The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice
Abstract
1. Introduction
2. Results
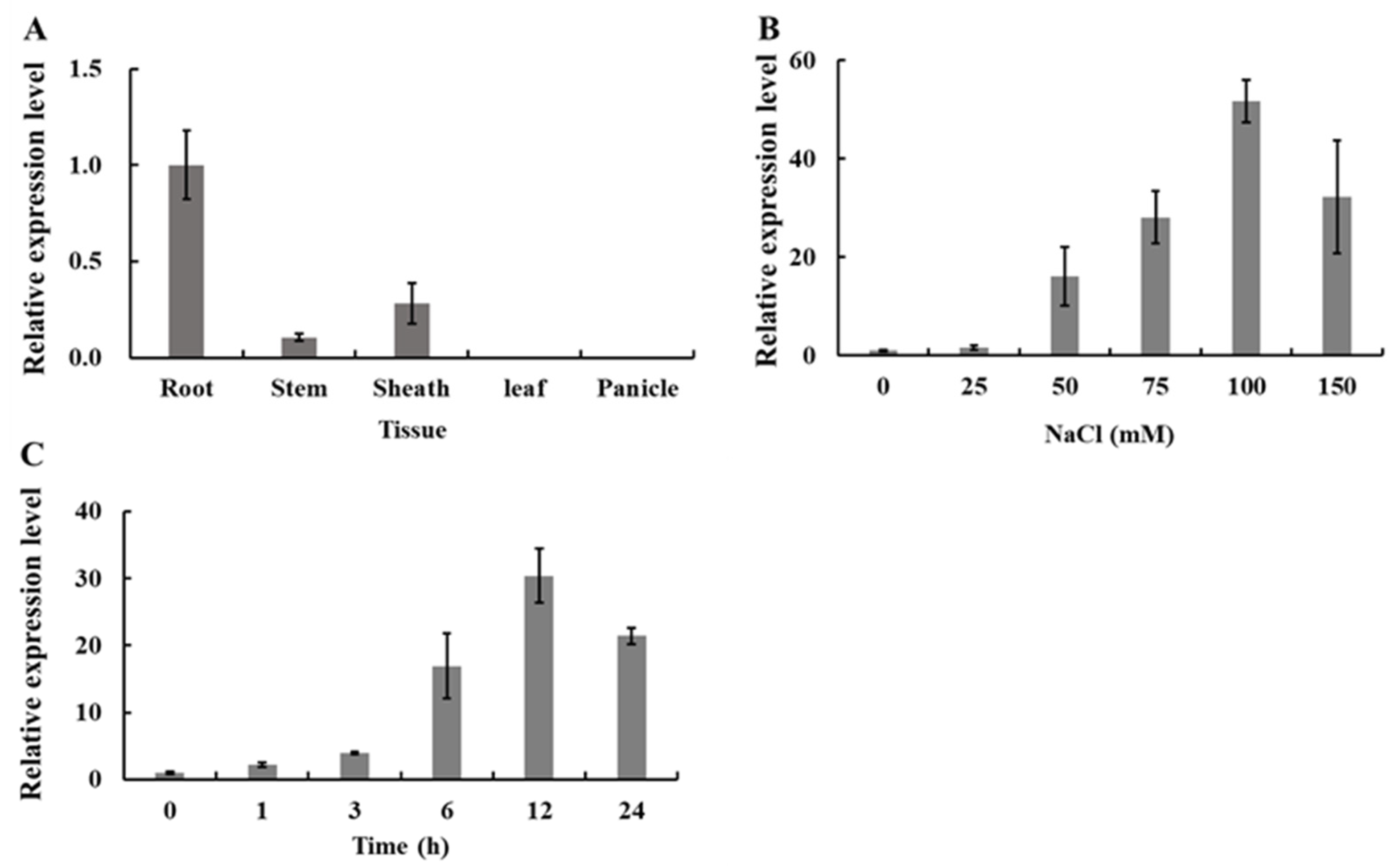
2.1. Expression Pattern of OsWRKY54
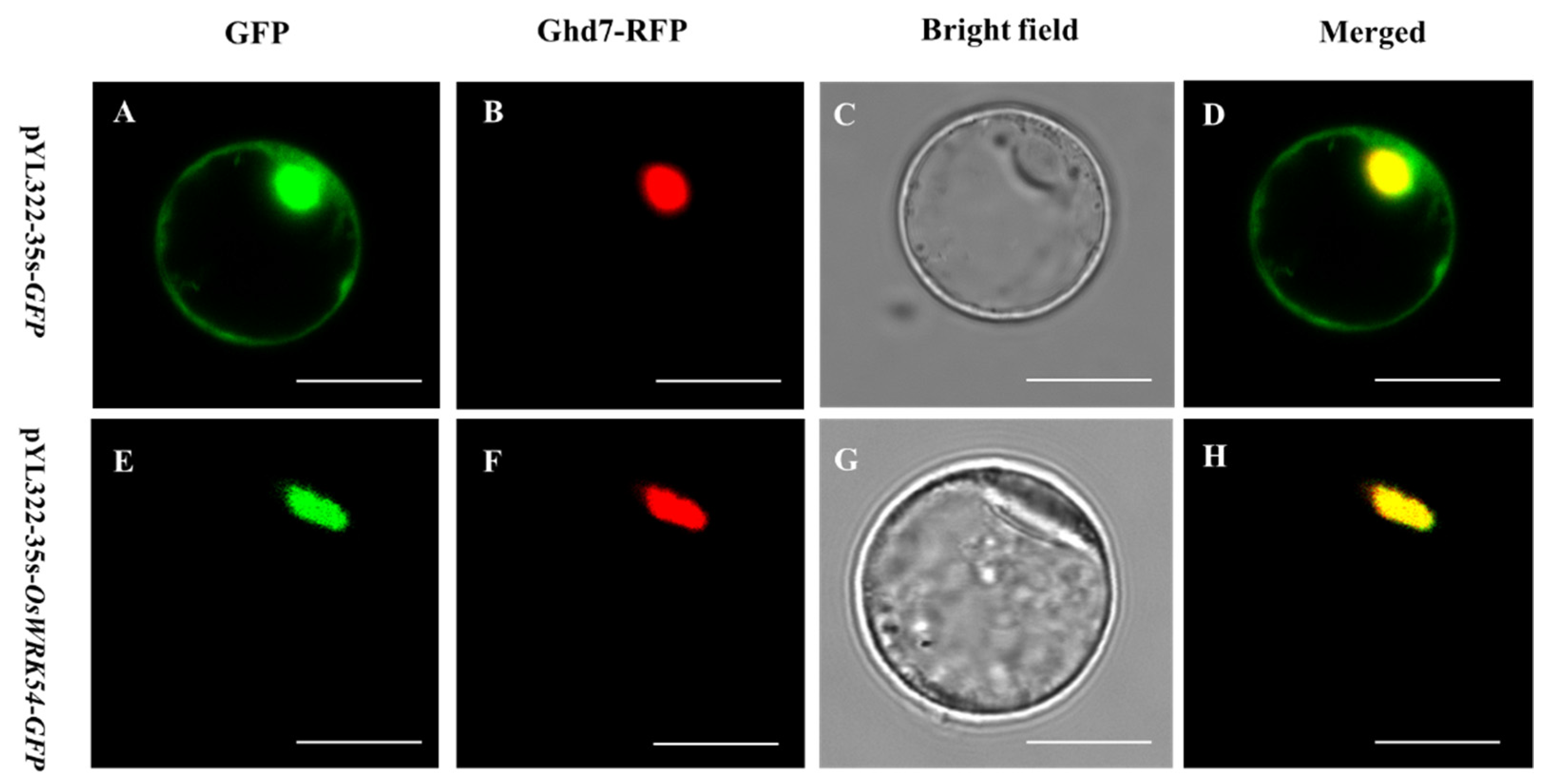
2.2. Subcellular Localization of OsWRKY54
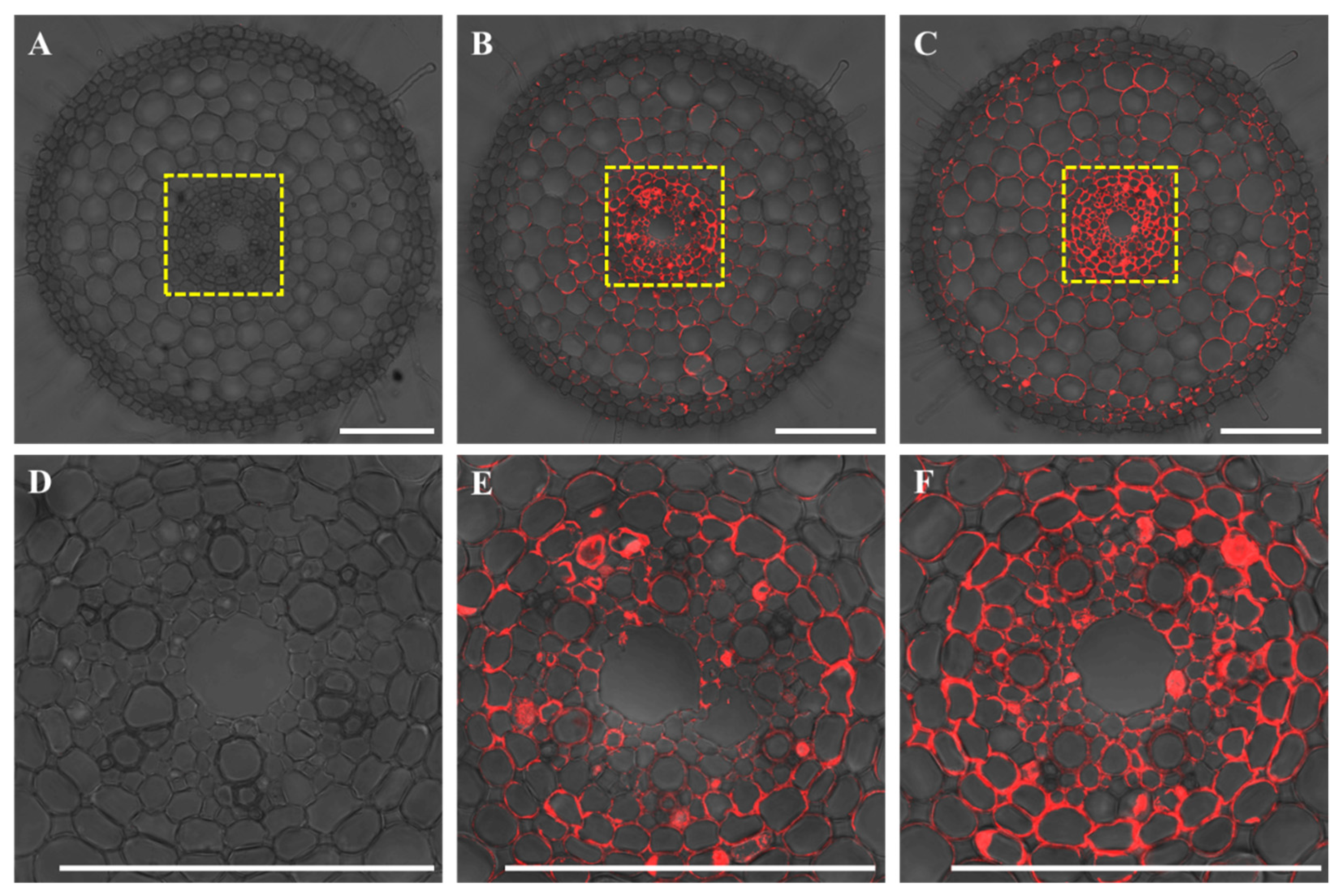
2.3. Cell-Specific Expression of OsWRKY54
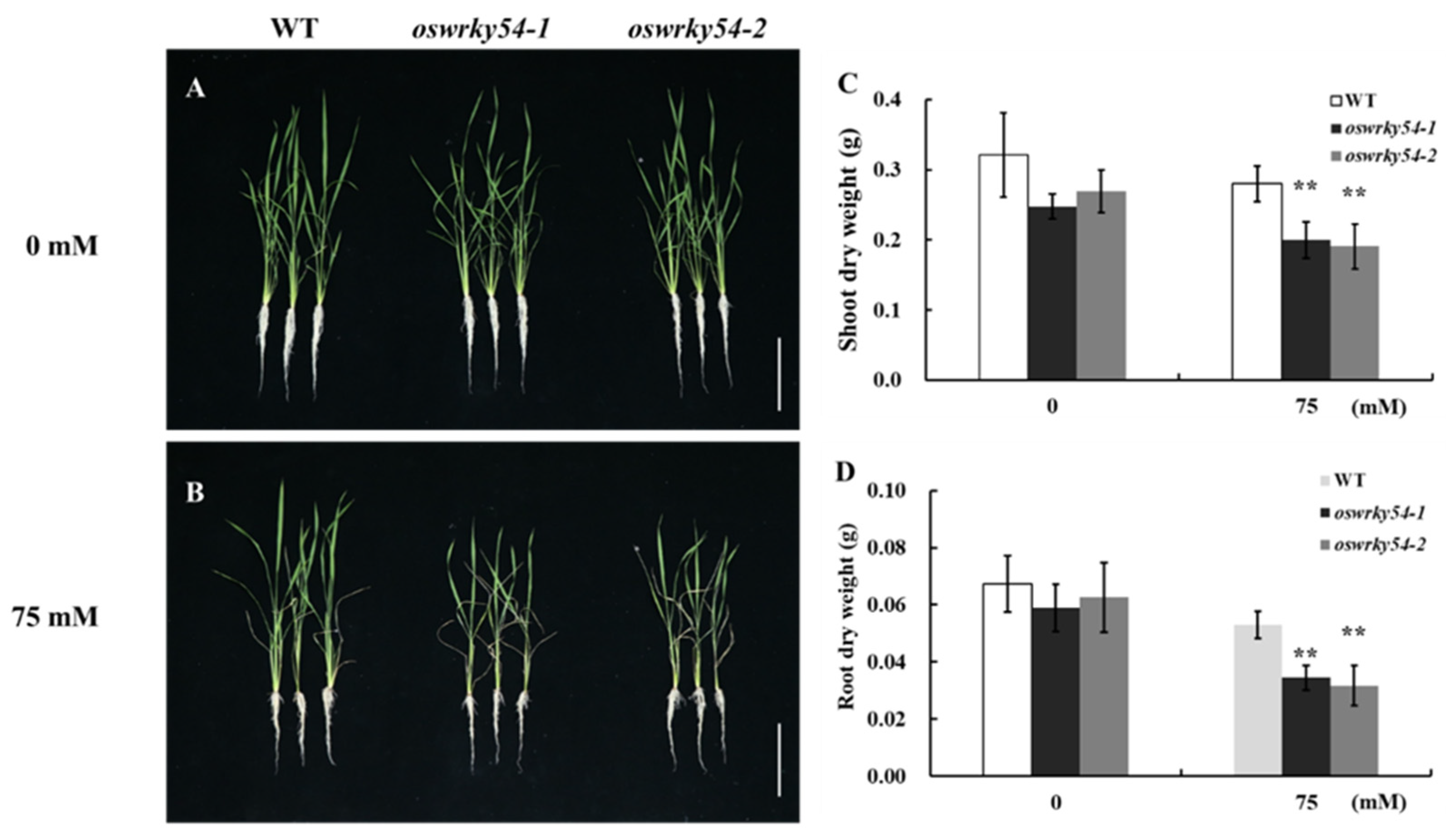
2.4. Knockout of OsWRKY54 Decreased Salt Tolerance of Rice
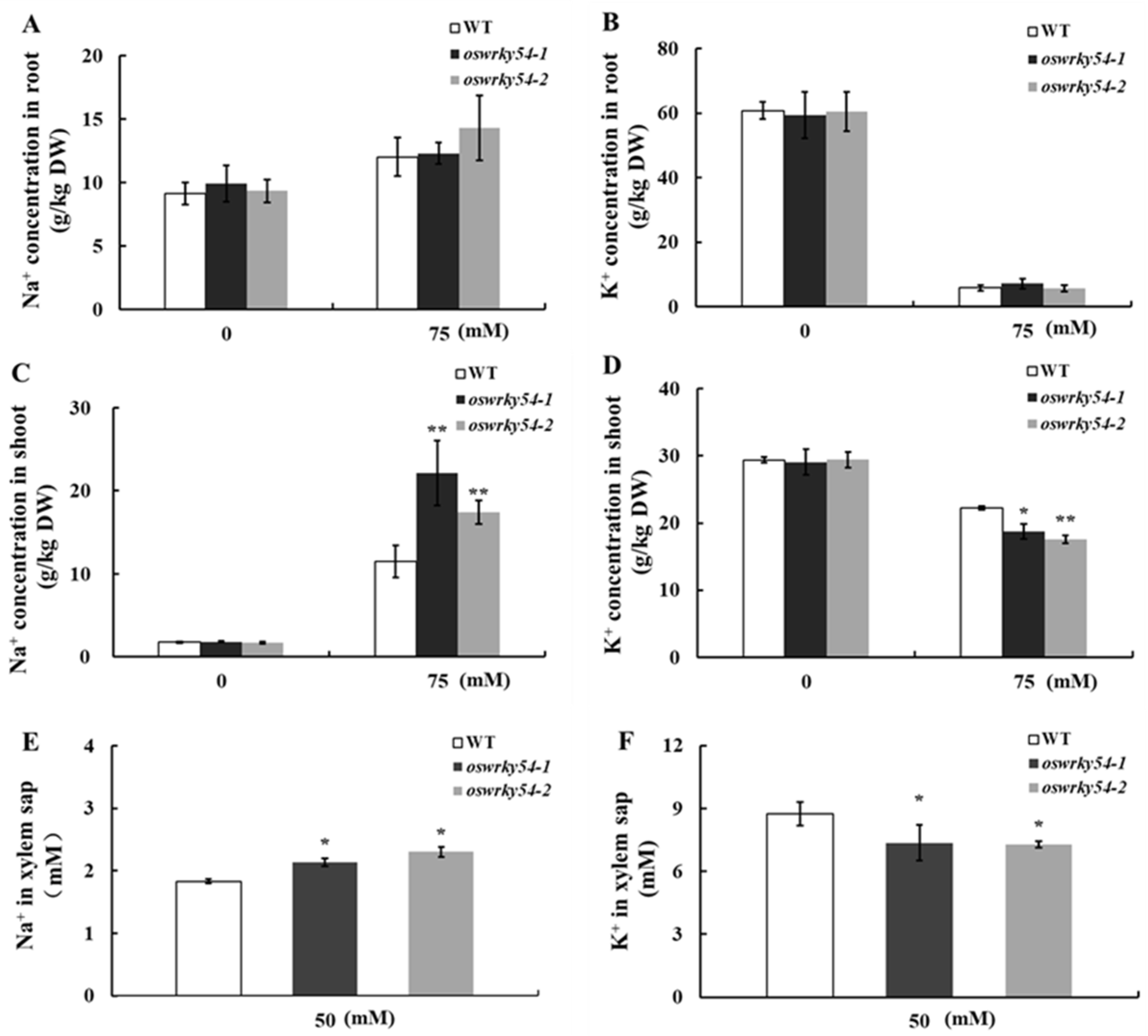
2.5. Loss of OsWRKY54 Alters Shoot Na+/K+ Homeostasis of Transgenic Rice
2.6. Transcriptional Activation Analysis of OsWRKY54
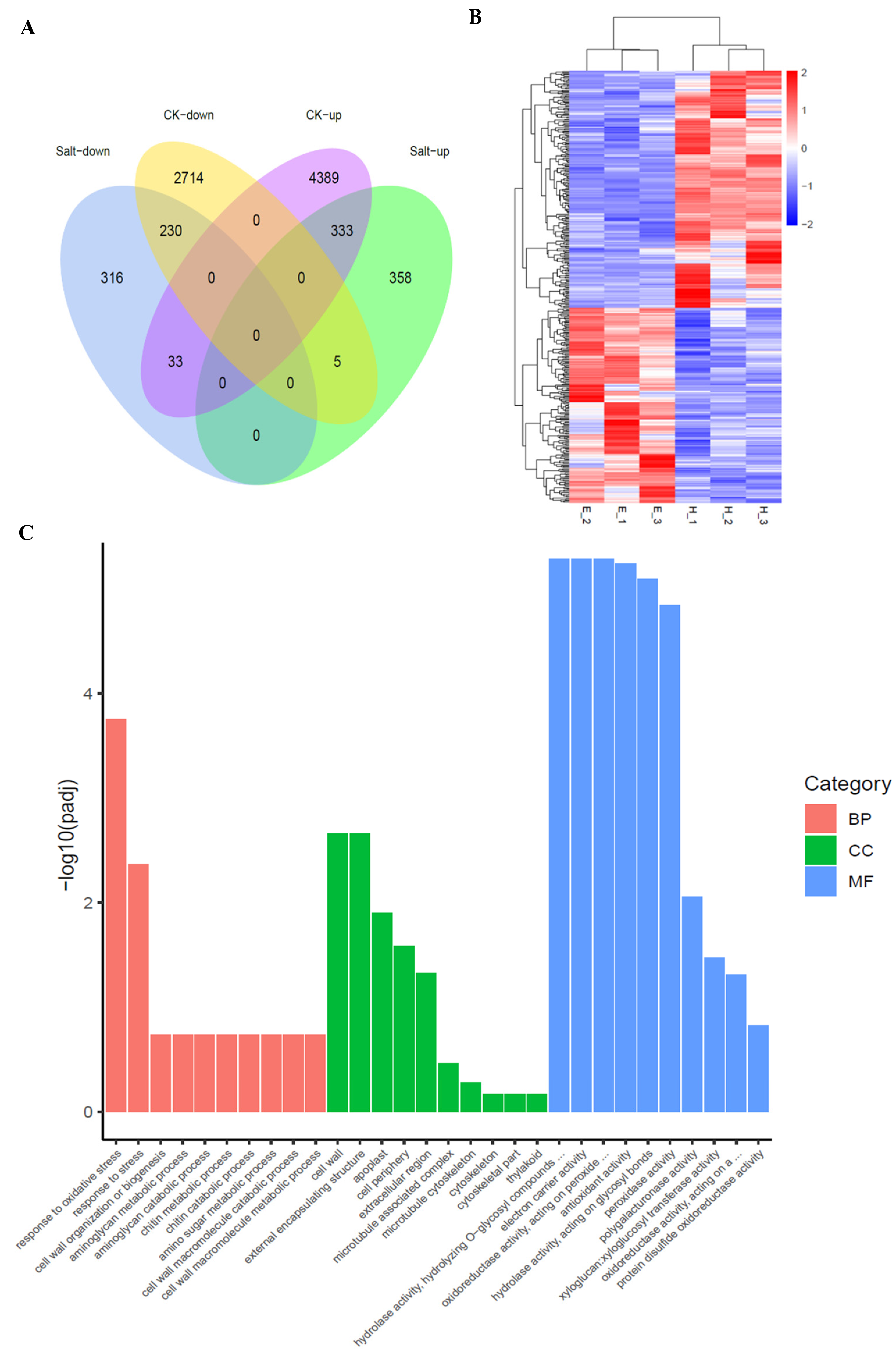
2.7. OsWRKY54 Affects the Expression Profiles of Many Stress-Related Genes
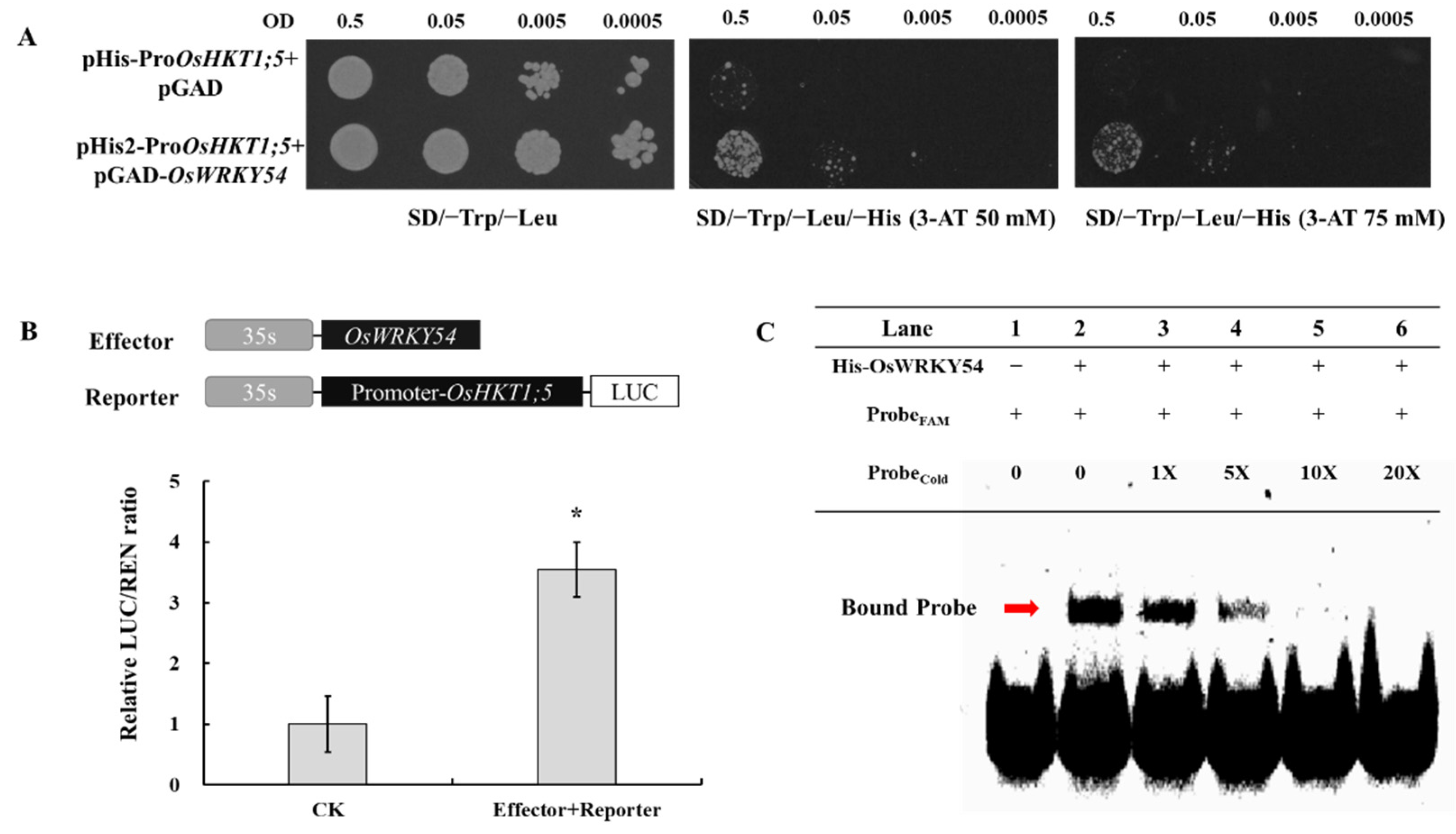
2.8. OsHKT1;5 Is a Direct Target of OsWRKY54
3. Discussion
4. Materials and Methods
4.1. Generation of OsWRKY54 Knockout Mutants
4.2. Plant Materials and Growth Conditions
4.3. Gene Expression Analysis
4.4. Subcellular Localization of OsWRKY54
4.5. Tissue Localization of OsWRKY54
4.6. Physiological Characterization of OsWRKY54 Mutant Lines
4.7. Na/K Concentration in Xylem Sap
4.8. Determination of Na/K Concentration
4.9. RNA-Seq Assay
4.10. Dual-LUC Transient Expression Assay
4.11. Yeast Assays
4.12. EMSA
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rengasamy, P. Soil processes affecting crop production in salt-affected soils. Funct. Plant Biol. 2010, 37, 613–620. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651. [Google Scholar] [CrossRef]
- Ismail, A.M.; Horie, T. Genomics, Physiology, and Molecular Breeding Approaches for Improving Salt Tolerance. Annu. Rev. Plant Biol. 2017, 68, 405–434. [Google Scholar] [CrossRef]
- Singh, K.; Foley, R.C.; Oñate-Sánchez, L. Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 2002, 5, 430–436. [Google Scholar] [CrossRef]
- Agarwal, P.; Reddy, M.P.; Chikara, J. WRKY: Its structure, evolutionary relationship, DNA-binding selectivity, role in stress tolerance and development of plants. Mol. Biol. Rep. 2011, 38, 3883–3896. [Google Scholar] [CrossRef] [PubMed]
- Bakshi, M.; Oelmüller, R. WRKY transcription factors: Jack of many trades in plants. Plant Signal. Behav. 2014, 9, e27700. [Google Scholar] [CrossRef]
- Ulker, B.; Somssich, I.E. WRKY transcription factors: From DNA binding towards biological function. Curr. Opin. Plant Biol. 2004, 7, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Eulgem, T.; Rushton, P.J.; Robatzek, S.; Somssich, I.E. The WRKY superfamily of plant transcription factors. Trends Plant Sci. 2000, 5, 199–206. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, L.; Wang, H.; Zhang, L.; Wang, F.; Yu, D. Arabidopsis transcription factor WRKY8 functions antagonistically with its interacting partner VQ9 to modulate salinity stress tolerance. Plant J. Cell Mol. Biol. 2013, 74, 730–745. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.F.; Liu, J.K.; Yang, F.M.; Zhang, G.Y.; Wang, D.; Zhang, L.; Ou, Y.B.; Yao, Y.A. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Plant Physiol. 2020, 168, 98–117. [Google Scholar] [CrossRef] [PubMed]
- Cai, R.; Dai, W.; Zhang, C.; Wang, Y.; Wu, M.; Zhao, Y.; Ma, Q.; Xiang, Y.; Cheng, B. The maize WRKY transcription factor ZmWRKY17 negatively regulates salt stress tolerance in transgenic Arabidopsis plants. Planta 2017, 246, 1215–1231. [Google Scholar] [CrossRef] [PubMed]
- Ye, H.; Qiao, L.; Guo, H.; Guo, L.; Ren, F.; Bai, J.; Wang, Y. Genome-Wide Identification of Wheat WRKY Gene Family Reveals That TaWRKY75-A Is Referred to Drought and Salt Resistances. Front. Plant Sci. 2021, 12, 663118. [Google Scholar] [CrossRef]
- Huang, S.; Hu, L.; Zhang, S.; Zhang, M.; Jiang, W.; Wu, T.; Du, X. Rice OsWRKY50 Mediates ABA-Dependent Seed Germination and Seedling Growth, and ABA-Independent Salt Stress Tolerance. Int. J. Mol. Sci. 2021, 22, 8625. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, B.; Li, J.; Sun, Z.; Chi, M.; Xing, Y.; Xu, B.; Yang, B.; Li, J.; Liu, J.; et al. A novel SAPK10-WRKY87-ABF1 biological pathway synergistically enhance abiotic stress tolerance in transgenic rice (Oryza sativa). Plant Physiol Biochem. 2021, 12, 252–262. [Google Scholar]
- Fu, S.; Fu, L.; Zhang, X.; Huang, J.; Yang, G.; Wang, Z.; Liu, Y.G.; Zhang, G.; Wu, D.; Xia, J. OsC2DP, a Novel C2 Domain-Containing Protein Is Required for Salt Tolerance in Rice. Plant Cell Physiol. 2019, 60, 2220–2230. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; Wang, G.; Hu, Y.; Liu, H.; Bai, X.; Qin, R.; Xing, Y. OsMFT1 increases spikelets per panicle and delays heading date in rice by suppressing Ehd1, FZP and SEPALLATA-like genes. J. Exp. Bot. 2018, 69, 4283–4293. [Google Scholar] [CrossRef]
- He, X.; Li, L.; Xu, H.; Xi, J.; Cao, X.; Xu, H.; Rong, S.; Dong, Y.; Wang, C.; Chen, R.; et al. A rice jacalin-related mannose-binding lectin gene, OsJRL, enhances Escherichia coli viability under high salinity stress and improves salinity tolerance of rice. J. Plant Biol. 2017, 19, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet. 2007, 115, 35–46. [Google Scholar] [CrossRef] [PubMed]
- Serra, T.S.; Figueiredo, D.D.; Cordeiro, A.M.; Almeida, D.M.; Lourenço, T.; Abreu, I.A.; Sebastián, A.; Fernandes, L.; Contreras-Moreira, B.; Oliveira, M.M.; et al. OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol. Biol. 2013, 82, 439–455. [Google Scholar] [CrossRef] [PubMed]
- Park, Y.C.; Chapagain, S.; Jang, C.S. The microtubule-associated RING finger protein 1 (OsMAR1) acts as a negative regulator for salt-stress response through the regulation of OCPI2 (O. sativa chymotrypsin protease inhibitor 2). Planta 2018, 247, 875–886. [Google Scholar] [CrossRef] [PubMed]
- Jadamba, C.; Kang, K.; Paek, N.C.; Lee, S.I.; Yoo, S.C. Overexpression of Rice Expansin7 (Osexpa7) Confers Enhanced Tolerance to Salt Stress in Rice. Int. J. Mol. Sci. 2020, 2, 454. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Hauser, F.; Schroeder, J.I. HKT transporter-mediated salinity resistance mechanisms in Arabidopsis and monocot crop plants. Trends Plant Sci. 2009, 14, 660–668. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zheng, C.; Kuang, B.; Wei, L.; Yan, L.; Wang, T. Receptor-Like Kinase RUPO Interacts with Potassium Transporters to Regulate Pollen Tube Growth and Integrity in Rice. PLoS Genet. 2016, 12, e1006085. [Google Scholar] [CrossRef]
- Fukuda, A.; Nakamura, A.; Hara, N.; Toki, S.; Tanaka, Y. Molecular and functional analyses of rice NHX-type Na+/H+ antiporter genes. Planta 2011, 233, 175–188. [Google Scholar] [CrossRef] [PubMed]
- Viana, V.E.; Carlos da Maia, L.; Busanello, C.; Pegoraro, C.; Costa de Oliveira, A. When rice gets the chills: Comparative transcriptome profiling at germination shows WRKY transcription factor responses. Plant Biol. 2021, 23, 100–112. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, T.; Hamamoto, S.; Uozumi, N. Sodium transport system in plant cells. Front. Plant Sci. 2013, 4, 410. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Gu, L.; Ringler, P.; Smith, S.; Rushton, P.J.; Shen, Q.J. Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. Int. J. Exp. Plant Biol. 2015, 236, 214–222. [Google Scholar] [CrossRef]
- Ye, M.; Glauser, G.; Lou, Y.; Erb, M.; Hu, L. Molecular Dissection of Early Defense Signaling Underlying Volatile-Mediated Defense Regulation and Herbivore Resistance in Rice. Plant Cell 2019, 31, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Gu, M.; Liang, R.; Shi, X.; Chen, L.; Hu, X.; Wang, S.; Dai, X.; Qu, H.; Li, H.; et al. OsWRKY21 and OsWRKY108 function redundantly to promote phosphate accumulation through maintaining the constitutive expression of OsPHT1;1 under phosphate-replete conditions. New Phytol. 2021, 229, 1598–1614. [Google Scholar] [CrossRef]
- Wei, X.; Zhou, H.; Xie, D.; Li, J.; Yang, M.; Chang, T.; Wang, D.; Hu, L.; Xie, G.; Wang, J.; et al. Genome-Wide Association Study in Rice Revealed a Novel Gene in Determining Plant Height and Stem Development, by Encoding a WRKY Transcription Factor. Int. J. Mol. Sci. 2021, 22, 8192. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A Robust CRISPR/Cas9 System for Convenient, High-Efficiency Multiplex Genome Editing in Monocot and Dicot Plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, N.; Ma, J.F. Spatial distribution and temporal variation of the rice silicon transporter Lsi1. Plant Physiol. 2007, 143, 1306–1313. [Google Scholar] [CrossRef]
- Wang, Z.; Zhang, B.; Chen, Z.; Wu, M.; Chao, D.; Wei, Q.; Xin, Y.; Li, L.; Ming, Z.; Xia, J. Three OsMYB36 members redundantly regulate Casparian strip formation at the root endodermis. Plant Cell 2022, 34, 2948–2968. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Liu, F.; Chao, D.; Xin, B.; Liu, K.; Cao, S.; Chen, X.; Peng, L.; Zhang, B.; Fu, S.; et al. The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 11999. https://doi.org/10.3390/ijms231911999
Huang J, Liu F, Chao D, Xin B, Liu K, Cao S, Chen X, Peng L, Zhang B, Fu S, et al. The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice. International Journal of Molecular Sciences. 2022; 23(19):11999. https://doi.org/10.3390/ijms231911999
Chicago/Turabian StyleHuang, Jingjing, Fuhang Liu, Dong Chao, Boning Xin, Kui Liu, Shuling Cao, Xingxiang Chen, Liyun Peng, Baolei Zhang, Shan Fu, and et al. 2022. "The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice" International Journal of Molecular Sciences 23, no. 19: 11999. https://doi.org/10.3390/ijms231911999
APA StyleHuang, J., Liu, F., Chao, D., Xin, B., Liu, K., Cao, S., Chen, X., Peng, L., Zhang, B., Fu, S., & Xia, J. (2022). The WRKY Transcription Factor OsWRKY54 Is Involved in Salt Tolerance in Rice. International Journal of Molecular Sciences, 23(19), 11999. https://doi.org/10.3390/ijms231911999




