P.; Kumawat, S.; Kumari, A.; Vinaykumar, R.; Vikram, A.; Jindal, S.K.; Kardile, H.; Kumar, R.; et al. Advances in Omics approaches for abiotic stress tolerance in tomato. Biology 2019, 8, 90. [Google Scholar] [CrossRef]
- Salava, H.; Thula, S.; Mohan, V.; Kumar, R.; Maghuly, F. Application of Genome Editing in Tomato Breeding: Mechanisms, Advances, and Prospects. Int. J. Mol. Sci. 2021, 22, 682. [Google Scholar] [CrossRef] [PubMed]
- Rahim, A.H.; Shafika, Z.K.; Bhuiyan, M.A.R. Evaluation and characterization of advanced rice mutant line of rice (Oryza sativa), MR219-4 and MR219-9 under drought condition. Res. Dev. Semin. 2012, 44, 26–28. Available online: https://inis.iaea.org/collection/NCLCollectionStore/_Public/44/096/44096860.pdf (accessed on 23 August 2021).
- Hallajian, M. Integration of mutation and conventional breeding approaches to develop new superior drought-tolerant plants in rice (Oryza sativa). Annu. Res. Rev. Biol. 2014, 4, 1173–1186. [Google Scholar] [CrossRef]
- Hay, S.; Oo, M.; Minn, M.; Linn, K.Z.; Mar, N.N.; Thin, P.P.; Student, P.D. Development of drought tolerant mutant from rice var. Manawthukha through mutation breeding technique using 60Co Gamma source. Int. J. Innov. Res. Sci. Eng. Technol. 2015, 4, 11205–11212. [Google Scholar]
- Efendi, B.; Sabaruddin, Z.; Lukman, H. Mutation with gamma rays irradiation to assemble green super rice tolerant to drought stress and high yield rice (Oryza sativa L.). Int. J. Adv. Sci. Eng. Tech. 2017, 5, 1–5. [Google Scholar]
- Pang, Y.; Chen, K.; Wang, X.; Xu, J.; Ali, J.; Li, Z. Recurrent selection breeding by dominant male sterility for multiple abiotic stresses tolerant rice cultivars. Euphytica 2017, 213, 268. [Google Scholar] [CrossRef]
- Dwivedi, S.L.; Stoddard, F.L.; Ortiz, R. Genomic-based root plasticity to enhance abiotic stress adaptation and edible yield in grain crops. Plant Sci. 2020, 295, 110365. [Google Scholar] [CrossRef]
- Choudhary, M.; Wani, S.H.; Kumar, P.; Bagaria, P.K.; Rakshit, S.; Roorkiwal, M.; Varshney, R.K. QTLian breeding for climate resilience in cereals: Progress and prospects. Funct. Integr. Genom. 2019, 19, 685–701. [Google Scholar] [CrossRef]
- Dixit, S.; Mallikarjuna Swamy, B.P.; Vikram, P.; Bernier, J.; Sta Cruz, M.T.; Amante, M.; Atri, D.; Kumar, A. Increased drought tolerance and wider adaptability of QDTY12.1 conferred by its interaction with QDTY2.3 and QDTY3.2. Mol. Breed. 2012, 30, 1767–1779. [Google Scholar] [CrossRef]
- Uga, Y.; Sugimoto, K.; Ogawa, S.; Rane, J.; Ishitani, M.; Hara, N.; Kitomi, Y.; Inukai, Y.; Ono, K.; Kanno, N.; et al. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat. Genet. 2013, 45, 1097–1102. [Google Scholar] [CrossRef] [PubMed]
- Arai-Sanoh, Y.; Takai, T.; Yoshinaga, S.; Nakano, H.; Kojima, M.; Sakakibara, H.; Kondo, M.; Uga, Y. Deep rooting conferred by DEEPER ROOTING 1 enhances rice yield in paddy fields. Sci. Rep. 2014, 4, 5563. [Google Scholar] [CrossRef] [PubMed]
- Shamsudin, N.A.A.; Swamy, B.P.M.; Ratnam, W.; Sta Cruz, M.T.; Raman, A.; Kumar, A. Marker assisted pyramiding of drought yield QTLs into a popular malaysian rice cultivar, MR219. BMC Genet. 2016, 17, 30. [Google Scholar] [CrossRef] [PubMed]
- Sahebi, M.; Hanafi, M.M.; Rafii, M.Y.; Mahmud, T.M.M.; Azizi, P.; Osman, M.; Abiri, R.; Taheri, S.; Kalhori, N.; Shabanimofrad, M.; et al. Improvement of drought tolerance in rice (Oryza sativa L.): Genetics, genomic tools, and the WRKY gene family. BioMed Res. Int. 2018, 2018, 3158474. [Google Scholar] [CrossRef] [PubMed]
- Linh, L.H.; Linh, T.H.; Xuan, T.D.; Ham, L.H.; Ismail, A.M.; Khanh, T.D. Molecular breeding to improve salt tolerance of rice (Oryza sativa L.) in the red river delta of vietnam. Int. J. Plant Genom. 2012, 2012, 949038. [Google Scholar] [CrossRef]
- Huyen, L.T.N.; Cuc, L.M.; Ismail, A.M.; Ham, L.H. Introgression the salinity tolerance QTLs Salto into AS996, the elite rice variety of Vietnam. Am. J. Plant Sci. 2012, 3, 981–987. [Google Scholar] [CrossRef]
- Das, G.; Rao, G.J.N. Molecular marker assisted gene stacking for biotic and abiotic stress resistance genes in an elite rice cultivar. Front. Plant Sci. 2015, 6, 698. [Google Scholar] [CrossRef]
- Babu, N.N.; Krishnan, S.G.; Vinod, K.K.; Krishnamurthy, S.L.; Singh, V.K.; Singh, M.P.; Singh, R.; Ellur, R.K.; Rai, V.; Bollinedi, H.; et al. Marker aided incorporation of Saltol, a Major QTL associated with seedling stage salt tolerance, into Oryza Sativa ‘Pusa Basmati 1121. Front. Plant Sci. 2017, 8, 41. [Google Scholar] [CrossRef]
- Singh, V.K.; Singh, B.D.; Kumar, A.; Maurya, S.; Krishnan, S.G.; Vinod, K.K.; Singh, M.P.; Ellur, R.K.; Bhowmick, P.K.; Singh, A.K. Marker-assisted introgression of Saltol QTL enhances seedling stage salt tolerance in the rice variety “Pusa Basmati 1”. Int. J. Genom. 2018, 2018, 8319879. [Google Scholar] [CrossRef]
- Zafar, S.A.; Hameed, A.; Nawaz, M.A.; MA, W.; Noor, M.A.; Hussain, M.; Mehboob-ur-Rahman. Mechanisms and molecular approaches for heat tolerance in rice (Oryza sativa L.) under climate change scenario. J. Integr. Agric. 2018, 17, 726–738. [Google Scholar] [CrossRef]
- Lang, N.T.; Ha, P.T.T.; Tru, P.C.; Toan, T.B.; Buu, B.C.; Cho, Y.-C. Breeding for heat tolerance rice based on marker-assisted backcrosing in Vietnam. Plant Breed. Biotech. 2015, 3, 274–281. [Google Scholar] [CrossRef]
- Li, X.M.; Chao, D.Y.; Wu, Y.; Huang, X.; Chen, K.; Cui, L.G.; Su, L.; Ye, W.W.; Chen, H.; Chen, H.C.; et al. Natural alleles of a Proteasome A2 subunit gene contribute to thermotolerance and adaptation of African rice. Nat. Genet. 2015, 47, 827–833. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Lei, J.; Huang, Y.; Zhu, S.; Chen, H.; Huang, R.; Peng, Z.; Tu, Q.; Shen, X.; Yan, S. Mapping quantitative trait loci for heat tolerance at anthesis in rice using chromosomal segment substitution lines. Breed. Sci. 2016, 66, 358–366. [Google Scholar] [CrossRef] [PubMed]
- Ps, S.; Sv, A.M.; Prakash, C.; Mk, R.; Tiwari, R.; Mohapatra, T.; Singh, N.K. High resolution mapping of QTLs for heat tolerance in rice using a 5K SNP array. Rice 2017, 10, 28. [Google Scholar] [CrossRef]
- Shimoyama, N.; Johnson, M.; Beaumont, A.; Schläppi, M. Multiple cold tolerance trait phenotyping reveals shared quantitative trait loci in Oryza sativa. Rice 2020, 13, 57. [Google Scholar] [CrossRef]
- Septiningsih, E.M.; Sanchez, D.L.; Singh, N.; Sendon, P.M.D.; Pamplona, A.M.; Heuer, S.; Mackill, D.J. Identifying novel QTLs for submergence tolerance in rice cultivars IR72 and Madabaru. Theor. Appl. Genet. 2012, 124, 867–874. [Google Scholar] [CrossRef] [PubMed]
- Septiningsih, E.M.; Hidayatun, N.; Sanchez, D.L.; Nugraha, Y.; Carandang, J.; Pamplona, A.M.; Collard, B.C.Y.; Ismail, A.M.; Mackill, D.J. Accelerating the development of new submergence tolerant rice varieties: The case of Ciherang-Sub1 and PSB Rc18-Sub1. Euphytica 2015, 202, 259–268. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Septiningsih, E.M.; Das, S.R.; Carandang, J.J.; Pamplona, A.M.; Sanchez, D.L.; Kato, Y.; Ye, G.; Reddy, J.N.; Singh, U.S.; et al. Developing new flood-tolerant varieties at the International Rice Research Institute (IRRI). Sabrao J. Breed. Genet. 2013, 45, 42–56. [Google Scholar]
- Luo, Y.; Yin, Z. Marker-assisted breeding of Thai Fragrance rice for semi-dwarf phenotype, submergence tolerance and disease resistance to Rice Blast and Bacterial Blight. Mol. Breed. 2013, 32, 709–721. [Google Scholar] [CrossRef]
- Ma, X.; Feng, F.; Wei, H.; Mei, H.; Xu, K.; Chen, S.; Li, T.; Liang, X.; Liu, H.; Luo, L. Genome-wide association study for plant height and grain yield in rice under contrasting moisture regimes. Front. Plant Sci. 2016, 7, 1801. [Google Scholar] [CrossRef]
- Tong, C.; Hill, C.B.; Zhou, G.; Zhang, X.-Q.; Jia, Y.; Li, C.; Zhang, G.; Jia, X.-Q.; Li, Y. Opportunities for improving waterlogging tolerance in cereal crops-physiological traits and genetic mechanisms opportunities for improving waterlogging tolerance in cereal. Crops-Physiological Traits and Genetic Mechanisms. Plants 2021, 10, 1560. [Google Scholar]
- Mohd Ikmal, A.; Noraziyah, A.A.S.; Wickneswari, R.; Amira, I.; Puteri Dinie Ellina, Z. Interéaction of submergence tolerance and drought yield QTLs (Sub1 and QDTYs) enhances morpho-physiological traits and survival of rice (Oryza Sativa L.) under submergence. Ann. Appl. Biol. 2021, 178, 355–366. [Google Scholar] [CrossRef]
- Ruengphayak, S.; Chaichumpoo, E.; Phromphan, S.; Kamolsukyunyong, W.; Sukhaket, W.; Phuvanartnarubal, E.; Korinsak, S.; Korinsak, S.; Vanavichit, A. Pseudo-backcrossing design for rapidly pyramiding multiple traits into a preferential rice variety. Rice 2015, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- Kuroha, T.; Nagai, K.; Gamuyao, R.; Wang, D.R.; Furuta, T.; Nakamori, M.; Kitaoka, T.; Adachi, K.; Minami, A.; Mori, Y.; et al. Ethylene-Gibberellin signaling underlies adaptation of rice to periodic flooding. Science 2018, 361, 181–186. [Google Scholar] [CrossRef] [PubMed]
- Xiao, N.; Gao, Y.; Qian, H.; Gao, Q.; Wu, Y.; Zhang, D.; Zhang, X.; Yu, L.; Li, Y.; Pan, C.; et al. Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol. 2018, 177, 1108–1123. [Google Scholar] [CrossRef]
- Yu, J.; Zhao, W.; Tong, W.; He, Q.; Yoon, M.Y.; Li, F.P.; Choi, B.; Heo, E.B.; Kim, K.W.; Park, Y.J. A Genome-Wide Association Study Reveals Candidate Genes Related to Salt Tolerance in Rice (Oryza sativa) at the Germination Stage. Int. J. Mol. Sci. 2018, 19, 3145. [Google Scholar] [CrossRef]
- Alseekh, S.; Kostova, D.; Bulut, M.; Fernie, A.R. Genome-wide association studies: Assessing trait characteristics in model and crop plants. Cell. Mol. Life Sci. 2021, 78, 5743–5754. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, S.; Mukherjee, A.; Das, P.; Bandyopadhyay, S.; Chattopadhyay, D.; Chatterjee, J.; Majumder, A.L. A Salt-tolerant chloroplastic FBPase from Oryza coarctata confers improved photosynthesis with higher yield and multi-stress tolerance to indica rice. Plant Cell Tissue Organ Cult. 2021, 145, 561–578. [Google Scholar] [CrossRef]
- Zeng, Y.; Wen, J.; Zhao, W.; Wang, Q.; Huang, W. Rational improvement of rice yield and cold tolerance by editing the three genes OsPIN5b, GS3, and OsMYB30 with the CRISPR–Cas9 System. Front. Plant Sci. 2020, 10, 1663. [Google Scholar] [CrossRef]
- Usman, B.; Nawaz, G.; Zhao, N.; Liao, S.; Liu, Y.; Li, R. Precise editing of the Ospyl9 gene by RNA-Guided Cas9 nuclease confers enhanced drought tolerance and grain yield in rice (Oryza Sativa L.) by regulating circadian rhythm and abiotic stress responsive proteins. Int. J. Mol. Sci. 2020, 21, 7854. [Google Scholar] [CrossRef]
- Ogata, T.; Ishizaki, T.; Fujita, M.; Fujita, Y. CRISPR/Cas9-Targeted mutagenesis of OsERA1 confers enhanced responses to abscisic acid and drought stress and increased primary root growth under nonstressed conditions in rice. PLoS ONE 2020, 15, e0243376. [Google Scholar] [CrossRef] [PubMed]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-Targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef]
- Santosh Kumar, V.V.; Verma, R.K.; Yadav, S.K.; Yadav, P.; Watts, A.; Rao, M.V.; Chinnusamy, V. CRISPR-Cas9 mediated genome editing of drought and salt tolerance (OsDST) gene in Indica mega rice cultivar MTU1010. Physiol. Mol. Biol. Plants 2020, 26, 1099–1110. [Google Scholar] [CrossRef]
- Zang, X.; Geng, X.; Wang, F.; Liu, Z.; Zhang, L.; Zhao, Y.; Tian, X.; Ni, Z.; Yao, Y.; Xin, M.; et al. Overexpression of wheat Ferritin gene TaFER-5B enhances tolerance to heat stress and other abiotic stresses associated with the ROS scavenging. BMC Plant Biol. 2017, 17, 14. [Google Scholar] [CrossRef] [PubMed]
- Mega, R.; Abe, F.; Kim, J.S.; Tsuboi, Y.; Tanaka, K.; Kobayashi, H.; Sakata, Y.; Hanada, K.; Tsujimoto, H.; Kikuchi, J.; et al. Tuning water-use efficiency and drought tolerance in wheat using abscisic acid receptors. Nat. Plants 2019, 5, 153–159. [Google Scholar] [CrossRef]
- Ayadi, M.; Brini, F.; Masmoudi, K. Overexpression of a wheat Aquaporin gene, Tdpip2;1, enhances salt and drought tolerance in transgenic durum wheat Cv. Maali. Int. J. Mol. Sci. 2019, 20, 2389. [Google Scholar] [CrossRef]
- Qin, N.; Xu, W.; Hu, L.; Li, Y.; Wang, H.; Qi, X.; Fang, Y.; Hua, X. Drought tolerance and proteomics studies of transgenic wheat containing the maize C4 Phosphoenolpyruvate Carboxylase (PEPC) gene. Protoplasma 2016, 253, 1503–1512. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Xu, W.; Zhang, J.; Guo, R.; Zhao, M.; Hu, L.; Wang, H.; Dong, H.; Li, Y. Physiological characteristics and metabolomics of transgenic wheat containing the maize C4 Phosphoenolpyruvate Carboxylase (PEPC) gene under high temperature stress. Protoplasma 2017, 254, 1017–1030. [Google Scholar] [CrossRef]
- Gao, H.; Wang, Y.; Xu, P.; Zhang, Z. Overexpression of a WRKY Transcription Factor TaWRKY2 enhances drought stress tolerance in transgenic wheat. Front. Plant Sci. 2018, 9, 997. [Google Scholar] [CrossRef]
- Cui, X.Y.; Gao, Y.; Guo, J.; Yu, T.F.; Zheng, W.J.; Liu, Y.W.; Chen, J.; Xu, Z.S.; Ma, Y.Z. BES/BZR Transcription Factor TaBZR2 positively regulates drought responses by activation of TaGST1. Plant Physiol. 2019, 180, 605–620. [Google Scholar] [CrossRef] [PubMed]
- Zang, X.; Geng, X.; He, K.; Wang, F.; Tian, X.; Xin, M.; Yao, Y.; Hu, Z.; Ni, Z.; Sun, Q.; et al. Overexpression of the wheat (Triticum aestivum L.) TaPEPKR2 gene enhances heat and dehydration tolerance in both wheat and Arabidopsis. Front. Plant Sci. 2018, 9, 1710. [Google Scholar] [CrossRef]
- le Roux, M.L.; Kunert, K.J.; van der Vyver, C.; Cullis, C.A.; Botha, A.M. Expression of a small Ubiquitin-like modifier protease increases drought tolerance in wheat (Triticum aestivum L.). Front. Plant Sci. 2019, 10, 266. [Google Scholar] [CrossRef]
- Kim, D.; Alptekin, B.; Budak, H. CRISPR/Cas9 Genome Editing in wheat. Funct. Integr. Genom. 2018, 18, 31–41. [Google Scholar] [CrossRef]
- Wei, A.Y.; He, C.M.; Li, B.; Li, N.; Zhang, J.R. The pyramid of transgenes TsVP and BetA effectively enhances the drought tolerance of maize plants. Plant Biotech. J. 2011, 9, 216–229. [Google Scholar] [CrossRef] [PubMed]
- Castiglioni, P.; Warner, D.; Bensen, R.J.; Anstrom, D.C.; Harrison, J.; Stoecker, M.; Abad, M.; Kumar, G.; Salvador, S.; D’Ordine, R.; et al. Bacterial RNA chaperones confer abiotic stress tolerance in plants and improved grain yield in maize under water-limited conditions. Plant Physiol. 2008, 147, 446–455. [Google Scholar] [CrossRef]
- Du, H.; Shen, X.; Huang, Y.; Huang, M.; Zhang, Z. Overexpression of Vitreoscilla Hemoglobin increases waterlogging tolerance in Arabidopsis and Maize. BMC Plant Biol. 2016, 16, 35. [Google Scholar] [CrossRef]
- Liu, Y.; Qin, L.; Han, L.; Xiang, Y.; Zhao, D. Overexpression of Maize SDD1 (ZmSDD1) improves drought resistance in Zea mays L. by reducing stomatal density. Plant Cell Tissue Organ Cult. 2015, 122, 147–159. [Google Scholar] [CrossRef]
- Casaretto, J.A.; El-kereamy, A.; Zeng, B.; Stiegelmeyer, S.M.; Chen, X.; Bi, Y.M.; Rothstein, S.J. Expression of OsMYB55 in Maize activates stress-responsive genes and enhances heat and drought tolerance. BMC Genom. 2016, 17, 312. [Google Scholar] [CrossRef]
- Yu, F.; Liang, K.; Fang, T.; Zhao, H.; Han, X.; Cai, M.; Qiu, F. A Group VII Ethylene Response Factor gene, ZmEREB180, coordinates waterlogging tolerance in maize seedlings. Plant Biotech. J. 2019, 17, 2286–2298. [Google Scholar] [CrossRef]
- Shi, J.; Gao, H.; Wang, H.; Lafitte, H.R.; Archibald, R.L.; Yang, M.; Hakimi, S.M.; Mo, H.; Habben, J.E. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotech. J. 2017, 15, 207–216. [Google Scholar] [CrossRef]
- Zhang, M.; Cao, Y.; Wang, Z.; Wang, Z.Q.; Shi, J.; Liang, X.; Song, W.; Chen, Q.; Lai, J.; Jiang, C. A Retrotransposon in an HKT1 family sodium transporter causes variation of leaf Na+ exclusion and salt tolerance in maize. New Phytol. 2018, 217, 1161–1176. [Google Scholar] [CrossRef] [PubMed]
- Roy, B.; Noren, S.K.; Mandal, A.B.; Basu, A.K. Genetic engineering for abiotic stress tolerance in agricultural crops. Biotechnology 2011, 10, 1–22. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.K.; Yoon, I.S.; Kim, K.H.; Kwon, T.R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Yasuda, H. Cross-tolerance to thermal stresses and its application to the development of cold tolerant rice. Jpn. Agric. Res. Q. 2017, 51, 99–105. [Google Scholar] [CrossRef]
- Baillo, E.H.; Kimotho, R.N.; Zhang, Z.; Xu, P. Transcription factors associated with abiotic and biotic stress tolerance and their potential for crops improvement. Genes 2019, 10, 771. [Google Scholar] [CrossRef]
- Kumar, V.; Datir, S.; Khare, T.; Shriram, V. Advances in biotechnological tools: Improving abiotic stress tolerance in rice. In Advances in Rice Research for Abiotic Stress Tolerance; Woodhead Publishing: Sawston, CA, USA, 2018; ISBN 9780128143322. [Google Scholar]
- Hima Kumari, P.; Venkatesh, K.; Krupanidhi, S.; Anil Kumar, S. Anil Kumar, S. An update on molecular strategies of transgenic rice tolerance to abiotic stresses. In Molecular Approaches in Plant Biology and Environmental Challenges. Energy, Environment, and Sustainability; Singh, S., Upadhyay, S., Pandey, A., Kumar, S., Eds.; Springer: Singapore, 2019. [Google Scholar]
- Tabassum, J.; Ahmad, S.; Hussain, B.; Mawia, A.M.; Zeb, A.; Ju, L. Applications and potential of genome-editing systems in rice improvement: Current and future perspectives. Agronomy 2021, 11, 1359. [Google Scholar] [CrossRef]
- Boonchai, C.; Udomchalothorn, T.; Sripinyowanich, S.; Comai, L.; Buaboocha, T.; Chadchawan, S. Rice overexpressing OsNUC1-s reveals differential gene expression leading to yield loss reduction after salt stress at the booting stage. Int. J. Mol. Sci. 2018, 19, 3936. [Google Scholar] [CrossRef]
- Wu, T.; Zhang, M.; Zhang, H.; Huang, K.; Chen, M.; Chen, C.; Yang, X.; Li, Z.; Chen, H.; Ma, Z.; et al. Identification and characterization of EDT1 conferring drought tolerance in rice. J. Plant Biol. 2019, 62, 39–47. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Jan, A.; Ishizaki, T.; Valencia, M.; Dedicova, B.; Maruyama, K.; Ogata, T.; Todaka, D.; Yamaguchi-Shinozaki, K.; Nakashima, K.; et al. Expression of the CCCH-Tandem Zinc Finger protein gene OsTZF5 under a stress-inducible promoter mitigates the effect of drought stress on rice grain yield under field conditions. Plant Biotech. J. 2020, 18, 1711–1721. [Google Scholar] [CrossRef]
- Qin, Q.; Wang, Y.; Huang, L.; Du, F.; Zhao, X.; Li, Z.; Wang, W.; Fu, B. A U-Box E3 Ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 2020, 102, 89–107. [Google Scholar] [CrossRef]
- Darwish, E.; Rehman, S.U.; Mao, X.; Jing, R. A Wheat Stress Induced WRKY Transcription factor TaWRKY32 confers drought stress tolerance in Oryza sativa. Asian J. Agric. Biol. 2021, 2021, 1–7. [Google Scholar] [CrossRef]
- Park, S.I.; Kwon, H.J.; Cho, M.H.; Song, J.S.; Kim, B.G.; Baek, J.H.; Kim, S.L.; Ji, H.S.; Kwon, T.R.; Kim, K.H.; et al. The OSERF115/AP2EREBP110 Transcription factor is involved in the multiple stress tolerance to heat and drought in rice plants. Int. J. Mol. Sci. 2021, 22, 7181. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Yu, C.; Liu, Y.; Yang, L.; Li, Y.; Yao, W.; Cai, Y.; Yan, X.; Li, S.; Cai, Y.; et al. GmFAD3A, A ω-3 Fatty acid desaturase gene, enhances cold tolerance and seed germination rate under low temperature in rice. Int. J. Mol. Sci. 2019, 20, 3796. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.; Feng, Q.; Lu, H.; Li, Y.; Wang, A.; Tian, Q.; Zhan, Q.; Lu, Y.; Zhang, L.; Huang, T.; et al. Pan-Genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat. Genet. 2018, 50, 278–284. [Google Scholar] [CrossRef]
- Wang, W.S.; Pan, Y.J.; Zhao, X.Q.; Dwivedi, D.; Zhu, L.H.; Ali, J.; Fu, B.Y.; Li, Z.K. Drought-induced site-specific DNA methylation and its association with drought tolerance in rice (Oryza Sativa L.). J. Exp. Bot. 2011, 62, 1951–1960. [Google Scholar] [CrossRef]
- Saraswat, S.; Yadav, A.K.; Sirohi, P.; Singh, N.K. Role of Epigenetics in crop improvement: Water and heat stress. J. Plant Biol. 2017, 60, 231–240. [Google Scholar] [CrossRef]
- Biswas, S.; Zhang, D.; Shi, J. CRISPR/Cas Systems: Opportunities and challenges for crop breeding. Plant Cell Rep. 2021, 40, 979–998. [Google Scholar] [CrossRef]
- Ansari, W.A.; Chandanshive, S.U.; Bhatt, V.; Nadaf, A.B.; Vats, S.; Katara, J.L.; Sonah, H.; Deshmukh, R. Genome Editing in Cereals: Approaches, Applications and Challenges. Int. J. Mol. Sci. 2020, 21, 4040. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; SkZ, A. Enhancement of drought stress tolerance in crops by plant growth promoting Rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Etesami, H.; Beattie, G.A. Mining halophytes for plant growth-promoting halotolerant bacteria to enhance the salinity tolerance of non-halophytic crops. Front. Microbiol. 2018, 9, 148. [Google Scholar] [CrossRef]
- Ji, J.; Yuan, D.; Jin, C.; Wang, G.; Li, X.; Guan, C. Enhancement of growth and salt tolerance of rice seedlings (Oryza Sativa L.) by regulating ethylene production with a novel halotolerant PGPR strain Glutamicibacter sp. YD01 containing ACC deaminase activity. Acta Physiol. Plant. 2020, 42, 42. [Google Scholar] [CrossRef]
- Joshi, B.; Chaudhary, A.; Singh, H.; Kumar, P.A. Prospective evaluation of individual and consortia plant growth promoting Rhizobacteria for drought stress amelioration in rice (Oryza Sativa L.). Plant Soil 2020, 457, 225–240. [Google Scholar] [CrossRef]
- Naik, K.; Mishra, S.; Srichandan, H.; Singh, P.K.; Choudhary, A. Microbial formulation and growth of cereals, pulses, oilseeds and vegetable crops. Sustain. Environ. Res. 2020, 30, 10. [Google Scholar] [CrossRef]
- Bonatelli, M.L.; Lacerda-Júnior, G.V.; dos Reis Junior, F.B.; Fernandes-Júnior, P.I.; Melo, I.S.; Quecine, M.C. Beneficial plant-associated microorganisms from semiarid regions and seasonally dry environments: A review. Front. Microbiol. 2021, 11, 553223. [Google Scholar] [CrossRef]
- Verma, H.; Kumar, D.; Kumar, V.; Kumari, M.; Singh, S.K.; Sharma, V.K.; Droby, S.; Santoyo, G.; White, J.F.; Kumar, A. The potential application of endophytes in management of stress from drought and salinity in crop plants. Microorganisms 2021, 9, 1729. [Google Scholar] [CrossRef]
- Islam, F.; Yasmeen, T.; Ali, Q.; Ali, S.; Arif, M.S.; Hussain, S.; Rizvi, H. Influence of Pseudomonas aeruginosa as PGPR on oxidative stress tolerance in wheat under Zn stress. Ecotoxicol. Environ. Saf. 2014, 104, 285–293. [Google Scholar]
- Timmusk, S.; Abd El-Daim, I.A.; Copolovici, L.; Tanilas, T.; Kännaste, A.; Behers, L.; Nevo, E.; Seisenbaeva, G.; Stenström, E.; Niinemets, Ü. Drought-tolerance of wheat improved by rhizosphere bacteria from harsh environments: Enhanced biomass production and reduced emissions of stress volatiles. PLoS ONE 2014, 9, e96086. [Google Scholar] [CrossRef]
- Barnawal, D.; Bharti, N.; Pandey, S.S.; Pandey, A.; Chanotiya, C.S.; Kalra, A. Plant Growth-Promoting Rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plant. 2017, 161, 502–514. [Google Scholar] [CrossRef]
- Nawaz, A.; Shahbaz, M.; Asadullah, M.; Imran, A.; Marghoob, M.U.; Imtiaz, M.; Mubeen, F. Potential of salt tolerant PGPR in growth and yield augmentation of wheat (Triticum aestivum L.) under saline conditions. Front. Microbiol. 2020, 11, 2019. [Google Scholar] [CrossRef]
- Rodríguez-Salazar, J.; Suárez, R.; Caballero-Mellado, J.; Iturriaga, G. Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett. 2009, 296, 52–59. [Google Scholar] [CrossRef]
- Curá, J.A.; Franz, D.R.; Filosofía, J.E.; Balestrasse, K.B.; Burgueño, L.E. Inoculation with Azospirillum sp. and Herbaspirillum sp. bacteria increases the tolerance of maize to drought stress. Microorganisms 2017, 5, 41. [Google Scholar] [CrossRef] [PubMed]
- Shirinbayan, S.; Khosravi, H.; Malakouti, M.J. Alleviation of drought stress in maize (Zea mays) by inoculation with Azotobacter strains isolated from semi-arid regions. Appl. Soil Ecol. 2019, 133, 138–145. [Google Scholar] [CrossRef]
- Checchio, M.V.; de Cássia Alves, R.; de Oliveira, K.R.; Moro, G.V.; dos Santos, D.M.M.; Gratão, P.L. Enhancement of salt tolerance in corn using Azospirillum brasilense: An approach on antioxidant systems. J. Plant Res. 2021, 134, 1279–1289. [Google Scholar] [CrossRef] [PubMed]
- International Wheat Genome Sequencing Consortium (IWGSC). Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [PubMed]
- Khadka, K.; Raizada, M.N.; Navabi, A. Recent progress in germplasm evaluation and gene mapping to enable breeding of drought-tolerant wheat. Front. Plant Sci. 2020, 11, 1149. [Google Scholar] [CrossRef] [PubMed]
- Barakat, M.N.; Saleh, M.S.; Al-Doss, A.A.; Moustafa, K.A.; Elshafei, A.A.; Zakri, A.M.; Al-Qurainy, F.H. Mapping of QTLs associated with abscisic acid and water stress in wheat. Biol. Plant. 2015, 59, 291–297. [Google Scholar] [CrossRef]
- Malik, S.; Malik, T.A. Genetic mapping of potential QTLs associated with drought tolerance in wheat. J. Anim. Plant Sci. 2015, 25, 1032–1040. [Google Scholar]
- Goel, S.; Singh, K.; Grewal, S.; Nath, M. Impact of “Omics” in improving drought tolerance in wheat. Crit. Rev. Plant Sci. 2020, 39, 222–235. [Google Scholar] [CrossRef]
- Colmer, T.D.; Flowers, T.J.; Munns, R. Use of wild relatives to improve salt tolerance in wheat. J. Exp. Bot. 2006, 57, 1059–1078. [Google Scholar] [CrossRef] [PubMed]
- Placido, D.F.; Campbell, M.T.; Folsom, J.J.; Cui, X.; Kruger, G.R.; Baenziger, P.S.; Walia, H. Introgression of novel traits from a wild wheat relative improves drought adaptation in wheat. Plant Physiol. 2013, 161, 1806–1819. [Google Scholar] [CrossRef] [PubMed]
- Raina, A.; Laskar, R.; Khursheed, S.; Amin, R.; Tantray, Y.; Parveen, K.; Khan, S. Role of Mutation breeding in crop improvement- past, present and future. Asian Res. J. Agric. 2016, 2, 1–13. [Google Scholar] [CrossRef]
- Sen, A.; Ozturk, I.; Yaycili, O.; Alikamanoglu, S. Drought tolerance in irradiated wheat mutants studied by genetic and biochemical markers. J. Plant Growth Regul. 2017, 36, 669–679. [Google Scholar] [CrossRef]
- Jankowicz-Cieslak, J.; Mba, C.; Till, B.J. Mutagenesis for crop breeding and functional genomics. In Biotechnologies for Plant Mutation Breeding: Protocols; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Todkar, L.; Singh, G.P.; Jain, N.; Singh, P.K.; Prabhu, K.V. Introgression of drought tolerance QTLs through marker assisted backcross breeding in wheat (Triticum aestivum L.). Indian J. Genet. Plant Breed. 2020, 80, 209–212. [Google Scholar] [CrossRef]
- Khalid, M.; Afzal, F.; Gul, A.; Amir, R.; Subhani, A.; Ahmed, Z.; Mahmood, Z.; Xia, X.; Rasheed, A.; He, Z. Molecular characterization of 87 functional genes in wheat diversity panel and their association with phenotypes under well-watered and water-limited conditions. Front. Plant Sci. 2019, 10, 717. [Google Scholar] [CrossRef]
- Ballesteros, D.C.; Mason, R.E.; Addison, C.K.; Andrea Acuña, M.; Nelly Arguello, M.; Subramanian, N.; Miller, R.G.; Sater, H.; Gbur, E.E.; Miller, D.; et al. Tolerance of wheat to vegetative stage soil waterlogging is conditioned by both constitutive and adaptive QTL. Euphytica 2015, 201, 329–343. [Google Scholar] [CrossRef]
- Khan, S.; Anwar, S.; Yu, S.; Sun, M.; Yang, Z.; Gao, Z.-Q. Development of drought-tolerant transgenic wheat: Achievements and limitations. Int. J. Mol. Sci. 2019, 20, 3350. [Google Scholar] [CrossRef]
- Abebe, T.; Guenzi, A.C.; Martin, B.; Cushman, J.C. Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiol. 2003, 131, 1748–1755. [Google Scholar] [CrossRef]
- Wang, G.P.; Hui, Z.; Li, F.; Zhao, M.R.; Zhang, J.; Wang, W. Improvement of heat and drought photosynthetic tolerance in wheat by overaccumulation of glycinebetaine. Plant Biotech. Rep. 2010, 4, 213–222. [Google Scholar] [CrossRef]
- He, C.; Zhang, W.; Gao, Q.; Yang, A.; Hu, X.; Zhang, J. Enhancement of drought resistance and biomass by increasing the amount of glycine betaine in wheat seedlings. Euphytica 2011, 177, 151–167. [Google Scholar] [CrossRef]
- Pavei, D.; Gonçalves-Vidigal, M.C.; Schuelter, A.R.; Schuster, I.; Vieira, E.S.N.; Vendruscolo, E.C.G.; Poletine, J.P. Response to water stress in transgenic (P5CS gene) wheat plants (Triticum aestivum L.). Aust. J. Crop Sci. 2016, 10, 776–783. [Google Scholar] [CrossRef]
- Tian, F.; Wang, W.; Liang, C.; Wang, X.; Wang, G.; Wang, W. Overaccumulation of glycine betaine makes the function of the thylakoid membrane better in wheat under salt stress. Crop J. 2017, 5, 73–82. [Google Scholar] [CrossRef]
- Maghsoudi, K.; Emam, Y.; Niazi, A.; Pessarakli, M.; Arvin, M.J. P5CS expression level and proline accumulation in the sensitive and tolerant wheat cultivars under control and drought stress conditions in the presence/absence of silicon and salicylic acid. J. Plant Interact. 2018, 13, 461–471. [Google Scholar] [CrossRef]
- Bahieldin, A.; Mahfouz, H.T.; Eissa, H.F.; Saleh, O.M.; Ramadan, A.M.; Ahmed, I.A.; Dyer, W.E.; El-Itriby, H.A.; Madkour, M.A. Field evaluation of transgenic wheat plants stably expressing the HVA1 gene for drought tolerance. Physiol. Plant. 2005, 123, 421–427. [Google Scholar] [CrossRef]
- Yu, T.F.; Xu, Z.S.; Guo, J.K.; Wang, Y.X.; Abernathy, B.; Fu, J.D.; Chen, X.; Zhou, Y.-B.; Chen, M.; Ye, X.G.; et al. Improved drought tolerance in wheat plants overexpressing a synthetic bacterial cold shock protein gene SeCspA. Sci. Rep. 2017, 7, 44050. [Google Scholar] [CrossRef] [PubMed]
- Ben-Saad, R.; Ben-Ramdhan, W.; Zouari, N.; Azaza, J.; Mieulet, D.; Guiderdoni, E.; Ellouz, R.; Hassairi, A. Marker-Free Transgenic Durum Wheat Cv. Karim Expressing the AlSAP gene exhibits a high level of tolerance to salinity and dehydration stresses. Mol. Breed. 2012, 30, 521–533. [Google Scholar] [CrossRef]
- Zhang, H.F.; Xu, W.G.; Wang, H.W.; Hu, L.; Li, Y.; Qi, X.L.; Zhang, L.; Li, C.X.; Hua, X. Pyramiding expression of maize genes encoding Phosphoenolpyruvate Carboxylase (PEPC) and Pyruvate Orthophosphate Dikinase (PPDK) synergistically improve the photosynthetic characteristics of transgenic wheat. Protoplasma 2014, 251, 1163–1173. [Google Scholar] [CrossRef] [PubMed]
- Hou, J.; Huang, X.; Sun, W.; Du, C.; Wang, C.; Xie, Y.; Ma, Y.; Ma, D. Accumulation of water-soluble carbohydrates and gene expression in wheat stems correlates with drought resistance. J. Plant Physiol. 2018, 231, 182–191. [Google Scholar] [CrossRef]
- Gao, S.; Xu, H.; Cheng, X.; Chen, M.; Xu, Z.; Li, L.; Ye, X.; Du, L.; Hao, X.; Ma, Y. Improvement of wheat drought and salt tolerance by expression of a stress-inducible transcription factor GmDREB of soybean (Glycine max). Chin. Sci. Bull. 2005, 50, 2714–2723. [Google Scholar] [CrossRef]
- Xue, G.P.; Way, H.M.; Richardson, T.; Drenth, J.; Joyce, P.A.; McIntyre, C.L. Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 2011, 4, 697–712. [Google Scholar] [CrossRef]
- Yadav, D.; Shavrukov, Y.; Bazanova, N.; Chirkova, L.; Borisjuk, N.; Kovalchuk, N.; Ismagul, A.; Parent, B.; Langridge, P.; Hrmova, M.; et al. Constitutive overexpression of the TaNF-YB4 gene in transgenic wheat significantly improves grain yield. J. Exp. Bot. 2015, 66, 6635–6650. [Google Scholar] [CrossRef]
- Li, L.; Zheng, M.; Deng, G.; Liang, J.; Zhang, H.; Pan, Z.; Long, H.; Yu, M. Overexpression of AtHDG11 enhanced drought tolerance in wheat (Triticum aestivum L.). Mol. Breed. 2016, 36, 23. [Google Scholar] [CrossRef]
- Bi, H.; Shi, J.; Kovalchuk, N.; Luang, S.; Bazanova, N.; Chirkova, L.; Zhang, D.; Shavrukov, Y.; Stepanenko, A.; Tricker, P.; et al. Overexpression of the TaSHN1 transcription factor in bread wheat leads to leaf surface modifications, improved drought tolerance, and no yield penalty under controlled growth conditions. Plant Cell Environ. 2018, 41, 2549–2566. [Google Scholar] [CrossRef]
- Luang, S.; Sornaraj, P.; Bazanova, N.; Jia, W.; Eini, O.; Hussain, S.S.; Kovalchuk, N.; Agarwal, P.K.; Hrmova, M.; Lopato, S. The Wheat TabZIP2 transcription factor is activated by the nutrient starvation-responsive SnRK3/CIPK protein kinase. Plant Mol. Biol. 2018, 96, 543–561. [Google Scholar] [CrossRef] [PubMed]
- Noor, S.; Ali, S.; Ali, G.M. Comparative study of transgenic (DREB1A) and non-transgenic wheat lines on relative water content, sugar, proline and chlorophyll under drought and salt stresses. Sarhad J. Agric. 2018, 34, 986–993. [Google Scholar] [CrossRef]
- Guerra, D.; Crosatti, C.; Khoshro, H.H.; Mastrangelo, A.M.; Mica, E.; Mazzucotelli, E. Post-Transcriptional and Post-Translational regulations of drought and heat response in plants: A Spider’s Web of mechanisms. Front. Plant Sci. 2015, 6, 57. [Google Scholar] [CrossRef]
- Cui, X.Y.; Du, Y.T.; Fu, J.d.; Yu, T.F.; Wang, C.T.; Chen, M.; Chen, J.; Ma, Y.Z.; Xu, Z.S. Wheat CBL-Interacting Protein Kinase 23 positively regulates drought stress and ABA responses. BMC Plant Biol. 2018, 18, 93. [Google Scholar] [CrossRef]
- Montenegro, J.D.; Golicz, A.A.; Bayer, P.E.; Hurgobin, B.; Lee, H.T.; Chan, C.K.K.; Visendi, P.; Lai, K.; Doležel, J.; Batley, J.; et al. The Pangenome of hexaploid bread wheat. Plant J. 2017, 90, 1007–1013. [Google Scholar] [CrossRef]
- Ribaut, J.M. Drought Adaptation in Cereals; The Haworth Press Inc.: Binghamton, NY, USA, 2006; p. 682. ISBN 9781560222781. [Google Scholar]
- Ribaut, J.M.; Ragot, M. Marker-assisted selection to improve drought adaptation in maize: The backcross approach, perspectives, limitations, and alternatives. J. Exp. Bot. 2007, 58, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Gong, F.; Takahashi, H.; Omori, F.; Wang, W.; Mano, Y.; Nakazono, M. QTLs for constitutive aerenchyma from Zea nicaraguensis improve tolerance of maize to root-zone oxygen deficiency. J. Exp. Bot. 2019, 70, 6475–6487. [Google Scholar] [CrossRef]
- Prasanna, B.M.; Cairns, J.E.; Zaidi, P.H.; Beyene, Y.; Makumbi, D.; Gowda, M.; Magorokosho, C.; Zaman-Allah, M.; Olsen, M.; Das, A.; et al. Beat the stress: Breeding for climate resilience in maize for the tropical rainfed environments. Theor. Appl. Genet. 2021, 134, 1729–1752. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Liu, H.; Wu, L.; Warburton, M.; Yan, J. Genome-Wide Association Studies in maize: Praise and stargaze. Mol. Plant 2017, 10, 359–374. [Google Scholar] [CrossRef]
- Lopes, M.S.; Araus, J.L.; van Heerden, P.D.R.; Foyer, C.H. Enhancing drought tolerance in C4 crops. J. Exp. Bot. 2011, 62, 3135–3153. [Google Scholar] [CrossRef]
- Shikha, M.; Kanika, A.; Rao, A.R.; Mallikarjuna, M.G.; Gupta, H.S.; Nepolean, T. Genomic selection for drought tolerance using Genome-Wide SNPs in Maize. Front. Plant Sci. 2017, 8, 550. [Google Scholar] [CrossRef]
- Abiko, T.; Kotula, L.; Shiono, K.; Malik, A.I.; Colmer, T.D.; Nakazono, M. Enhanced formation of aerenchyma and induction of a barrier to radial oxygen loss in adventitious roots of Zea Nicaraguensis contribute to its waterlogging tolerance as compared with Maize (Zea mays ssp. mays). Plant Cell Environ. 2012, 35, 1618–1630. [Google Scholar] [CrossRef]
- Sharma, D.; Khulbe, R.K.; Pal, R.S.; Bettanaika, J.; Kant, L. Wild progenitor and landraces led genetic gain in the modern-day Maize (Zea mays L.). In Landraces-Traditional Variety and Natural Breed; IntechOpen: London, UK, 2021; pp. 1–16. [Google Scholar]
- Hirsch, C.N.; Foerster, J.M.; Johnson, J.M.; Sekhon, R.S.; Muttoni, G.; Vaillancourt, B.; Peñagaricano, F.; Lindquist, E.; Pedraza, M.A.; Barry, K.; et al. Insights into the Maize Pan-Genome and Pan-Transcriptome. Plant Cell 2014, 26, 121–135. [Google Scholar] [CrossRef]
- Bayer, P.E.; Golicz, A.A.; Scheben, A.; Batley, J.; Edwards, D. Plant Pan-Genomes are the new reference. Nat. Plants 2020, 6, 914–920. [Google Scholar] [CrossRef]
- Woodhouse, M.R.; Cannon, E.K.; Portwood, J.L.; Harper, L.C.; Gardiner, J.M.; Schaeffer, M.L.; Andorf, C.M. A Pan-Genomic approach to genome databases using maize as a model system. BMC Plant Biol. 2021, 21, 385. [Google Scholar] [CrossRef]
- Li, C.; Sun, B.; Li, Y.; Liu, C.; Wu, X.; Zhang, D.; Shi, Y.; Song, Y.; Buckler, E.S.; Zhang, Z.; et al. Numerous genetic loci identified for drought tolerance in the maize nested association mapping populations. BMC Genom. 2016, 17, 894. [Google Scholar] [CrossRef]
- Kaur, B.; Sandhu, K.S.; Kamal, R.; Kaur, K.; Singh, J.; Röder, M.S.; Muqaddasi, Q.H. Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: Applications, challenges, and prospects. Plants 2021, 10, 1989. [Google Scholar] [CrossRef]
- Shou, H.; Bordallo, P.; Wang, K. Expression of the Nicotiana Protein Kinase (NPK1) enhanced drought tolerance in transgenic Maize. J. Exp. Bot. 2004, 55, 1013–1019. [Google Scholar] [CrossRef]
- Nelson, D.E.; Repetti, P.P.; Adams, T.R.; Creelman, R.A.; Wu, J.; Warner, D.C.; Anstrom, D.C.; Bensen, R.J.; Castiglioni, P.P.; Donnarummo, M.G.; et al. Plant Nuclear Factor Y (NF-Y) B subunits confer drought tolerance and lead to improved corn yields on water-limited acres. Proc. Natl. Acad. Sci. USA 2007, 104, 16450–16455. [Google Scholar] [CrossRef] [PubMed]
- Huanca-Mamani, W.; Arias-Carrasco, R.; Cárdenas-Ninasivincha, S.; Rojas-Herrera, M.; Sepúlveda-Hermosilla, G.; Caris-Maldonado, J.C.; Bastías, E.; Maracaja-Coutinho, V. Long Non-Coding RNAs responsive to salt and boron stress in the hyper-arid Lluteño Maize from Atacama Desert. Genes 2018, 9, 170. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.J.; Jian, L.; Xu, J.; Zhang, Q.; Zhang, M.; Jin, M.; Peng, Y.; Yan, J.; Han, B.; Liu, J.; et al. High-Throughput CRISPR/Cas9 mutagenesis streamlines trait gene identification in maize. Plant Cell 2020, 32, 1397–1413. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, C.; Lenderts, B.; Feigenbutz, L.; Barone, P.; Llaca, V.; Fengler, K.; Svitashev, S. CRISPR–Cas9-Mediated 75.5-Mb inversion in maize. Nat. Plants 2020, 6, 1427–1431. [Google Scholar] [CrossRef]
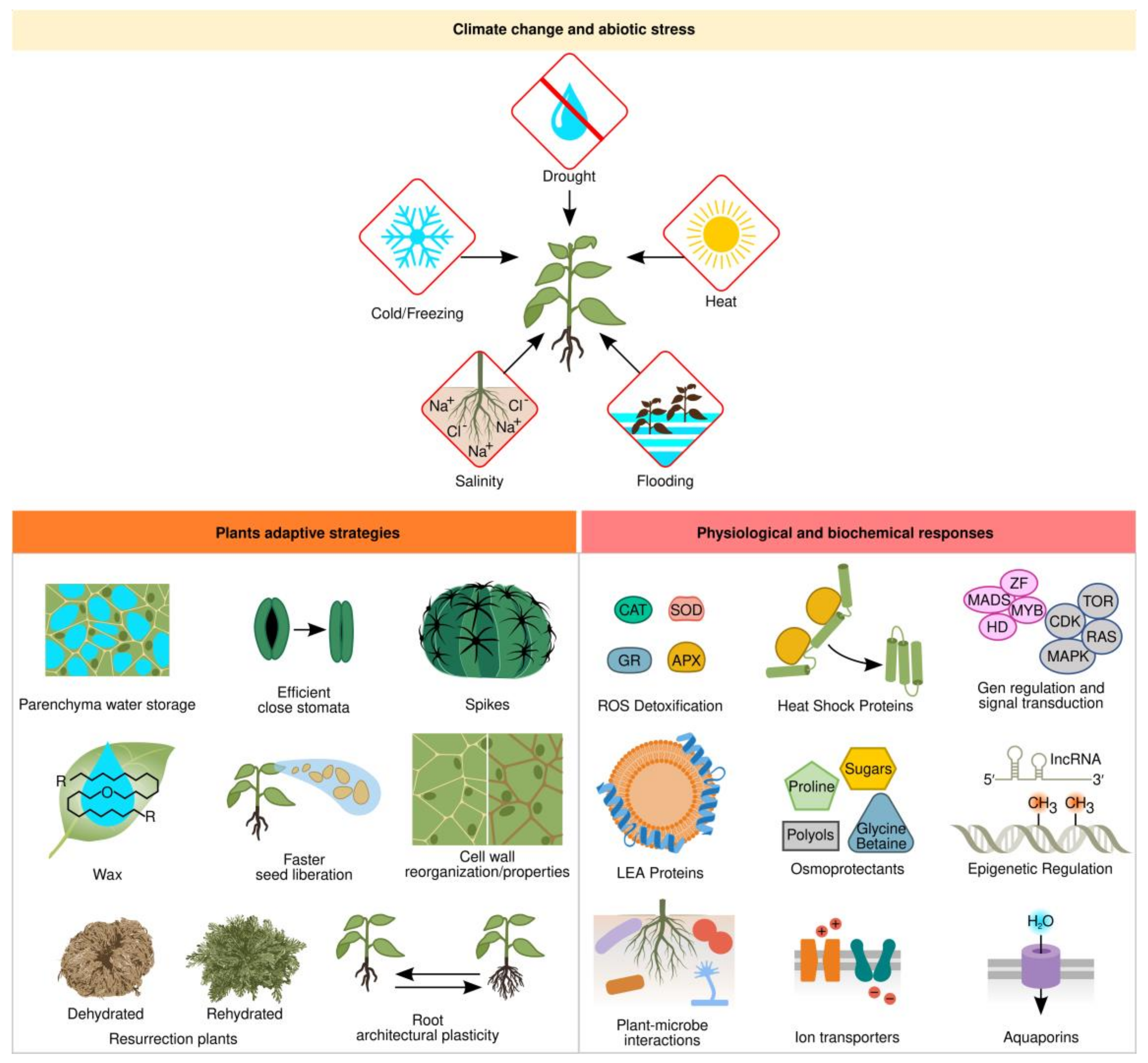
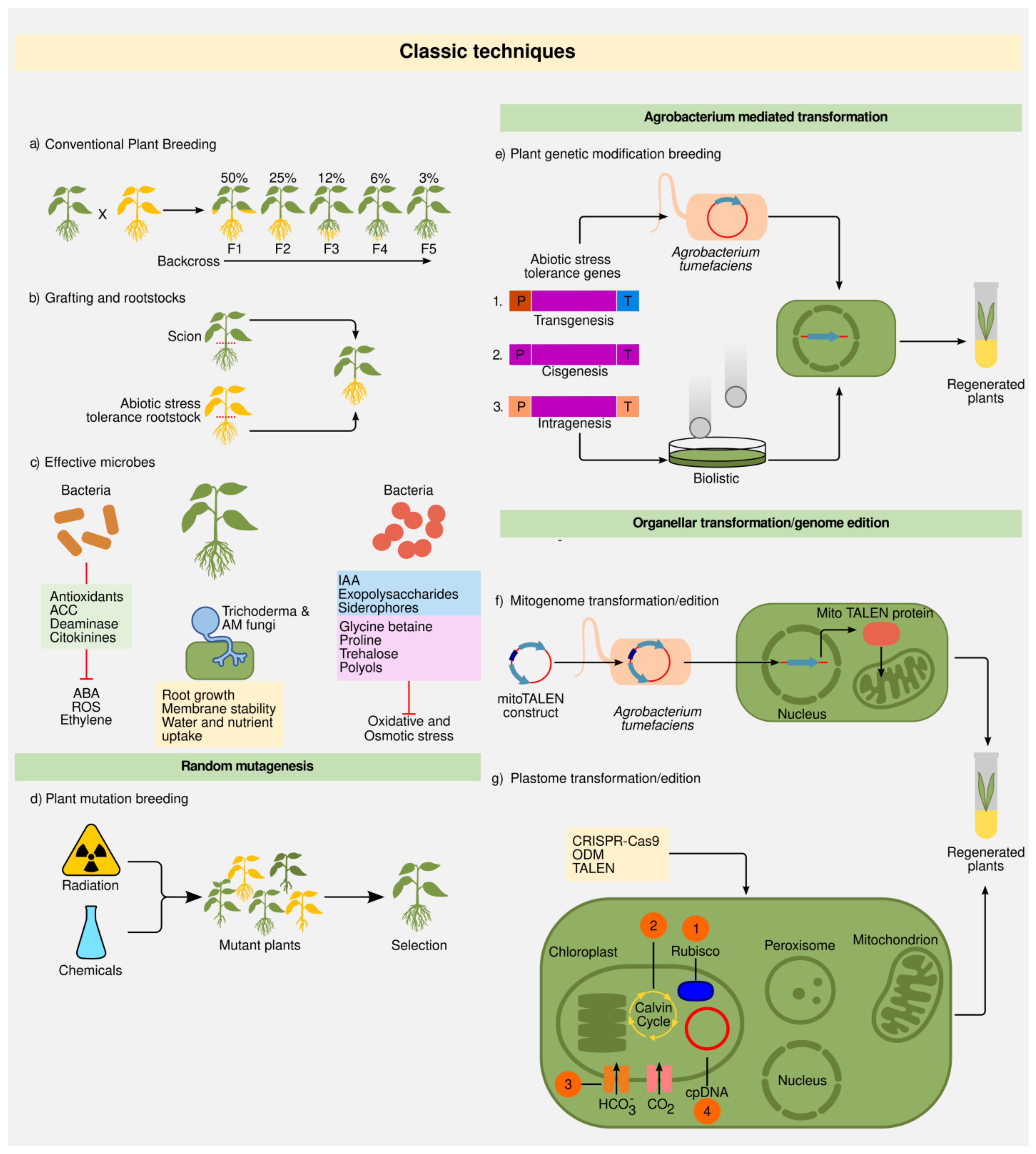
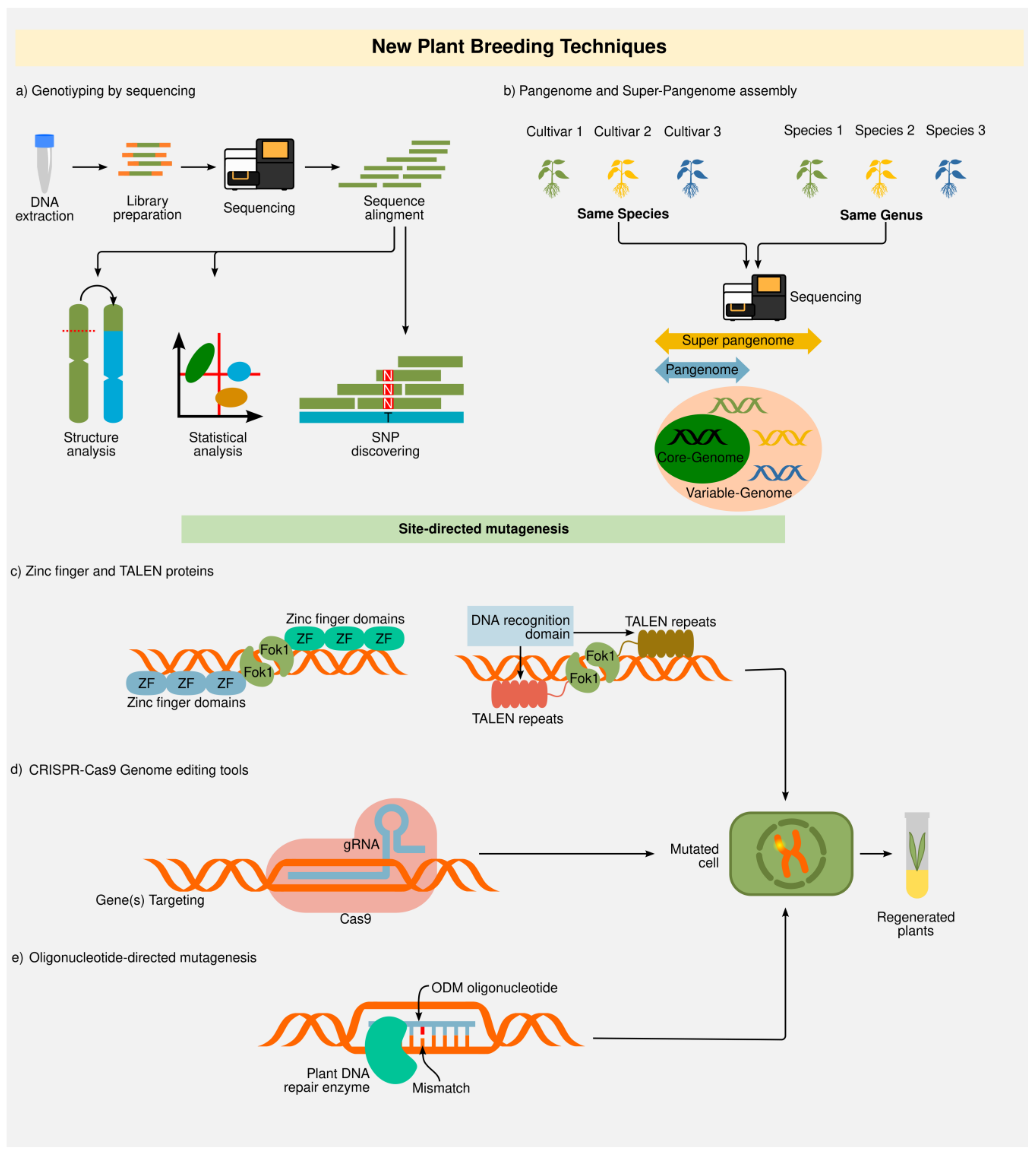
| Cas Enzyme | Tools | Uses | References |
|---|---|---|---|
| Cas9 and Cas9 engineered versions | CRISPR-Cas9 | Generates DBS on DNA (blunt end). DNA repair mechanisms provoke frameshift mutations resulting in gene knock-out. Use of more than one sgRNA can induce longer deletions or multiplex gene targeting | [290,314,315] |
| DNA-free | Requires delivery of gRNA/Cas9 ribonucleoproteins (RNPs) to editing without transgene integration to the genome | [320,350] | |
| IGE-XVE | Cas9 expression system inducible by estrogens (17-β-estradiol). | [345] | |
| IGE-HS | Cas9 expression system inducible by heat shock | [346] | |
| TSKO | Tissue-specific knockout system. A cell or tissue specific promoter controls the expression of Cas9, provoking spatial regulation of gene editing | [347] | |
| TSKO + IGE | This combination provides spatial- and temporal-regulated genome editing | [345] | |
| CBE | Mediates G-C to A-T base conversion in target DNA strand | [328] | |
| ABE | Induce A-T to G-C base changeover in target DNA strand | [332] | |
| STEME | Used in a high-throughput manner to modify cis regulatory elements and genome wide screening | [333] | |
| STOP | Facilitates gene silencing by creating stop codon without the need of DBS | [356] | |
| SMART | Based in the rescue of lethal mutations to quickly assess the efficiency of base editing. | [331] | |
| SL | Provokes alterations in start codon (ATG to ATA, ACG, or GTG) | [349] | |
| SKIP | Mutates G at the end of an intron, which can lead to exon skipping into mature transcripts | [348] | |
| PASS | Convert the three possible PTCs (TAA, TAG, and TGA) into TGG (tryptophan) | [357] | |
| dCas9-SunTag-TET1cd | Epigenome editing through TET1-cd demethylase, allowing for specific gene up-regulation | [341] | |
| dCas9-SunTag-DRMcd | Epigenome editing through DRM methyltransferase, enabling specific gene down-regulation | [342] | |
| Prime editing | Creates new genetic changes (or repairing) at the target DNA without DSB or DRT | [339,340] | |
| Cas12a (former Cpf1) | CRISPR-Cas12a | Cas12a targets T-rich regions of the genome where Cas9 is not suitable to use, facilitates multiplexing, assists for precise DNA repair by exogenous DRT. Cas12a generates staggered ends with 4–5 nucleotide overhangs, which is advantageous for genetic insertions or specificity during NHEJ or HDR. Moreover, Cas12a offers future modifications at the same target site, because it cuts DNA strands distal to the PAM sequence | [324,325] |
| Cas13 and Cas13 engineered versions | CRISPR-Cas13 | Cas13 has ribonuclease activity capable of targeting and cleaving ssRNA. Potential applications in plant virus interference or repression of eukaryotic gene expression | [306,308] |
| m6A | RNA epigenome editing. Edits the methylation stage of target transcripts | [343] | |
| REPAIR | RNA editing. For A to I (G) base substitution at RNA level | [334,337] | |
| RESCUE | RNA editing. For C to U base replacement at RNA level | [337,338] |
| Crop | Molecular Strategy | Gene | Improved Stress Tolerance | References |
|---|---|---|---|---|
| Rice | Haplotype analysis with GWAS | SEMIDWARF1 | flooding | [442] |
| GWAS | LOC_Os10g34840 | cold | [443] | |
| QTLs and MAS | TT1 | heat | [430] | |
| Overexpression | PcCFR | salinity, drought, and cold stress | [446] | |
| CRISPR | OsMYB30 | cold | [447] | |
| CRISPR | OsPYL9 | drought | [448] | |
| CRISPR | OsERA1 | drought | [449] | |
| CRISPR | OsRR22 | salinity and osmotic stress | [450] | |
| CRISPR | OsDST | drought, salinity, and osmotic stress | [451] | |
| CRISPR | OsMPK5 | various abiotic (and biotic) stresses | [297] | |
| Wheat | Overexpression | TaFER-5B | heat, cold, and drought | [452] |
| Overexpression | TaPYL4 | drought | [453] | |
| Overexpression | TdPIP2 | salinity and drought | [454] | |
| Overexpression | ZmPEPC | drought and high temperature | [455,456] | |
| Overexpression | TaWRKY2 | drought | [457] | |
| Overexpression | TaBZR2 | drought | [458] | |
| Overexpression | TaPEPKR2 | drought, osmotic, and heat stress | [459] | |
| Overexpression | AtOTS1 | drought | [460] | |
| CRISPR | TaERF3 | drought | [461] | |
| CRISPR | TaDREB2 | drought | [461] | |
| Maize | Overexpression | betA | drought | [462] |
| Overexpression | TsVP | drought | [462] | |
| Overexpression | CSPS | drought | [463] | |
| Overexpression | TPP | drought | [33] | |
| Overexpression | VHb | waterlogging | [464] | |
| Overexpression | SDD1 | drought | [465] | |
| Overexpression | OsMYB55 | drought and high temperature | [466] | |
| Overexpression | ZmERB180 | waterlogging | [467] | |
| CRISPR | ARGOS8 | drought | [468] | |
| CRISPR | ZmHKT1 | salinity | [469] |
| Crop | Rice | Wheat | Maize |
|---|---|---|---|
| Growth promoting rhizobacteria or fungi species/strain | Acinetobacter lwoffii | Acinetobacter sp. | Alcaligenes faecalis (AF3) |
| Arthrobacter defluvii | Arthrobacter protophormiae (SA3) | Arthrobacter pascens | |
| Azospirillum brasilense AZ39 | Azospirillum brasilense Sp245 | A. brasilense | |
| Azotobacter vinellandii (SRI Az 3) | A. brasilense NO40 | Azospirillum lipoferum | |
| Arthrobacter nitroguajacolicus (YB3 and YB5) | Azotobacter chrocoocum (E1) | Azotobacter sp. | |
| Bacillus haynesii | Bacillus amyloliquefaciens 5113 | Bacillus amyloliquefaciens | |
| Bacillus megaterium (NBRI 20M) | Bacillus aquimaris | B. licheniformis | |
| Bacillus paralicheniformis | B. insolitus | Bacillus megaterium | |
| Glutamicibacter sp. YD01 | Bacillus licheniformis | B. subtilis | |
| Jeotgalicoccus huakuii | Bacillus pumilus | B. thuringiensis | |
| Lysinibacillus fusiformis | Bacillus subtilis (LDR2) | Bukholderia phytofirmans (psJN) | |
| Oceanobacillus picturae | Bacillus thuringiensis AZP2 | Enterobacter sp. (FD17) | |
| Pantoea sp. | Dietzia natronolimnaea (STR1) | Herbaspirillum sp. | |
| Phyllobacterium brassicacearum | Enterobacter ludwigii | Klebsiella variicola F2 | |
| Pseudomonas jessenii R62 | Enterobacter sp. | Massilia sp. RK4 | |
| Pseudomonas pseudoalcaligenes | Exiguobacterium aurantiacum | Paenibacillus favisporus | |
| Pseudomonas putida | Flavobacterium sp. | Pantoea sp. | |
| Pseudomonas synxantha R81 | Klebsiella sp. | Pseudomonas aeruginosa (Pa2) | |
| Staphylococcus cohnii | Marinobacterium sp. | Pseudomonas entomophila | |
| Glomus intraradices | Mesorhizobium ciceri (CR-30 and CR-39) | P. fluorescens N3 | |
| Glomus coronatum | Microbacterium spp. | P. fluorescens YX2 | |
| Glomus constrictum | Paenibacillus polymyxa | Pseudomonas monteilii | |
| Glomus claroideum | Pantoea sp. | P. putida (Q7, GAP-P45, UW4) | |
| Streptomyces sp. strains | Pseudomonas aeruginosa | Pseudomonas stutzeri | |
| Trichoderma harzianum | Pseudomonas fluorescence | P. syringae | |
| P. syringae | Proteus penneri (Pp1) | ||
| Pseudomonas sp. (E2) | Raoultella planticola YL2 | ||
| Rhizobium leguminosarum (LR-30) | Rhizobium sp. | ||
| Rhizobium phaseoli (MR-2) | Rhizoglomus intraradices | ||
| Serratia sp. | Streptomyces sp. | ||
| Sinorhizobium sp. | Trichoderma atroviride | ||
| Stenotrophomonas sp. strains | |||
| References | [77,79,85,489,490,491,492,493,494,495] | [77,79,489,490,494,495,496,497,498,499] | [77,79,84,489,490,494,495,500,501,502,503] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Villalobos-López, M.A.; Arroyo-Becerra, A.; Quintero-Jiménez, A.; Iturriaga, G. Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops. Int. J. Mol. Sci. 2022, 23, 12053. https://doi.org/10.3390/ijms231912053
Villalobos-López MA, Arroyo-Becerra A, Quintero-Jiménez A, Iturriaga G. Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops. International Journal of Molecular Sciences. 2022; 23(19):12053. https://doi.org/10.3390/ijms231912053
Chicago/Turabian StyleVillalobos-López, Miguel Angel, Analilia Arroyo-Becerra, Anareli Quintero-Jiménez, and Gabriel Iturriaga. 2022. "Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops" International Journal of Molecular Sciences 23, no. 19: 12053. https://doi.org/10.3390/ijms231912053
APA StyleVillalobos-López, M. A., Arroyo-Becerra, A., Quintero-Jiménez, A., & Iturriaga, G. (2022). Biotechnological Advances to Improve Abiotic Stress Tolerance in Crops. International Journal of Molecular Sciences, 23(19), 12053. https://doi.org/10.3390/ijms231912053







