Ectopic Expression of Arabidopsis thaliana zDof1.3 in Tomato (Solanum lycopersicum L.) Is Associated with Improved Greenhouse Productivity and Enhanced Carbon and Nitrogen Use
Abstract
1. Introduction
2. Results
2.1. Selection and Initial Characterization of the Transgenic Line
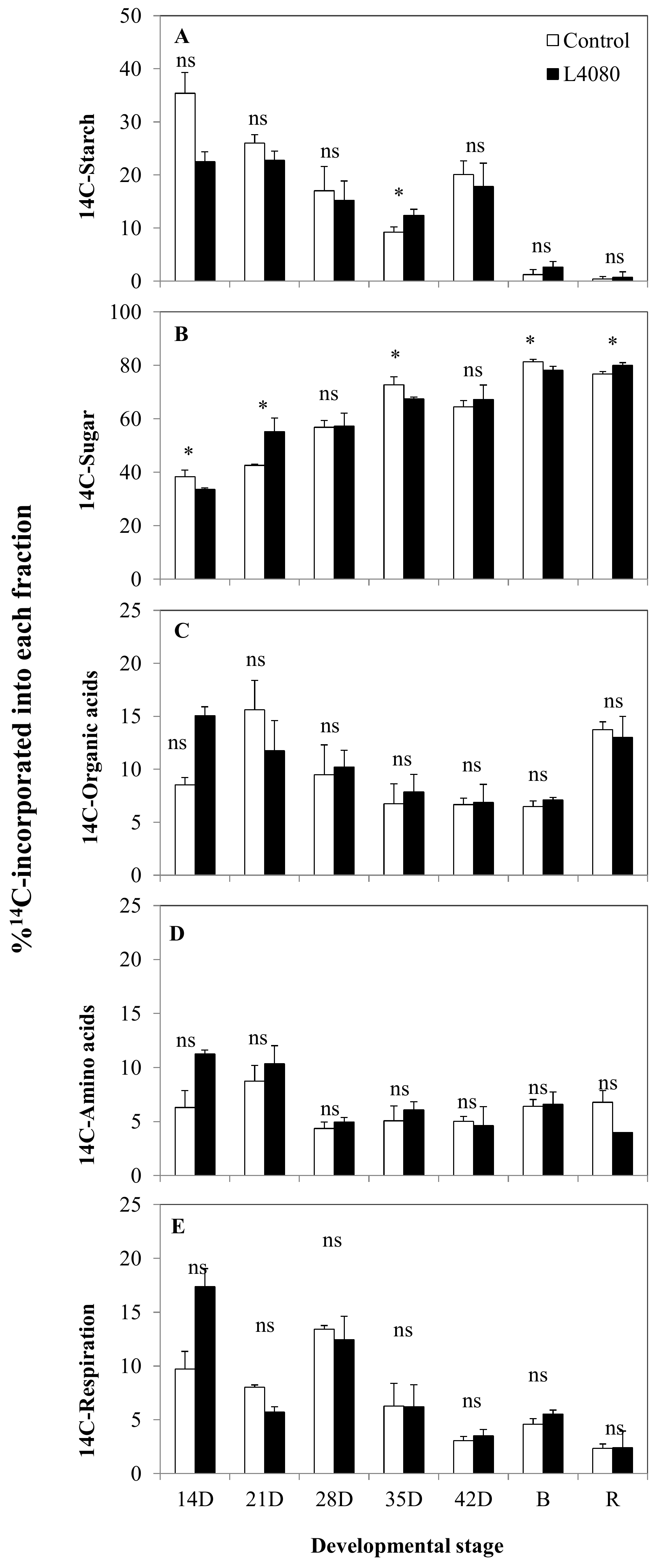
2.2. Sugar and Starch Accumulation and Partitioning in Leaf and Fruit
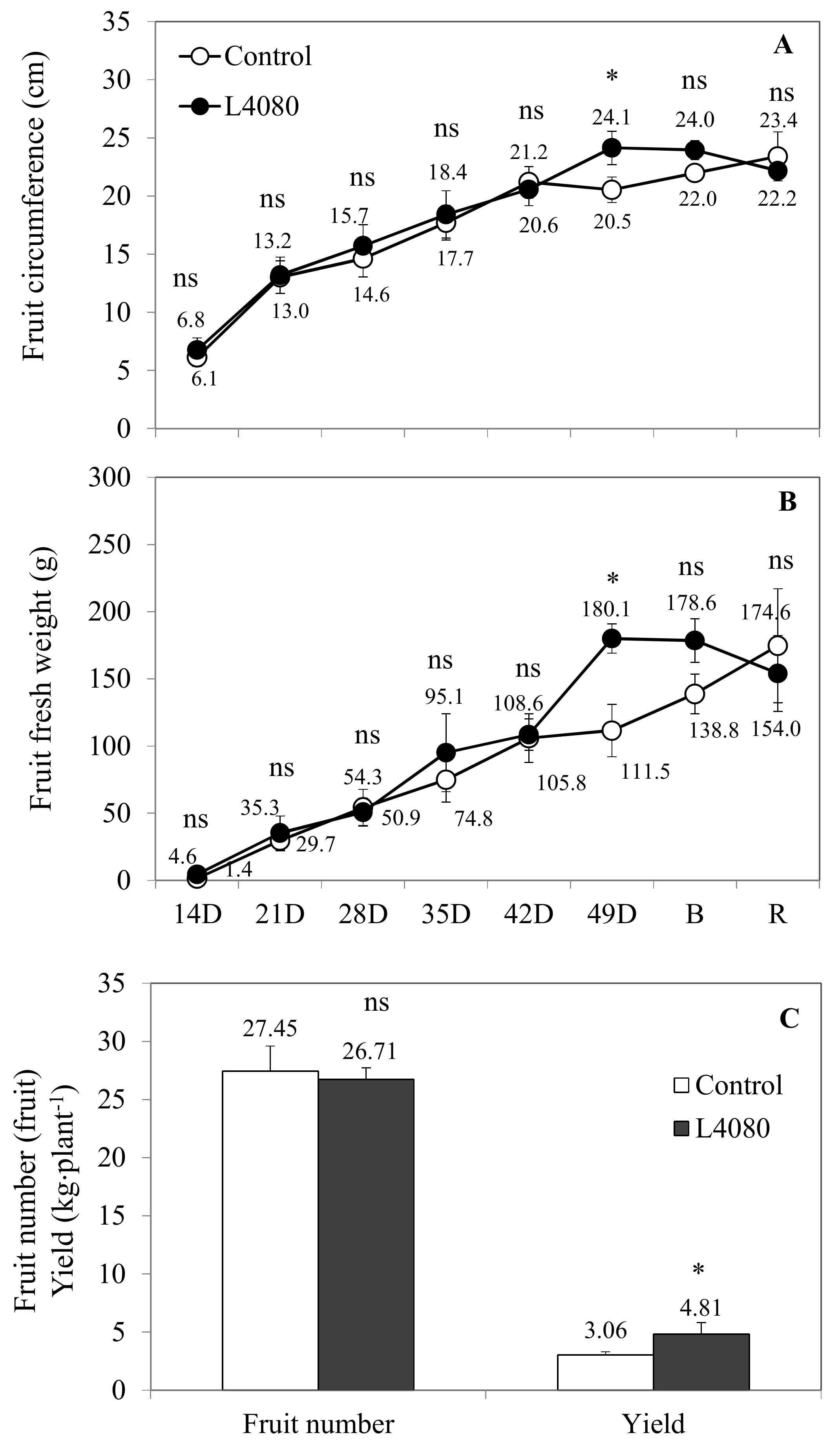
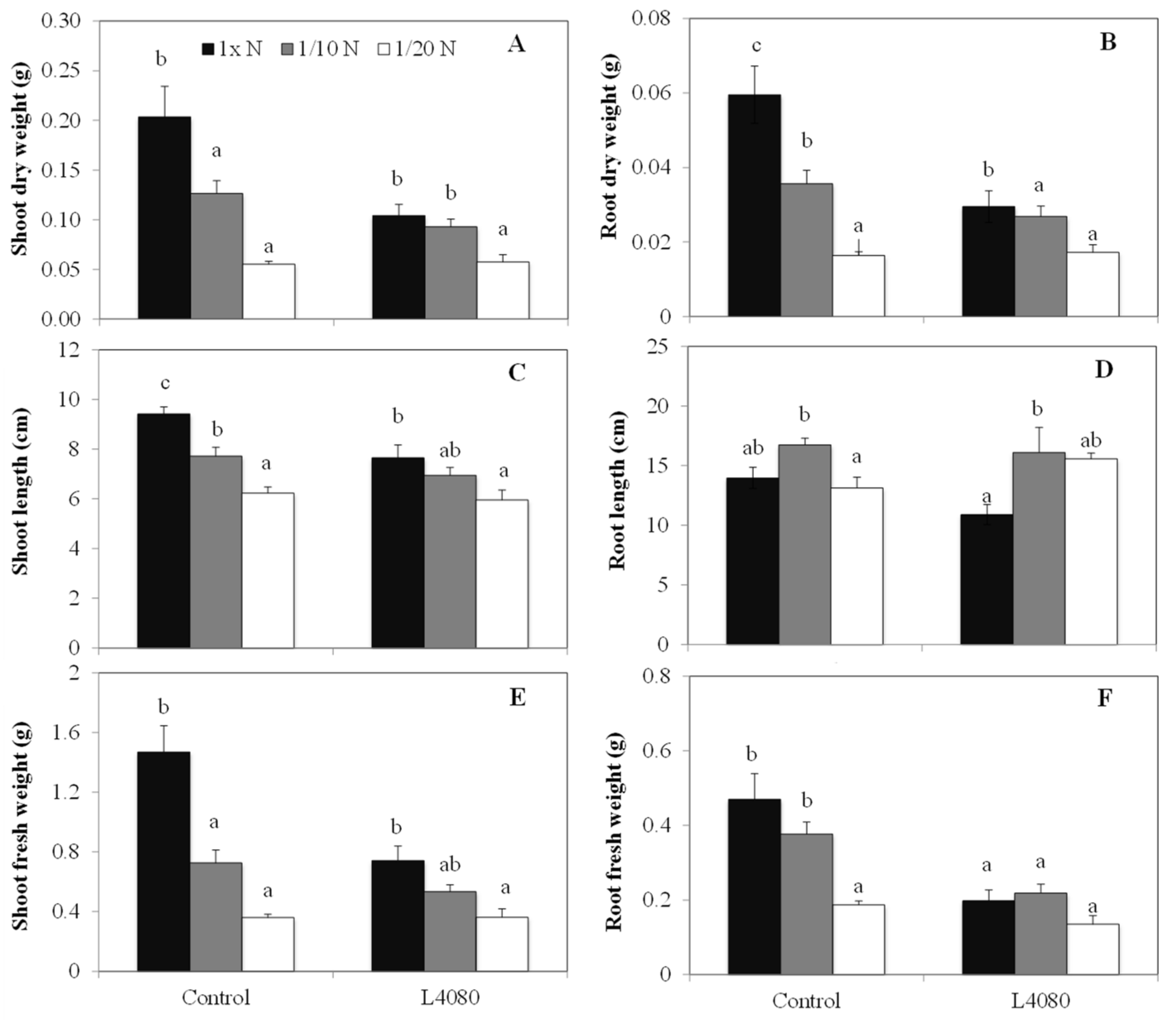
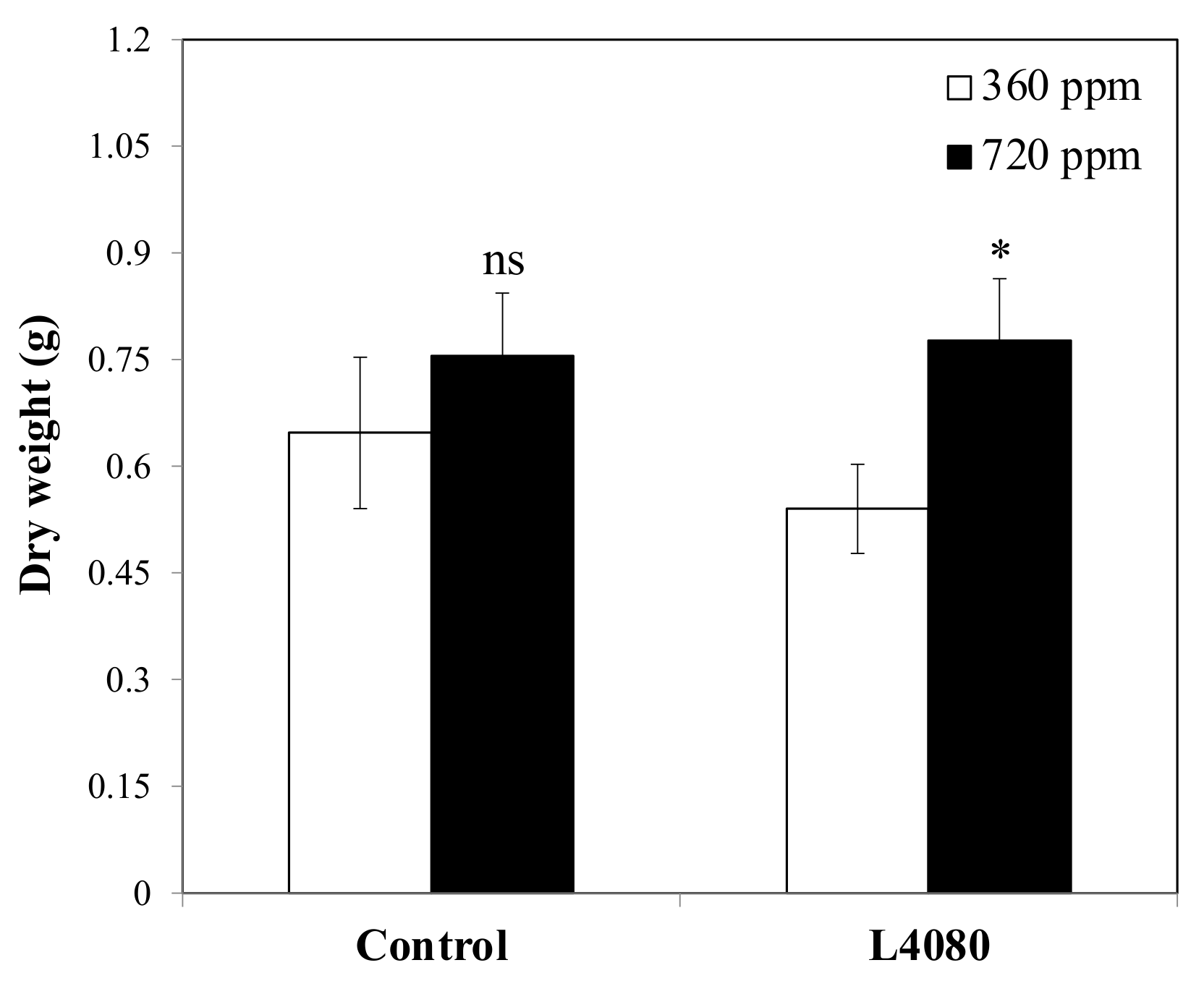
2.3. Plant Productivity under Limited Nitrogen and Higher Carbon Dioxide (CO2)
2.4. Eco-Physiological Parameters
2.5. Metabolite Profiling of Leaf and Fruit
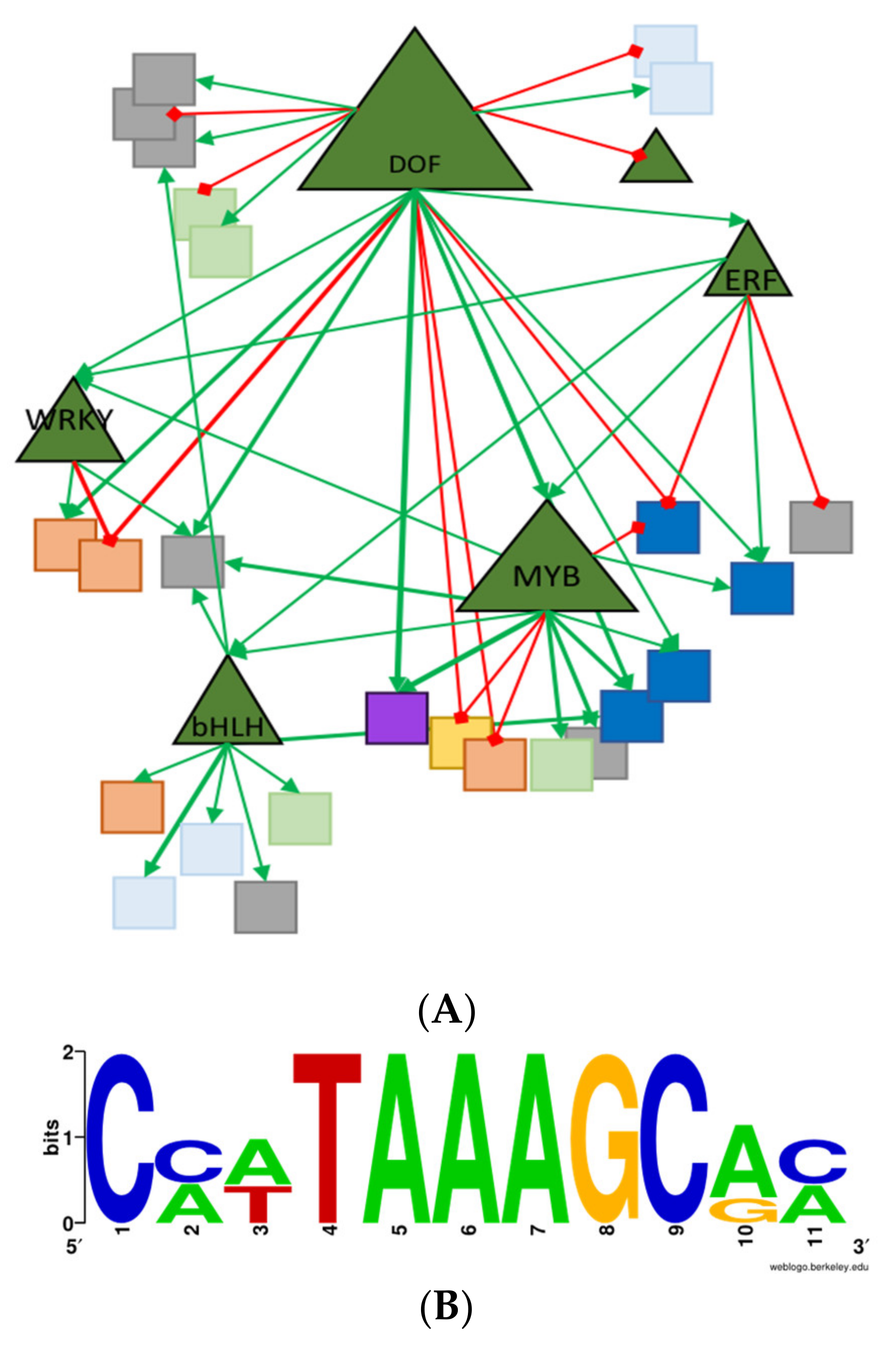
2.6. Transcriptomic Analysis of Leaf Tissue
2.7. Connecting The Novel Traits to AtzDof1.3 Expression
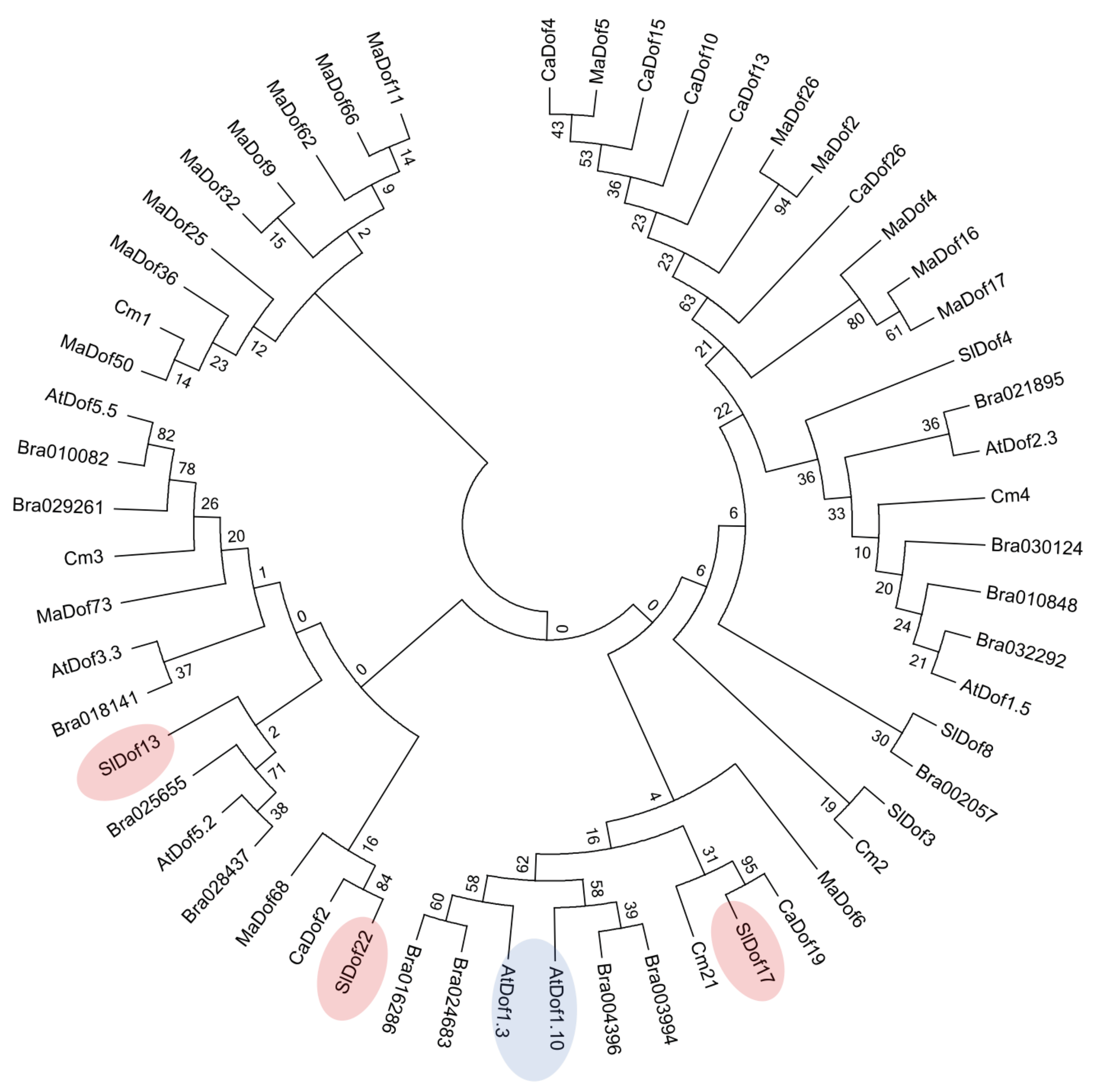
2.8. Sequence, Evolutionary, and Comparative Analysis of AtzDof1.3
2.9. Sequence Analysis of AtzDof1.3 Homologues in Tomato
2.10. The Role of AtzDof1.3 in Arabidopsis
3. Discussion
3.1. L4080 Was Most Likely Altered in Source–Sink Relations
3.2. L4080 Varied in N-Use and Root Growth in Juvenile Plants
3.3. Post-transcriptional Modification May Be an Important Driver in The Phenotypic Changes in L4080
3.4. The Evolutionary History of AtzDof1.3 and Homologues in Tomato Differ
3.5. AtzDof1.3 May Be Important for Germination of Arabidopsis
3.6. Summary
4. Materials and Methods
4.1. Generation of Transgenic Lines
4.2. Growth Conditions
4.3. Detection of The AtzDof1.3 Transgene in Tomato
4.4. Semi-Quantitative Reverse Transcription Polymerase Chain Reaction (Semi-qRT-PCR)
4.5. Eco-Physiology and Biochemical Measurements
4.6. GC-MS-TOF Metabolite Profiling
4.7. Analysis Using the Affymetrix GeneChip
4.8. A. thaliana zDof1.3 Construct Assembly and Transient Expression
4.9. Analysis of the Predicted Amino Acid Sequence of AtDof1.3 and Its Homologues
4.10. Analysis of AtDof1.3 in Arabidopsis Using T-DNA Knockouts Lines
4.11. Statistical and Network Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Beckles, D.M. Factors affecting the postharvest soluble solids and sugar content of tomato (Solanum lycopersicum L.) fruit. Postharvest Biol. Technol. 2012, 63, 129–140. [Google Scholar] [CrossRef]
- Godfray, H.C.J.; Beddington, J.R.; Crute, I.R.; Haddad, L.; Lawrence, D.; Muir, J.F.; Pretty, J.; Robinson, S.; Thomas, S.M.; Toulmin, C. Food Security: The Challenge of Feeding 9 Billion People. Science 2010, 327, 812–818. [Google Scholar] [CrossRef] [PubMed]
- Gur, A.; Zamir, D. Mendelizing all Components of a Pyramid of Three Yield QTL in Tomato. Front. Plant Sci. 2015, 6, 1096. [Google Scholar] [CrossRef] [PubMed]
- Krieger, U.; Lippman, Z.B.; Zamir, D. The flowering gene SINGLE FLOWER TRUSS drives heterosis for yield in tomato. Nat. Genet. 2010, 42, 459–463. [Google Scholar] [CrossRef]
- Tanksley, S.D.; Fulton, T.M. Dissecting quantitative trait variation—Examples from the tomato. Euphytica 2007, 154, 365–370. [Google Scholar] [CrossRef]
- Cohen, I.; Rapaport, T.; Berger, R.T.; Rachmilevitch, S. The effects of elevated CO2 and nitrogen nutrition on root dynamics. Plant Sci. 2018, 272, 294–300. [Google Scholar] [CrossRef]
- Gur, A.; Semel, Y.; Osorio, S.; Friedmann, M.; Seekh, S.; Ghareeb, B.; Mohammad, A.; Pleban, T.; Gera, G.; Fernie, A.R.; et al. Yield quantitative trait loci from wild tomato are predominately expressed by the shoot. Theor. Appl. Genet. 2011, 122, 405–420. [Google Scholar] [CrossRef]
- Baldazzi, V.; Bertin, N.; Genard, M. A Model of Fruit Growth Integrating Cell Division and Expansion Processes. In IV International Symposium on Models for Plant Growth, Environmental Control and Farm Management in Protected Cultivation—Hortimodel; International Society for Horticultural Science: Leuven, Belgium, 2012; Volume 957, pp. 191–196. [Google Scholar]
- Bertin, N.; Causse, M.; Brunel, B.; Tricon, D.; Genard, M. Identification of growth processes involved in QTLs for tomato fruit size and composition. J. Exp. Bot. 2009, 60, 237–248. [Google Scholar] [CrossRef]
- Hanson, P.M.; Sitathani, K.; Sadashiva, A.T.; Yang, R.Y.; Graham, E.; Ledesma, D. Performance of Solanum habrochaites LA1777 introgression line hybrids for marketable tomato fruit yield in Asia. Euphytica 2007, 158, 167–178. [Google Scholar] [CrossRef]
- Soyk, S.; Benoit, M.; Lippman, Z.B. New Horizons for Dissecting Epistasis in Crop Quantitative Trait Variation. Annu. Rev. Genet. 2020, 54, 287–307. [Google Scholar] [CrossRef]
- Zsogon, A.; Cermak, T.; Voytas, D.; Peres, L.E.P. Genome editing as a tool to achieve the crop ideotype and de novo domestication of wild relatives: Case study in tomato. Plant Sci. 2017, 256, 120–130. [Google Scholar] [CrossRef]
- Rodriguez-Leal, D.; Lemmon, Z.H.; Man, J.; Bartlett, M.E.; Lippman, Z.B. Engineering Quantitative Trait Variation for Crop Improvement by Genome Editing. Cell 2017, 171, 470–480. [Google Scholar] [CrossRef]
- Soyk, S.; Muller, N.A.; Park, S.J.; Schmalenbach, I.; Jiang, K.; Hayama, R.; Zhang, L.; Van Eck, J.; Jimenez-Gomez, J.M.; Lippman, Z.B. Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat. Genet. 2017, 49, 162–168. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef]
- Sun, Z.F.; Guo, T.T.; Liu, Y.; Liu, Q.; Fang, Y.D. The Roles of Arabidopsis CDF2 in Transcriptional and Posttranscriptional Regulation of Primary MicroRNAs. PLoS Genet. 2015, 11, e1005598. [Google Scholar] [CrossRef]
- Iwase, A.; Matsui, K.; Ohme-Takagi, M. Manipulation of plant metabolic pathways by transcription factors. Plant Biotechnol. 2009, 26, 29–38. [Google Scholar] [CrossRef]
- Century, K.; Reuber, T.L.; Ratcliffe, O.J. Regulating the regulators: The future prospects for transcription-factor-based agricultural biotechnology products. Plant Physiol. 2008, 147, 20–29. [Google Scholar] [CrossRef]
- Doebley, J.; Lukens, L. Transcriptional regulators and the evolution of plant form. Plant Cell 1998, 10, 1075–1082. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Y.; Turečková, V.; Xue, G.-P.; Fernie, A.R.; Mueller-Roeber, B.; Balazadeh, S. The NAC Transcription Factor SlNAP2 Regulates Leaf Senescence and Fruit Yield in Tomato. Plant Physiol. 2018, 177, 1286–1302. [Google Scholar] [CrossRef]
- Abraham, Z.; del Pozo, J.C. Ectopic Expression of E2FB, a Cell Cycle Transcription Factor, Accelerates Flowering and Increases Fruit Yield in Tomato. J. Plant Growth Regul. 2012, 31, 11–24. [Google Scholar] [CrossRef]
- Shahzad, R.; Jamil, S.; Ahmad, S.; Nisar, A.; Amina, Z.; Saleem, S.; Iqbal, M.Z.; Atif, R.M.; Wang, X.K. Harnessing the potential of plant transcription factors in developing climate resilient crops to improve global food security: Current and future perspectives. Saudi J. Biol. Sci. 2021, 28, 2323–2341. [Google Scholar] [CrossRef] [PubMed]
- Sulpice, R.; Pyl, E.T.; Ishihara, H.; Trenkamp, S.; Steinfath, M.; Witucka-Wall, H.; Gibon, Y.; Usadel, B.; Poree, F.; Piques, M.C.; et al. Starch as a major integrator in the regulation of plant growth. Proc. Natl. Acad. Sci. USA 2009, 106, 10348–10353. [Google Scholar] [CrossRef] [PubMed]
- Stitt, M.; Zeeman, S.C. Starch turnover: Pathways, regulation and role in growth. Curr. Opin. Plant Biol. 2012, 15, 282–292. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, S. The Dof family of plant transcription factors. Trends Plant Sci. 2002, 7, 555–560. [Google Scholar] [CrossRef]
- Yanagisawa, S. Dof proteins: Involvement of transcription factors with a novel DNA-binding domain in tissue-specific and signal-responsive gene expression. Seikagaku 1998, 70, 280–285. [Google Scholar]
- Kumar, R.; Taware, R.; Gaur, V.S.; Guru, S.K.; Kumar, A. Influence of nitrogen on the expression of TaDof1 transcription factor in wheat and its relationship with photo synthetic and ammonium assimilating efficiency. Mol. Biol. Rep. 2009, 36, 2209–2220. [Google Scholar] [CrossRef]
- Kumar, A.; Kanwal, P.; Gupta, A.K.; Singh, B.R.; Gaur, V.S. A Full-Length Dof1 Transcription Factor of Finger Millet and its Response to a Circadian Cycle. Plant Mol. Biol. Report. 2014, 32, 419–427. [Google Scholar] [CrossRef]
- Gupta, S.; Gupta, S.M.; Gupta, A.K.; Gaur, V.S.; Kumar, A. Fluctuation of Dof1/Dof2 expression ratio under the influence of varying nitrogen and light conditions: Involvement in differential regulation of nitrogen metabolism in two genotypes of finger millet (Eleusine coracana L.). Gene 2014, 546, 327–335. [Google Scholar] [CrossRef]
- Yanagisawa, S.; Akiyama, A.; Kisaka, H.; Uchimiya, H.; Miwa, T. Metabolic engineering with Dof1 transcription factor in plants: Improved nitrogen assimilation and growth under low-nitrogen conditions. Proc. Natl. Acad. Sci. USA 2004, 101, 7833–7838. [Google Scholar] [CrossRef]
- Santos, L.A.; de Souza, S.R.; Fernandes, M.S. OsDof25 expression alters carbon and nitrogen metabolism in Arabidopsis under high N-supply. Plant Biotechnol. Rep. 2012, 6, 327–337. [Google Scholar] [CrossRef]
- Corrales, A.R.; Carrillo, L.; Lasierra, P.; Nebauer, S.G.; Dominguez-Figueroa, J.; Renau-Morata, B.; Pollmann, S.; Granell, A.; Molina, R.V.; Vicente-Carbajosa, J.; et al. Multifaceted role of cycling DOF factor 3 (CDF3) in the regulation of flowering time and abiotic stress responses in Arabidopsis. Plant Cell Environ. 2017, 40, 748–764. [Google Scholar] [CrossRef]
- Domínguez-Figueroa, J.; Carrillo, L.; Renau-Morata, B.; Yang, L.; Molina, R.-V.; Marino, D.; Canales, J.; Weih, M.; Vicente-Carbajosa, J.; Nebauer, S.G.; et al. The Arabidopsis Transcription Factor CDF3 Is Involved in Nitrogen Responses and Improves Nitrogen Use Efficiency in Tomato. Front. Plant Sci. 2020, 11, 1825. [Google Scholar] [CrossRef]
- Renau-Morata, B.; Molina, R.V.; Carrillo, L.; Cebolla-Comejo, J.; Sanchez-Perales, M.; Pollmann, S.; Dominguez-Figueroa, J.; Corrales, A.R.; Flexas, J.; Vicente-Carbajosa, J.; et al. Ectopic Expression of CDF3 Genes in Tomato Enhances Biomass Production and Yield under Salinity Stress Conditions. Front. Plant Sci. 2017, 8, 660. [Google Scholar] [CrossRef]
- Wang, H.W.; Zhang, B.; Hao, Y.J.; Huang, J.; Tian, A.G.; Liao, Y.; Zhang, J.S.; Chen, S.Y. The soybean Dof-type transcription factor genes, GmDof4 and GmDof11, enhance lipid content in the seeds of transgenic Arabidopsis plants. Plant J. 2007, 52, 716–729. [Google Scholar] [CrossRef]
- Kurai, T.; Wakayama, M.; Abiko, T.; Yanagisawa, S.; Aoki, N.; Ohsugi, R. Introduction of the ZmDof1 gene into rice enhances carbon and nitrogen assimilation under low-nitrogen conditions. Plant Biotechnol. J. 2011, 9, 826–837. [Google Scholar] [CrossRef]
- Wang, Y.L.; Fu, B.; Pan, L.F.; Chen, L.M.; Fu, X.H.; Li, K.Z. Overexpression of Arabidopsis Dof1, GS1 and GS2 Enhanced Nitrogen Assimilation in Transgenic Tobacco Grown Under Low-Nitrogen Conditions. Plant Mol. Biol. Rep. 2013, 31, 886–900. [Google Scholar] [CrossRef]
- Deng, W.; Yan, F.; Zhang, X.L.; Tang, Y.W.; Yuan, Y.J. Transcriptional profiling of canola developing embryo and identification of the important roles of BnDof5.6 in embryo development and fatty acids synthesis. Plant Cell Physiol. 2015, 56, 1624–1640. [Google Scholar] [CrossRef]
- Tanaka, M.; Takahata, Y.; Nakayama, H.; Nakatani, M.; Tahara, M. Altered carbohydrate metabolism in the storage roots of sweetpotato plants overexpressing the SRF1 gene, which encodes a Dof zinc finger transcription factor. Planta 2009, 230, 737–746. [Google Scholar] [CrossRef]
- Huang, X.L.; Zhang, Y.Y.; Wang, L.L.; Dong, X.Y.; Hu, W.X.; Jiang, M.; Chen, G.; An, G.; Xiong, F.; Wu, Y.F. OsDOF11 Affects Nitrogen Metabolism by Sucrose Transport Signaling in Rice (Oryza sativa L.). Front. Plant Sci. 2021, 12, 1788. [Google Scholar] [CrossRef]
- Renau-Morata, B.; Carrillo, L.; Cebolla-Cornejo, J.; Molina, R.V.; Marti, R.; Dominguez-Figueroa, J.; Vicente-Carbajosa, J.; Medina, J.; Nebauer, S.G. The targeted overexpression of SlCDF4 in the fruit enhances tomato size and yield involving gibberellin signalling. Sci. Rep. 2020, 10, 10645. [Google Scholar] [CrossRef]
- Xu, X.M.; Li, F.; Wang, Y.Z.; Tang, S.W.; Dai, Q.Z.; Zhu, S.Y.; Liu, T.M. Identification of Dof transcription factors in ramie (Boehmeria nivea L. Gaud) and their expression in response to different nitrogen treatments. 3 Biotech 2018, 8, 496. [Google Scholar] [CrossRef] [PubMed]
- Lam Cheng, K.-L. Golden2-Like (GLK2) Transcription Factor: Developmental Control of Tomato Fruit Photosynthesis and Its Contribution to Ripe Fruit Characteristics; University of California Davis: Davis, CA, USA, 2013. [Google Scholar]
- Smith, A.M.; Stitt, M. Coordination of carbon supply and plant growth. Plant Cell Environ. 2007, 30, 1126–1149. [Google Scholar] [CrossRef] [PubMed]
- Blair, E.J.; Goralogia, G.S.; Lincoln, M.J.; Imaizumi, T.; Nagel, D.H. Clock-Controlled and Cold-Induced CYCLING DOF FACTOR6 Alters Growth and Development in Arabidopsis. Front. Plant Sci. 2022, 13, 2458. [Google Scholar] [CrossRef] [PubMed]
- Krahmer, J.; Goralogia, G.S.; Kubota, A.; Zardilis, A.; Johnson, R.S.; Song, Y.H.; MacCoss, M.J.; Le Bihan, T.; Halliday, K.J.; Imaizumi, T.; et al. Time-resolved interaction proteomics of the GIGANTEA protein under diurnal cycles in Arabidopsis. FEBS Lett. 2019, 593, 319–338. [Google Scholar] [CrossRef]
- Sharkey, T.D.; Berry, J.A.; Raschke, K. Starch and Sucrose Synthesis in Phaseolus-Vulgaris as Affected by Light, CO2, and Abscisic-Acid. Plant Physiol. 1985, 77, 617–620. [Google Scholar] [CrossRef]
- Dong, S.Y.; Beckles, D.M. Dynamic changes in the starch-sugar interconversion within plant source and sink tissues promote a better abiotic stress response. J. Plant Physiol. 2019, 234, 80–93. [Google Scholar] [CrossRef]
- Dong, S.Y.; Zhang, J.; Beckles, D.M. A pivotal role for starch in the reconfiguration of C-14-partitioning and allocation in Arabidopsis thaliana under short-term abiotic stress. Sci. Rep. 2018, 8, 9314. [Google Scholar] [CrossRef]
- Luengwilai, K.; Fiehn, O.E.; Beckles, D.M. Comparison of Leaf and Fruit Metabolism in Two Tomato (Solanum lycopersicum L.) Genotypes Varying in Total Soluble Solids. J. Agric. Food Chem. 2010, 58, 11790–11800. [Google Scholar] [CrossRef]
- Luengwilai, K.; Beckles, D.M. Starch Granules in Tomato Fruit Show a Complex Pattern of Degradation. J. Agric. Food Chem. 2009, 57, 8480–8487. [Google Scholar] [CrossRef]
- Luengwilai, K.; Tananuwong, K.; Shoemaker, C.F.; Beckles, D.M. Starch Molecular Structure Shows Little Association with Fruit Physiology and Starch Metabolism in Tomato. J. Agric. Food Chem. 2010, 58, 1275–1282. [Google Scholar] [CrossRef]
- Sweetlove, L.J.; Tomlinson, K.L.; Hill, S.A. The effect of exogenous sugars on the control of flux by adenosine 5′-diphosphoglucose pyrophosphorylase in potato tuber discs. Planta 2002, 214, 741–750. [Google Scholar] [CrossRef]
- N′tchobo, H.; Dali, N.; Nguyen-Quoc, B.; Foyer, C.H.; Yelle, S. Starch synthesis in tomato remains constant throughout fruit development and is dependent on sucrose supply and sucrose synthase activity. J. Exp. Bot. 1999, 50, 1457–1463. [Google Scholar] [CrossRef]
- VicenteCarbajosa, J.; Moose, S.P.; Parsons, R.L.; Schmidt, R.J. A maize zinc-finger protein binds the prolamin box in zein gene promoters and interacts with the basic leucine zipper transcriptional activator Opaque2. Proc. Natl. Acad. Sci. USA 1997, 94, 7685–7690. [Google Scholar] [CrossRef]
- Marzabal, P.; Gas, E.; Fontanet, P.; Vicente-Carbajosa, J.; Torrent, M.; Ludevid, M.D. The maize Dof protein PBF activates transcription of gamma-zein during maize seed development. Plant Mol. Biol. 2008, 67, 441–454. [Google Scholar] [CrossRef]
- Tsujimoto-Inui, Y.; Naito, Y.; Sakurai, N.; Suzuki, H.; Sasaki, R.; Takahashi, H.; Ohtsuki, N.; Nakano, T.; Yanagisawa, S.; Shibata, D.; et al. Functional genomics of the Dof transcription factor family genes in suspension-cultured cells of Arabidopsis thaliana. Plant Biotechnol. 2009, 26, 15–28. [Google Scholar] [CrossRef]
- Cavalar, M.; Phlippen, Y.; Kreuzaler, F.; Peterhaensel, C. A drastic reduction in DOF1 transcript levels does not affect C-4-specific gene expression in maize. J. Plant Physiol. 2007, 164, 1665–1674. [Google Scholar] [CrossRef]
- Walker, R.L.; Burns, I.G.; Moorby, J. Responses of plant growth rate to nitrogen supply: A comparison of relative addition and N interruption treatments. J. Exp. Bot. 2001, 52, 309–317. [Google Scholar] [CrossRef]
- Hirose, T. Nitrogen use efficiency revisited. Oecologia 2011, 166, 863–867. [Google Scholar] [CrossRef]
- Crush, J.R.; Care, D.A.; Gourdin, A.; Woodfield, D.R. Root growth media effects on root morphology and architecture in white clover. N. Z. J. Agric. Res. 2005, 48, 255–263. [Google Scholar] [CrossRef]
- Bloom, A.J.; Smart, D.R.; Nguyen, D.T.; Searles, P.S. Nitrogen assimilation and growth of wheat under elevated carbon dioxide. Proc. Natl. Acad. Sci. USA 2002, 99, 1730–1735. [Google Scholar] [CrossRef]
- Stamova, B.S.; Roessner, U.; Suren, S.; Laudencia-Chingcuanco, D.; Bacic, A.; Beckles, D.M. Metabolic profiling of transgenic wheat over-expressing the high-molecular-weight Dx5 glutenin subunit. Metabolomics 2009, 5, 239–252. [Google Scholar] [CrossRef]
- Singh, B.K. Plant Amino Acids: Biochemistry and Biotechnology; CRC Press: Boca Raton, FL, USA, 1999; p. 611. [Google Scholar]
- Weckwerth, W.; Loureiro, M.E.; Wenzel, K.; Fiehn, O. Differential metabolic networks unravel the effects of silent plant phenotypes. Proc. Natl. Acad. Sci. USA 2004, 101, 7809–7814. [Google Scholar] [CrossRef]
- Waters, B.M.; Blevins, D.G.; Eide, D.J. Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol. 2002, 129, 85–94. [Google Scholar] [CrossRef]
- Borlotti, A.; Vigani, G.; Zocchi, G. Iron deficiency affects nitrogen metabolism in cucumber (Cucumis sativus L.) plants. BMC Plant Biol. 2012, 12, 189. [Google Scholar] [CrossRef]
- Liu, X.Y.; Cui, H.Q.; Li, A.N.; Zhang, M.; Teng, Y.B. The nitrate transporter NRT1.1 is involved in iron deficiency responses in Arabidopsis. J. Plant Nutr. Soil Sci. 2015, 178, 601–608. [Google Scholar] [CrossRef]
- Schmitz, G.; Reinhold, T.; Gobel, C.; Feussner, I.; Neuhaus, H.E.; Conrath, U. Limitation of Nocturnal ATP Import into Chloroplasts Seems to Affect Hormonal Crosstalk, Prime Defense, and Enhance Disease Resistance in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2010, 23, 1584–1591. [Google Scholar] [CrossRef]
- Porri, A.; Torti, S.; Romera-Branchat, M.; Coupland, G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 2012, 139, 2198–2209. [Google Scholar] [CrossRef] [PubMed]
- Chen, H. Stability of plant defense proteins in the gut of insect herbivores. Plant Physiol. 2007, 144, 1233. [Google Scholar] [CrossRef]
- Gupta, N.; Gupta, A.K.; Singh, N.K.; Kumar, A. Differential Expression of PBF Dof Transcription Factor in Different Tissues of Three Finger Millet Genotypes Differing in Seed Protein Content and Color. Plant Mol. Biol. Report. 2011, 29, 69–76. [Google Scholar] [CrossRef]
- Corrales, A.R.; Nebauer, S.G.; Carrillo, L.; Fernandez-Nohales, P.; Marques, J.; Renau-Morata, B.; Granell, A.; Pollmann, S.; Vicente-Carbajosa, J.; Molina, R.V.; et al. Characterization of tomato Cycling Dof Factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. J. Exp. Bot. 2014, 65, 995–1012. [Google Scholar] [CrossRef]
- Yang, X.H.; Tuskan, G.A.; Cheng, Z.M. Divergence of the Dof gene families in poplar, Arabidopsis, and rice suggests multiple modes of gene evolution after duplication. Plant Physiol. 2006, 142, 820–830. [Google Scholar] [CrossRef] [PubMed]
- Cai, X.F.; Zhang, Y.Y.; Zhang, C.J.; Zhang, T.Y.; Hu, T.X.; Ye, J.; Zhang, J.H.; Wang, T.T.; Li, H.X.; Ye, Z.B. Genome-wide Analysis of Plant-specific Dof Transcription Factor Family in Tomato. J. Integr. Plant Biol. 2013, 55, 552–566. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef]
- Osorio, S.; Ruan, Y.L.; Fernie, A.R. An update on source-to-sink carbon partitioning in tomato. Front. Plant Sci. 2014, 5, 516. [Google Scholar] [CrossRef]
- Bertin, N.; Tchamitchian, M.; Baldet, P.; Devaux, C.; Brunel, B.; Gary, C. Contribution of carbohydrate pools to the variations in leaf mass per area within a tomato plant. New Phytol. 1999, 143, 53–61. [Google Scholar] [CrossRef]
- Prudent, M.; Causse, M.; Genard, M.; Tripodi, P.; Grandillo, S.; Bertin, N. Genetic and physiological analysis of tomato fruit weight and composition: Influence of carbon availability on QTL detection. J. Exp. Bot. 2009, 60, 923–937. [Google Scholar] [CrossRef]
- Bloom, A.J.; Burger, M.; Rubio-Asensio, J.S.; Cousins, A.B. Carbon Dioxide Enrichment Inhibits Nitrate Assimilation in Wheat and Arabidopsis. Science 2010, 328, 899–903. [Google Scholar] [CrossRef]
- Bloom, A.J. The increasing importance of distinguishing among plant nitrogen sources. Curr. Opin. Plant Biol. 2015, 25, 10–16. [Google Scholar] [CrossRef]
- Chan, Z.L.; Bigelow, P.J.; Loescher, W.; Grumet, R. Comparison of salt stress resistance genes in transgenic Arabidopsis thaliana indicates that extent of transcriptomic change may not predict secondary phenotypic or fitness effects. Plant Biotechnol. J. 2012, 10, 284–300. [Google Scholar] [CrossRef]
- Ricroch, A.E.; Berge, J.B.; Kuntz, M. Evaluation of Genetically Engineered Crops Using Transcriptomic, Proteomic, and Metabolomic Profiling Techniques. Plant Physiol. 2011, 155, 1752–1761. [Google Scholar] [CrossRef]
- Guyer, D.; Tuttle, A.; Rouse, S.; Volrath, S.; Johnson, M.; Potter, S.; Gorlach, J.; Goff, S.; Crossland, L.; Ward, E. Activation of latent transgenes in Arabidopsis using a hybrid transcription factor. Genetics 1998, 149, 633–639. [Google Scholar] [CrossRef]
- Haseloff, J.; Hodge, S. Gene Expresssion. U.S. Patent US6255558B1; Application number US09/133,321, 13 August 1998. [Google Scholar]
- Powell, A.L.T.; Nguyen, C.V.; Hill, T.; Cheng, K.L.; Figueroa-Balderas, R.; Aktas, H.; Ashrafi, H.; Pons, C.; Fernandez-Munoz, R.; Vicente, A.; et al. Uniform ripening Encodes a Golden 2-like Transcription Factor Regulating Tomato Fruit Chloroplast Development. Science 2012, 336, 1711–1715. [Google Scholar] [CrossRef]
- Luengwilai, K.; Beckles, D.M. Structural Investigations and Morphology of Tomato Fruit Starch. J. Agric. Food Chem. 2009, 57, 282–291. [Google Scholar] [CrossRef]
- McClean, E.O.; Adams, D.; Franklin, R.E. Cation exchange capacities of plant roots as related to their nitrogen contents. Soil Sci. Soc. Proc. 1956, 20, 345–347. [Google Scholar] [CrossRef]
- Leterrier, M.; Holappa, L.D.; Broglie, K.E.; Beckles, D.M. Cloning, characterisation and comparative analysis of a starch synthase IV gene in wheat: Functional and evolutionary implications. BMC Plant Biol. 2008, 8, 98. [Google Scholar] [CrossRef]
- Fernandez-Pozo, N.; Menda, N.; Edwards, J.D.; Saha, S.; Tecle, I.Y.; Strickler, S.R.; Bombarely, A.; Fisher-York, T.; Pujar, A.; Foerster, H.; et al. The Sol Genomics Network (SGN)-from genotype to phenotype to breeding. Nucleic Acids Res. 2015, 43, D1036–D1041. [Google Scholar] [CrossRef]
- Arnon, D.I. Copper Enzymes in Isolated Chloroplasts—Polyphenoloxidase in Beta-Vulgaris. Plant Physiol. 1949, 24, 1–15. [Google Scholar] [CrossRef]
- AOAC. Official Methods of Analysis of AOAC International; AOAC: Rockville, MA, USA, 1997. [Google Scholar]
- Jacobs, A.; Lunde, C.; Bacic, A.; Tester, M.; Roessner, U. The impact of constitutive heterologous expression of a moss Na+ transporter on the metabolomes of rice and barley. Metabolomics 2007, 3, 307–317. [Google Scholar] [CrossRef]
- Mtunguja, M.K.; Thitisaksakul, M.; Muzanila, Y.C.; Chen, G.; Wansuksri, R.; Piyachomkwan, K.; Laswai, H.; Chen, G.; Shoemaker, C.F.; Sinha, N.; et al. Assessing variation in physicochemical, structural, and functional properties of root starches from novel Tanzanian cassava (Manihot esculenta Crantz.) landraces. Starch-Stärke 2016, 68, 514–527. [Google Scholar] [CrossRef][Green Version]
- Gregg, J.P.; Lit, L.; Baron, C.A.; Hertz-Picciotto, I.; Walker, W.; Davis, R.A.; Croen, L.A.; Ozonoff, S.; Hansen, R.; Pessah, I.N.; et al. Gene expression changes in children with autism. Genomics 2008, 91, 22–29. [Google Scholar] [CrossRef]
- Lin, M.; Wei, L.-J.; Sellers, W.R.; Lieberfarb, M.; Wong, W.-H.; Li, C. dChipSNP: Significance curve and clustering of SNP-Array-Based Loss-of-Heterozygosity Data. Bioinformatics 2004, 20, 1233–1240. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The Neighbor-Joining Method—A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Felsenstein, J. Confidence-Limits on Phylogenies—An Approach Using the Bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Zuckerkandl, E.; Pauling, L. (Eds.) Evolutionary Divergence and Convergence in Proteins; Academic Press: New York, NY, USA, 1965; pp. 97–166. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- TAIR. The Arabidopsis Information Resource. Available online: http://www.arabidopsis.org/servlets/TairObject?id=137136&type=locus (accessed on 20 December 2018).
- Kleinboelting, N.; Huep, G.; Kloetgen, A.; Viehoever, P.; Weisshaar, B. GABI-Kat SimpleSearch: New features of the Arabidopsis thaliana T-DNA mutant database. Nucleic Acids Res. 2012, 40, D1211–D1215. [Google Scholar] [CrossRef]
- Rosso, M.G.; Li, Y.; Strizhov, N.; Reiss, B.; Dekker, K.; Weisshaar, B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Mol. Biol. 2003, 53, 247–259. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Lopez-Vidriero, I.; Carrasco, J.L.; Godoy, M.; Vera, P.; Solano, R. DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. USA 2014, 111, 2367–2372. [Google Scholar] [CrossRef]
- O′Malley, R.C.; Huang, S.S.C.; Song, L.; Lewsey, M.G.; Bartlett, A.; Nery, J.R.; Galli, M.; Gallavotti, A.; Ecker, J.R. Cistrome and Epicistrome Features Shape the Regulatory DNA Landscape. Cell 2016, 165, 1280–1292. [Google Scholar] [CrossRef]
- Weirauch, M.T.; Yang, A.; Albu, M.; Cote, A.G.; Montenegro-Montero, A.; Drewe, P.; Najafabadi, H.S.; Lambert, S.A.; Mann, I.; Cook, K.; et al. Determination and Inference of Eukaryotic Transcription Factor Sequence Specificity. Cell 2014, 158, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Grant, C.E.; Bailey, T.L.; Noble, W.S. FIMO: Scanning for occurrences of a given motif. Bioinformatics 2011, 27, 1017–1018. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hao, Q.; Bai, L.; Xu, J.; Yin, W.; Song, L.; Xu, L.; Guo, X.; Fan, C.; Chen, Y. Overexpression of the soybean transcription factor GmDof4 significantly enhances the lipid content of Chlorella ellipsoidea. Biotechnol. biofuels 2014, 7, 128. [Google Scholar] [CrossRef]
- Wu, J.; Chen, L.; Chen, M.; Zhou, W.; Dong, Q.; Jiang, H.; Cheng, B. The DOF-Domain Transcription Factor ZmDOF36 Positively Regulates Starch Synthesis in Transgenic Maize. Frontiers in Plant Science 2019, 10, 465. [Google Scholar] [CrossRef]
- Zhang, A.D.; Wang, W.Q.; Tong, Y.; Li, M.J.; Grierson, D.; Ferguson, I.; Chen, K.S.; Yin, X.R. Transcriptome Analysis Identifies a Zinc Finger Protein Regulating Starch Degradation in Kiwifruit. Plant Physiol 2018, 178, 850–863. [Google Scholar] [CrossRef]


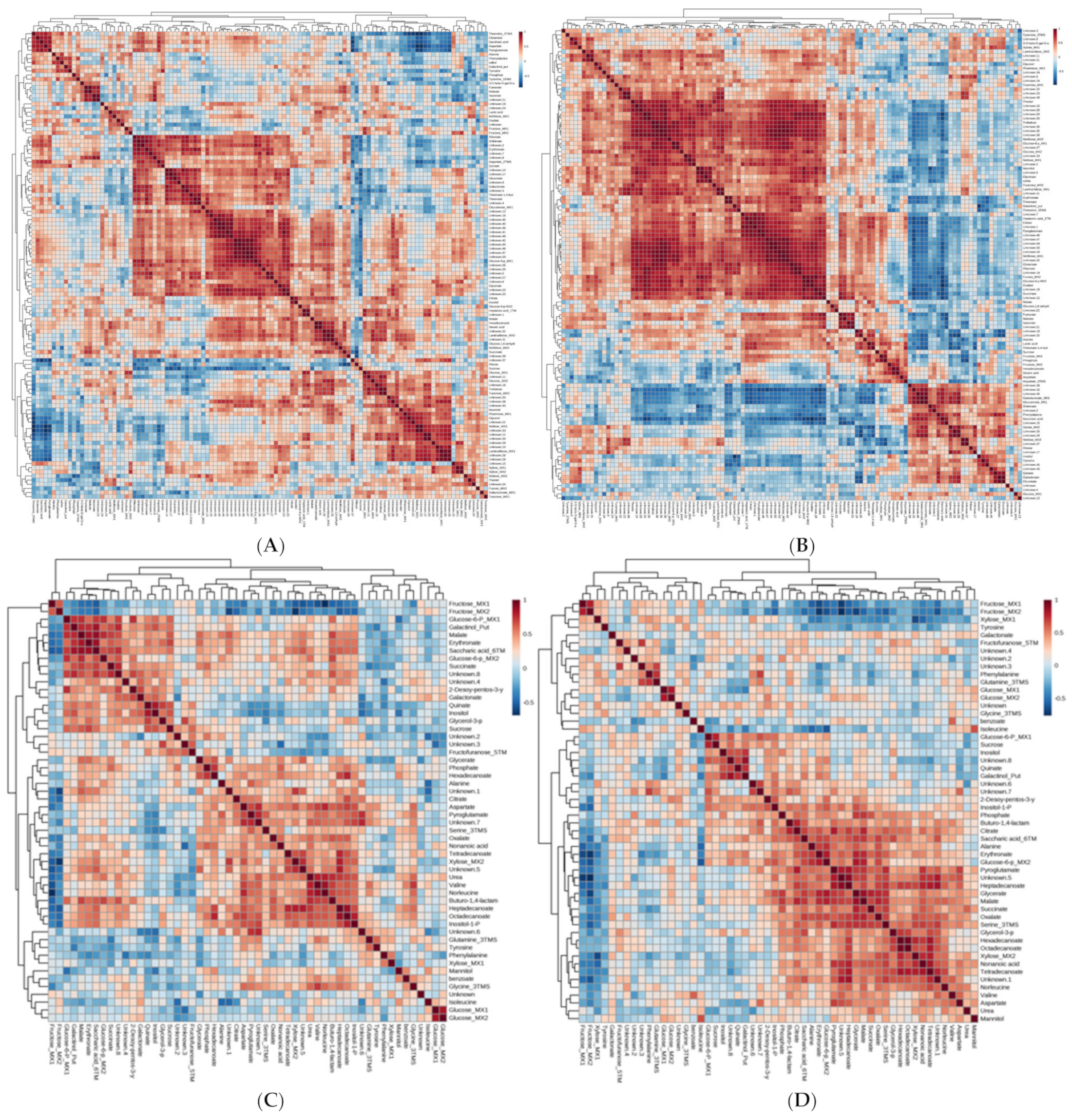

| Metabolites | L4080 vs. WT | |||
|---|---|---|---|---|
| 21 DPA | Breaker | |||
| x-fold | t-test | x-fold | t-test | |
| Aspartate | 0.818 | 0.005 | 3.841 | 0.002 |
| Citrate | 0.696 | 0.014 | 1.093 | 0.179 |
| Fucose_MX2 | 2.556 | 0.706 | 1.832 | 0.039 |
| Galactonate | 1.155 | 0.000 | 1.165 | 0.176 |
| Galacturonate_MX1 | 83.124 | 0.002 | 3.627 | 0.052 |
| Gluconate | 1.255 | 0.023 | 1.189 | 0.257 |
| Glucose_MX2 | 2.069 | 0.215 | 3.219 | 0.020 |
| Glucose-6-p_MX1 | 0.806 | 0.298 | 2.029 | 0.011 |
| Glucose-6-p-MX2 | 0.913 | 0.158 | 1.775 | 0.04 |
| Glucuronate_MX1 | 3.589 | 0.003 | 1.713 | 0.456 |
| Glutamate | 0.599 | 0.799 | 2.157 | 0.005 |
| Heptanoic acid_1TMS | 0.540 | 0.008 | 0.926 | 0.929 |
| Inositol | 0.901 | 0.603 | 0.736 | 0.046 |
| Lactic acid | 0.739 | 0.035 | 1.042 | 0.897 |
| Maltose_MX1 | 1.350 | 0.002 | 0.838 | 0.833 |
| Mannitol | 0.840 | 0.238 | 1.061 | 0.883 |
| Melibiose_MX1 | 0.530 | 0.176 | 3.198 | 0.012 |
| Oxalate | 0.459 | 0.027 | 1.494 | 0.063 |
| Phenylalanine | 1.934 | 0.000 | 1.828 | 0.021 |
| Pyroglutamate | 0.630 | 0.491 | 1.651 | 0.000 |
| Ribonate | 0.853 | 0.008 | 1.829 | 0.009 |
| Saccharic acid | 0.869 | 0.000 | 1.140 | 0.524 |
| Shikimate | 1.212 | 0.002 | 1.618 | 0.007 |
| Threonate-1,4-lactone | 0.092 | 0.000 | 0.584 | 0.189 |
| Threonine 3TMS | 0.685 | 0.028 | 3.307 | 0.001 |
| Tyramine 3TMS | 1.270 | 0.000 | 1.835 | 0.016 |
| Tyrosine | 1.043 | 0.117 | 0.267 | 0.016 |
| Xylose_MX2 | 2.946 | 0.004 | 1.940 | 0.224 |
| Unknown | 0.502 | 0.045 | 1.587 | 0.070 |
| Unknown | 0.521 | 0.005 | 0.866 | 0.489 |
| Unknown | 0.550 | 0.427 | 2.812 | 0.003 |
| Unknown | 0.585 | 0.311 | 2.707 | 0.001 |
| Unknown | 0.632 | 0.038 | 3.259 | 0.181 |
| Unknown | 0.638 | 0.095 | 2.164 | 0.051 |
| Unknown | 0.695 | 0.090 | 2.168 | 0.022 |
| Unknown | 0.784 | 0.133 | 1.643 | 0.001 |
| Unknown | 0.831 | 0.879 | 1.911 | 0.040 |
| Unknown | 0.837 | 0.049 | 1.772 | 0.021 |
| Unknown | 0.848 | 0.060 | 2.279 | 0.000 |
| Unknown | 0.948 | 0.000 | 1.478 | 0.002 |
| Unknown | 0.962 | 0.16 | 1.681 | 0.043 |
| Unknown | 0.986 | 0.003 | 1.295 | 0.010 |
| Unknown | 1.180 | 0.078 | 1.514 | 0.100 |
| Unknown | 1.447 | 0.003 | 1.223 | 0.173 |
| Unknown | 1.518 | 0.006 | 0.597 | 0.265 |
| Unknown | 1.605 | 0.007 | 0.838 | 0.833 |
| Unknown | 1.967 | 0.015 | 1.522 | 0.123 |
| Unknown | 2.129 | 0.007 | 1.849 | 0.785 |
| Unknown | 2.788 | 0.027 | 0.943 | 0.737 |
| Unknown | 3.318 | 0.021 | 0.190 | 0.065 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luengwilai, K.; Yu, J.; Jiménez, R.C.; Thitisaksakul, M.; Vega, A.; Dong, S.; Beckles, D.M. Ectopic Expression of Arabidopsis thaliana zDof1.3 in Tomato (Solanum lycopersicum L.) Is Associated with Improved Greenhouse Productivity and Enhanced Carbon and Nitrogen Use. Int. J. Mol. Sci. 2022, 23, 11229. https://doi.org/10.3390/ijms231911229
Luengwilai K, Yu J, Jiménez RC, Thitisaksakul M, Vega A, Dong S, Beckles DM. Ectopic Expression of Arabidopsis thaliana zDof1.3 in Tomato (Solanum lycopersicum L.) Is Associated with Improved Greenhouse Productivity and Enhanced Carbon and Nitrogen Use. International Journal of Molecular Sciences. 2022; 23(19):11229. https://doi.org/10.3390/ijms231911229
Chicago/Turabian StyleLuengwilai, Kietsuda, Jingwei Yu, Randi C. Jiménez, Maysaya Thitisaksakul, Andrea Vega, Shaoyun Dong, and Diane M. Beckles. 2022. "Ectopic Expression of Arabidopsis thaliana zDof1.3 in Tomato (Solanum lycopersicum L.) Is Associated with Improved Greenhouse Productivity and Enhanced Carbon and Nitrogen Use" International Journal of Molecular Sciences 23, no. 19: 11229. https://doi.org/10.3390/ijms231911229
APA StyleLuengwilai, K., Yu, J., Jiménez, R. C., Thitisaksakul, M., Vega, A., Dong, S., & Beckles, D. M. (2022). Ectopic Expression of Arabidopsis thaliana zDof1.3 in Tomato (Solanum lycopersicum L.) Is Associated with Improved Greenhouse Productivity and Enhanced Carbon and Nitrogen Use. International Journal of Molecular Sciences, 23(19), 11229. https://doi.org/10.3390/ijms231911229







