Advancements in MAFLD Modeling with Human Cell and Organoid Models
Abstract
1. Introduction
1.1. MAFLD Epidemiology
1.2. Genetic and Environmental Factors Driving MAFLD
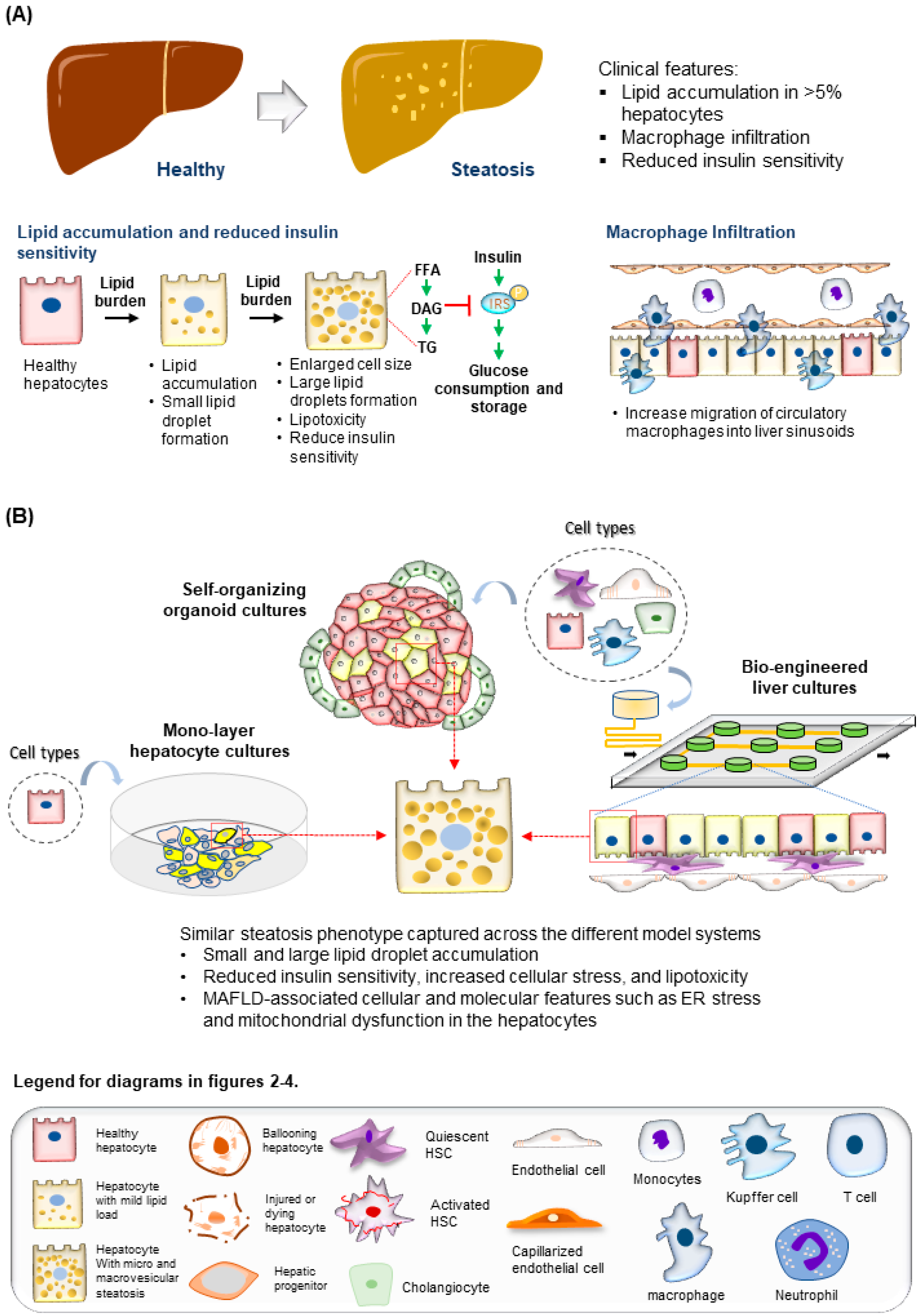
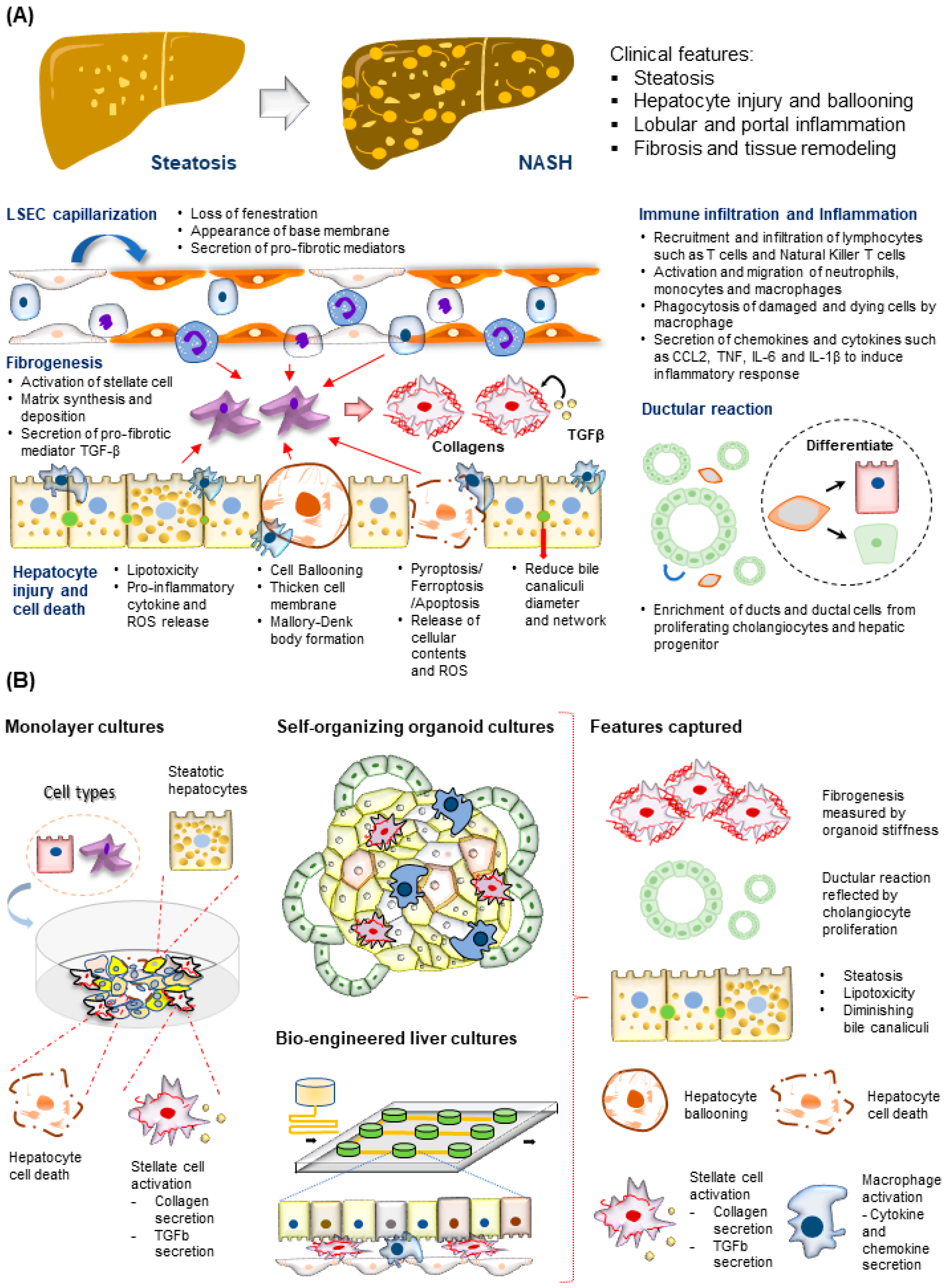
1.3. Molecular and Cellular Features of MAFLD Progression
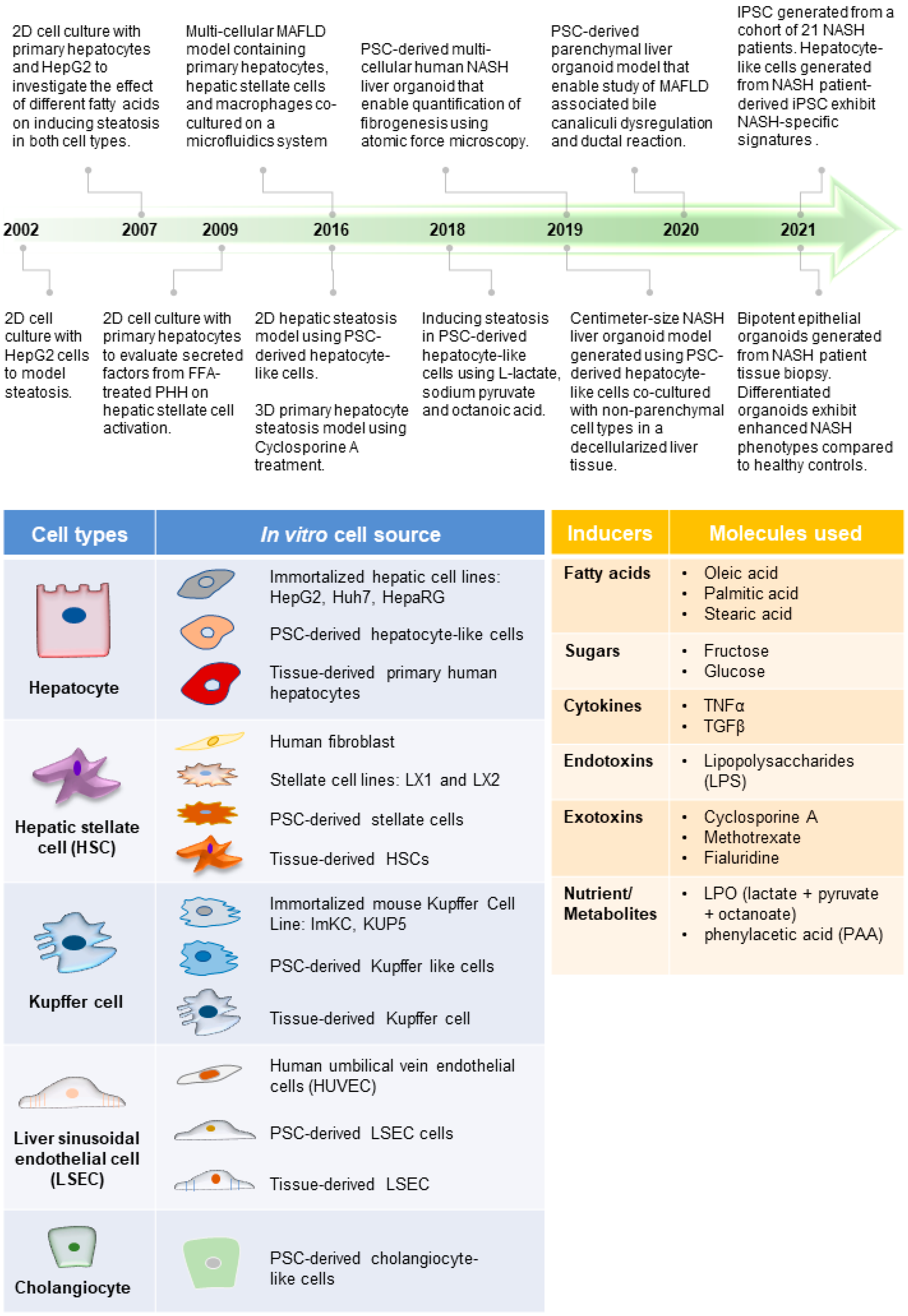
1.4. In Vitro Human Models for MAFLD
2. Human Immortalized and Primary Cell Lines for Modeling MAFLD
2.1. Immortalized Hepatic Cell Lines for Modeling MAFLD
2.2. Liver-Tissue-Derived Primary Cells for Modeling MAFLD
2.3. PSC-Derived Hepatic Cells for MAFLD Modeling
2.4. Liver Non-Hepatic Parenchymal Cell Types
3. Human Multi-Cellular 3D MAFLD Models
Self-Organizing Liver Organoid Model
| Key Approach | Cell Culture Method | Major Features |
|---|---|---|
| Self-organizing liver organoid models | Step-wise differentiation from pluripotent stem cells (PSC) |
|
| ||
| Co-culture of parenchymal and non- parenchymal liver cell types |
| |
| Bio-engineered liver models | Microfluidics culture | |
| ||
| Precision Cut Tissue Slice (PCTS) |
| |
| Organ scaffolds |
|
4. Bioengineered Liver Models
4.1. MAFLD Models with Microfluidics Platform
4.2. MAFLD Tissue-like Culture with Liver Tissue Slices and Organ Scaffolds
5. Current Challenges and Future of MAFLD Modeling
5.1. Modeling Immune Landscape of MAFLD Progression
5.2. Recapitulating MAFLD Stage Transition and Structural Features of Late MAFLD Development
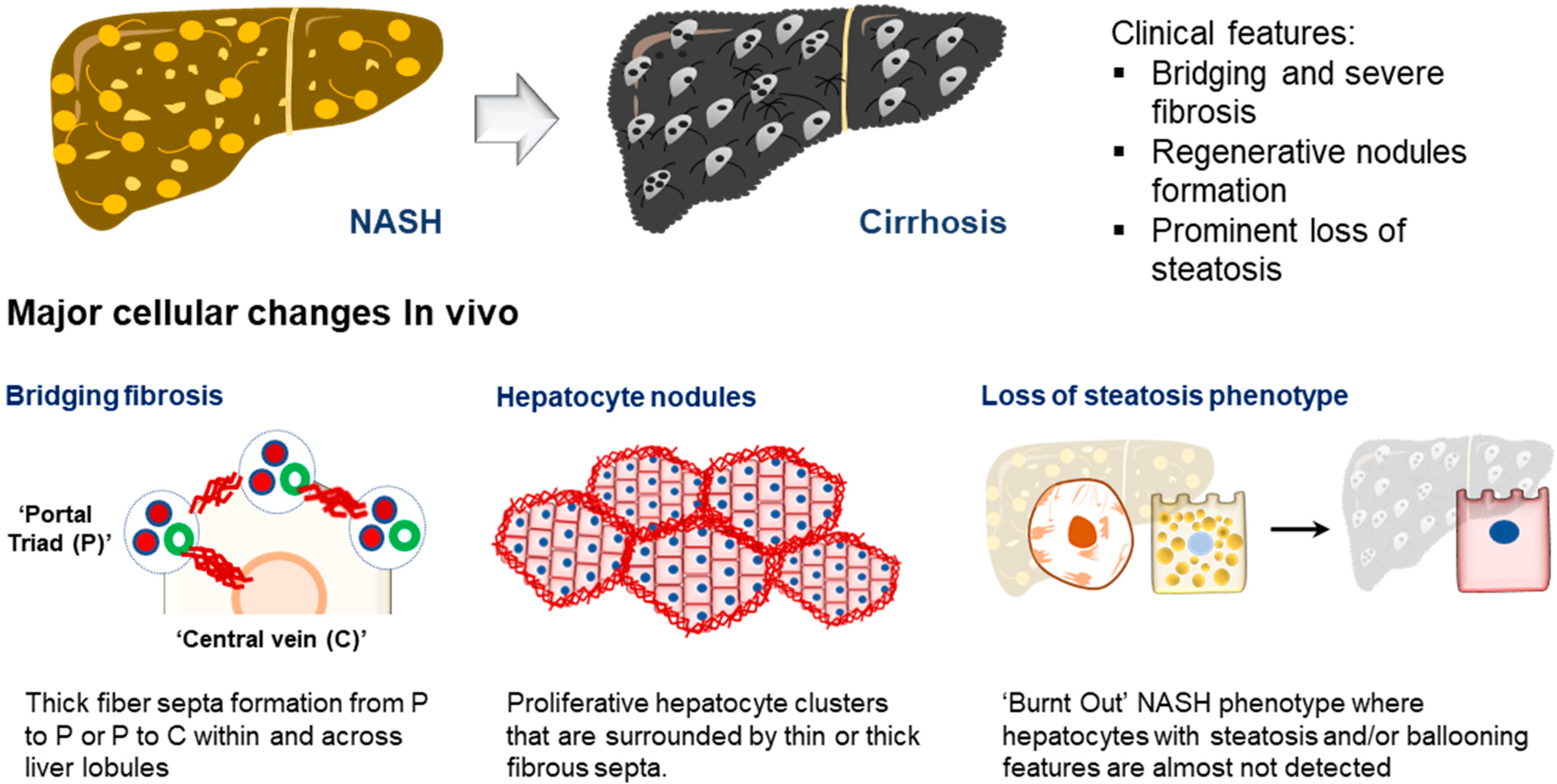
5.3. Modeling Organ Crosstalk in MAFLD
6. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| List of Abbreviations Used | |||
| Abbreviation | Full Name | Gene Name | Full Name |
| 2D | 2-dimensional | ABCB11 | ATP Binding Cassette subfamily B member 11 |
| 3D | 3-dimensional | ABCB4 | ATP Binding Cassette subfamily B member 4 |
| AAA | Aromatic Amino Acid | ALB | Albumin |
| BCAA | Branched-Chain Amino Acid | APOC3 | Apolipoprotein C3 |
| BMP-8B | Bone Morphogenetic Protein-8B | CK7 | Keratin 7 |
| CCL | C-C motif Chemokine Ligand | CK8 | Keratin 8 |
| CDK | Cyclin-dependent Kinase | CLEC4F | C-type lectin domain family 4 member F |
| CKD | Chronic Kidney Disease | CYP3A4 | Cytochrome P450 family 3 subfamily A member 4 |
| CTRP9 | C1q/TNF-Related Protein 9 | FGF19 | Fibroblast Growth Factor 19 |
| CVD | Cardiovascular Disease | GCKR | Glucokinase Regulator |
| CXCL | Chemokine (C-X-C motif) Ligand | HNF4A | Hepatocyte Nuclear Factor 4 alpha |
| DAMP | Death-associated Molecular Pattern | HSD17B13 | Hydroxysteroid 17-beta Dehydrogenase 13 |
| DNL | De Novo Lipogenesis | MBOAT7 | Membrane-Bound O-acyltransferase Domain-Containing 7 |
| ECM | Extra-cellular Matrix | NR0B2 | Nuclear Receptor subfamily 0 group B member 2 |
| ER | Endoplasmic Reticulum | PNPLA3 | Patatin-like Phospholipase Domain-Containing 3 |
| FFA | Free Fatty Acid | TM6SF2 | Transmembrane 6 Superfamily Member 2 |
| FXR | Farnesoid X Receptor | ||
| GLP1R | Glucagon-Like Peptide 1 Receptor | ||
| GWAS | Genome-wide Association Studies | ||
| HCC | Hepatocellular Carcinoma | ||
| HLC | Hepatocyte-like Cell | ||
| hLiMT | Human Liver Microtissues | ||
| HSC | Hepatic Stellate Cell | ||
| IL | Interleukin | ||
| iPSC | Induced Pluripotent Stem Cell | ||
| IRS2 | Insulin Receptor Substrate-2 | ||
| LSEC | Liver Sinusoidal Endothelial Cell | ||
| MAFLD | Metabolic (Dysfunction) Associated Fatty Liver Disease | ||
| MMP | Matrix Metalloproteinase | ||
| NAFLD | Non-alcoholic Fatty Liver Disease | ||
| NASH | Non-alcoholic Steatohepatitis | ||
| NEFA | Non-esterified Fatty Acid | ||
| NK | Natural Killer | ||
| NPC | Non-parenchymal Cell | ||
| OA | Oleic Acid | ||
| OCA | Obeticholic Acid | ||
| P3NP | N-terminal pro-peptide of collagen type III | ||
| PA | Palmitic Acid | ||
| PAMP | Pathogen-associated Molecular Pattern | ||
| PCTS | Precision-Cut Tissue Slice | ||
| PDGF | Platelet-Derived Growth Factor | ||
| PHH | Primary Human Hepatocyte | ||
| PI3K | Phosphatidylinositol-3-OH Kinase | ||
| PKC | Protein Kinase C | ||
| PSC | Pluripotent Stem Cell | ||
| ROS | Reactive Oxygen Species | ||
| SA | Stearic Acid | ||
| SGLT2 | Sodium–Glucose Cotransporter-2 | ||
| SNP | Single-Nucleotide Polymorphism | ||
| T2DM | Type II Diabetes Mellitus | ||
| TH17 | T helper 17 (TH17) cells | ||
| TGF | Transforming Growth Factor | ||
| TNF | Tumor Necrosis Factor | ||
| Treg | Regulatory T cells | ||
| VLDL | Very-low-density lipoprotein. | ||
| α-SMA | Alpha-Smooth Muscle Actin | ||
References
- Anstee, Q.M.; Targher, G.; Day, C.P. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat. Rev. Gastroenterol. Hepatol. 2013, 10, 330–344. [Google Scholar] [CrossRef] [PubMed]
- Yeh, M.M.; Brunt, E.M. Pathological features of fatty liver disease. Gastroenterology 2014, 147, 754–764. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.Y.; Wang, R.F.; Bu, Z.Y.; Targher, G.; Byrne, C.D.; Sun, D.Q.; Zheng, M.H. Association of metabolic dysfunction-associated fatty liver disease with kidney disease. Nat. Rev. Nephrol. 2022, 18, 259–268. [Google Scholar] [CrossRef] [PubMed]
- Younossi, Z.M.; Golabi, P.; de Avila, L.; Paik, J.M.; Srishord, M.; Fukui, N.; Qiu, Y.; Burns, L.; Afendy, A.; Nader, F. The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: A systematic review and meta-analysis. J. Hepatol. 2019, 71, 793–801. [Google Scholar] [CrossRef]
- Hardy, T.; Oakley, F.; Anstee, Q.M.; Day, C.P. Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu. Rev. Pathol. 2016, 11, 451–496. [Google Scholar] [CrossRef]
- Eslam, M.; Sanyal, A.J.; George, J.; International Consensus Panel. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014.e1. [Google Scholar] [CrossRef]
- Le Garf, S.; Negre, V.; Anty, R.; Gual, P. Metabolic Fatty Liver Disease in Children: A Growing Public Health Problem. Biomedicines 2021, 9, 1915. [Google Scholar] [CrossRef]
- Lazarus, J.V.; Mark, H.E.; Villota-Rivas, M.; Palayew, A.; Carrieri, P.; Colombo, M.; Ekstedt, M.; Esmat, G.; George, J.; Marchesini, G.; et al. The global NAFLD policy review and preparedness index: Are countries ready to address this silent public health challenge? J. Hepatol. 2022, 76, 771–780. [Google Scholar] [CrossRef]
- Estes, C.; Razavi, H.; Loomba, R.; Younossi, Z.; Sanyal, A.J. Modeling the epidemic of nonalcoholic fatty liver disease demonstrates an exponential increase in burden of disease. Hepatology 2018, 67, 123–133. [Google Scholar] [CrossRef]
- Sanyal, A.J.; Van Natta, M.L.; Clark, J.; Neuschwander-Tetri, B.A.; Diehl, A.; Dasarathy, S.; Loomba, R.; Chalasani, N.; Kowdley, K.; Hameed, B.; et al. Prospective Study of Outcomes in Adults with Nonalcoholic Fatty Liver Disease. N. Engl. J. Med. 2021, 385, 1559–1569. [Google Scholar] [CrossRef]
- Ferguson, D.; Finck, B.N. Emerging therapeutic approaches for the treatment of NAFLD and type 2 diabetes mellitus. Nat. Rev. Endocrinol. 2021, 17, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Dufour, J.F.; Anstee, Q.M.; Bugianesi, E.; Harrison, S.; Loomba, R.; Paradis, V.; Tilg, H.; Wong, V.W.; Zelber-Sagi, S. Current therapies and new developments in NASH. Gut 2022, 71, 2123–2134. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Valenti, L.; Romeo, S. Genetics and epigenetics of NAFLD and NASH: Clinical impact. J. Hepatol. 2018, 68, 268–279. [Google Scholar] [CrossRef] [PubMed]
- Loomba, R.; Schork, N.; Chen, C.H.; Bettencourt, R.; Bhatt, A.; Ang, B.; Nguyen, P.; Hernandez, C.; Richards, L.; Salotti, J.; et al. Heritability of Hepatic Fibrosis and Steatosis Based on a Prospective Twin Study. Gastroenterology 2015, 149, 1784–1793. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; George, J. Genetic contributions to NAFLD: Leveraging shared genetics to uncover systems biology. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 40–52. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Seth, D.; Day, C.P. Genetic Factors That Affect Risk of Alcoholic and Nonalcoholic Fatty Liver Disease. Gastroenterology 2016, 150, 1728–1744.e7. [Google Scholar] [CrossRef]
- Abul-Husn, N.S.; Cheng, X.; Li, A.H.; Xin, Y.; Schurmann, C.; Stevis, P.; Liu, Y.; Kozlitina, J.; Stender, S.; Wood, G.C.; et al. A Protein-Truncating HSD17B13 Variant and Protection from Chronic Liver Disease. N. Engl. J. Med. 2018, 378, 1096–1106. [Google Scholar] [CrossRef]
- Huang, Y.; Cohen, J.C.; Hobbs, H.H. Expression and characterization of a PNPLA3 protein isoform (I148M) associated with nonalcoholic fatty liver disease. J. Biol. Chem. 2011, 286, 37085–37093. [Google Scholar] [CrossRef]
- Tilson, S.G.; Morell, C.M.; Lenaerts, A.S.; Park, S.B.; Hu, Z.; Jenkins, B.; Koulman, A.; Liang, T.J.; Vallier, L. Modeling PNPLA3-Associated NAFLD Using Human-Induced Pluripotent Stem Cells. Hepatology 2021, 74, 2998–3017. [Google Scholar] [CrossRef]
- Basantani, M.K.; Sitnick, M.T.; Cai, L.; Brenner, D.S.; Gardner, N.P.; Li, J.Z.; Schoiswohl, G.; Yang, K.; Kumari, M.; Gross, R.W.; et al. Pnpla3/Adiponutrin deficiency in mice does not contribute to fatty liver disease or metabolic syndrome. J. Lipid Res. 2011, 52, 318–329. [Google Scholar] [CrossRef]
- Ochi, T.; Munekage, K.; Ono, M.; Higuchi, T.; Tsuda, M.; Hayashi, Y.; Okamoto, N.; Toda, K.; Sakamoto, S.; Oben, J.A.; et al. Patatin-like phospholipase domain-containing protein 3 is involved in hepatic fatty acid and triglyceride metabolism through X-box binding protein 1 and modulation of endoplasmic reticulum stress in mice. Hepatol. Res. 2016, 46, 584–592. [Google Scholar] [CrossRef]
- Anstee, Q.M.; Goldin, R.D. Mouse models in non-alcoholic fatty liver disease and steatohepatitis research. Int. J. Exp. Pathol. 2006, 87, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, X.; Cirillo, P.; Sautin, Y.; McCall, S.; Bruchette, J.L.; Diehl, A.M.; Johnson, R.J.; Abdelmalek, M.F. Fructose consumption as a risk factor for non-alcoholic fatty liver disease. J. Hepatol. 2008, 48, 993–999. [Google Scholar] [CrossRef] [PubMed]
- Abdelmalek, M.F.; Suzuki, A.; Guy, C.; Unalp-Arida, A.; Colvin, R.; Johnson, R.J.; Diehl, A.M.; Nonalcoholic Steatohepatitis Clinical Research Network. Increased fructose consumption is associated with fibrosis severity in patients with nonalcoholic fatty liver disease. Hepatology 2010, 51, 1961–1971. [Google Scholar] [CrossRef] [PubMed]
- Jegatheesan, P.; De Bandt, J.P. Fructose and NAFLD: The Multifaceted Aspects of Fructose Metabolism. Nutrients 2017, 9, 230. [Google Scholar] [CrossRef]
- Day, C.P.; James, O.F. Steatohepatitis: A tale of two “hits”? Gastroenterology 1998, 114, 842–845. [Google Scholar] [CrossRef]
- Tilg, H.; Adolph, T.E.; Moschen, A.R. Multiple Parallel Hits Hypothesis in Nonalcoholic Fatty Liver Disease: Revisited After a Decade. Hepatology 2021, 73, 833–842. [Google Scholar] [CrossRef]
- Loomba, R.; Seguritan, V.; Li, W.; Long, T.; Klitgord, N.; Bhatt, A.; Dulai, P.S.; Caussy, C.; Bettencourt, R.; Highlander, S.K.; et al. Gut Microbiome-Based Metagenomic Signature for Non-invasive Detection of Advanced Fibrosis in Human Nonalcoholic Fatty Liver Disease. Cell Metab. 2017, 25, 1054–1062.e5. [Google Scholar] [CrossRef]
- Hoyles, L.; Fernandez-Real, J.M.; Federici, M.; Serino, M.; Abbott, J.; Charpentier, J.; Heymes, C.; Luque, J.L.; Anthony, E.; Barton, R.H.; et al. Molecular phenomics and metagenomics of hepatic steatosis in non-diabetic obese women. Nat. Med. 2018, 24, 1070–1080. [Google Scholar] [CrossRef]
- Caussy, C.; Tripathi, A.; Humphrey, G.; Bassirian, S.; Singh, S.; Faulkner, C.; Bettencourt, R.; Rizo, E.; Richards, L.; Xu, Z.Z.; et al. A gut microbiome signature for cirrhosis due to nonalcoholic fatty liver disease. Nat. Commun. 2019, 10, 1406. [Google Scholar] [CrossRef]
- Trefts, E.; Gannon, M.; Wasserman, D.H. The liver. Curr. Biol. 2017, 27, R1147–R1151. [Google Scholar] [CrossRef] [PubMed]
- Tandra, S.; Yeh, M.M.; Brunt, E.M.; Vuppalanchi, R.; Cummings, O.W.; Ünalp-Arida, A.; Wilson, L.A.; Chalasani, N. Presence and significance of microvesicular steatosis in nonalcoholic fatty liver disease. J. Hepatol. 2011, 55, 654–659. [Google Scholar] [CrossRef] [PubMed]
- Marrache, M.K.; Rockey, D.C. Statins for treatment of chronic liver disease. Curr. Opin. Gastroenterol. 2021, 37, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Marchesini, G.; Brizi, M.; Morselli-Labate, A.M.; Bianchi, G.; Bugianesi, E.; McCullough, A.J.; Forlani, G.; Melchionda, N. Association of nonalcoholic fatty liver disease with insulin resistance. Am. J. Med. 1999, 107, 450–455. [Google Scholar] [CrossRef]
- Perry, R.J.; Samuel, V.T.; Petersen, K.F.; Shulman, G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature 2014, 510, 84–91. [Google Scholar] [CrossRef]
- Petersen, M.C.; Madiraju, A.K.; Gassaway, B.M.; Marcel, M.; Nasiri, A.R.; Butrico, G.; Marcucci, M.J.; Zhang, D.; Abulizi, A.; Zhang, X.M.; et al. Insulin receptor Thr1160 phosphorylation mediates lipid-induced hepatic insulin resistance. J. Clin. Investig. 2016, 126, 4361–4371. [Google Scholar] [CrossRef]
- Magkos, F.; Su, X.; Bradley, D.; Fabbrini, E.; Conte, C.; Eagon, J.C.; Varela, J.E.; Brunt, E.M.; Patterson, B.W.; Klein, S. Intrahepatic diacylglycerol content is associated with hepatic insulin resistance in obese subjects. Gastroenterology 2012, 142, 1444–1446.e2. [Google Scholar] [CrossRef]
- Samuel, V.T.; Liu, Z.X.; Wang, A.; Beddow, S.A.; Geisler, J.G.; Kahn, M.; Zhang, X.M.; Monia, B.P.; Bhanot, S.; Shulman, G.I. Inhibition of protein kinase Cepsilon prevents hepatic insulin resistance in nonalcoholic fatty liver disease. J. Clin. Investig. 2007, 117, 739–745. [Google Scholar] [CrossRef]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef]
- Assaily, W.; Rubinger, D.A.; Wheaton, K.; Lin, Y.; Ma, W.; Xuan, W.; Brown-Endres, L.; Tsuchihara, K.; Mak, T.W.; Benchimol, S. ROS-mediated p53 induction of Lpin1 regulates fatty acid oxidation in response to nutritional stress. Mol. Cell 2011, 44, 491–501. [Google Scholar] [CrossRef]
- Bell, C.C.; Hendriks, D.F.; Moro, S.M.; Ellis, E.; Walsh, J.; Renblom, A.; Fredriksson Puigvert, L.; Dankers, A.C.; Jacobs, F.; Snoeys, J.; et al. Characterization of primary human hepatocyte spheroids as a model system for drug-induced liver injury, liver function and disease. Sci. Rep. 2016, 6, 25187. [Google Scholar] [CrossRef] [PubMed]
- Lyall, M.J.; Cartier, J.; Thomson, J.P.; Cameron, K.; Meseguer-Ripolles, J.; O’Duibhir, E.; Szkolnicka, D.; Villarin, B.L.; Wang, Y.; Blanco, G.R.; et al. Modelling non-alcoholic fatty liver disease in human hepatocyte-like cells. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170362. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, Y.; Tanaka, S.; Haga, Y. Enhanced GLUT2 gene expression in an oleic acid-induced in vitro fatty liver model. Hepatol. Res. 2002, 23, 138–144. [Google Scholar] [CrossRef]
- Gomez-Lechon, M.J.; Donato, M.T.; Martinez-Romero, A.; Jimenez, N.; Castell, J.V.; O’Connor, J.E. A human hepatocellular in vitro model to investigate steatosis. Chem. Biol. Interact. 2007, 165, 106–116. [Google Scholar] [CrossRef]
- Wobser, H.; Dorn, C.; Weiss, T.S.; Amann, T.; Bollheimer, C.; Buttner, R.; Scholmerich, J.; Hellerbrand, C. Lipid accumulation in hepatocytes induces fibrogenic activation of hepatic stellate cells. Cell Res. 2009, 19, 996–1005. [Google Scholar] [CrossRef]
- McCarron, S.; Bathon, B.; Conlon, D.M.; Abbey, D.; Rader, D.J.; Gawronski, K.; Brown, C.D.; Olthoff, K.M.; Shaked, A.; Raabe, T.D. Functional Characterization of Organoids Derived From Irreversibly Damaged Liver of Patients With NASH. Hepatology 2021, 74, 1825–1844. [Google Scholar] [CrossRef] [PubMed]
- Ramli, M.N.B.; Lim, Y.S.; Koe, C.T.; Demircioglu, D.; Tng, W.; Gonzales, K.A.U.; Tan, C.P.; Szczerbinska, I.; Liang, H.; Soe, E.L.; et al. Human Pluripotent Stem Cell-Derived Organoids as Models of Liver Disease. Gastroenterology 2020, 159, 1471–1486.e12. [Google Scholar] [CrossRef]
- Graffmann, N.; Ring, S.; Kawala, M.A.; Wruck, W.; Ncube, A.; Trompeter, H.I.; Adjaye, J. Modeling Nonalcoholic Fatty Liver Disease with Human Pluripotent Stem Cell-Derived Immature Hepatocyte-Like Cells Reveals Activation of PLIN2 and Confirms Regulatory Functions of Peroxisome Proliferator-Activated Receptor Alpha. Stem Cells Dev. 2016, 25, 1119–1133. [Google Scholar] [CrossRef] [PubMed]
- Duwaerts, C.C.; Le Guillou, D.; Her, C.L.; Phillips, N.J.; Willenbring, H.; Mattis, A.N.; Maher, J.J. Induced Pluripotent Stem Cell-derived Hepatocytes From Patients With Nonalcoholic Fatty Liver Disease Display a Disease-specific Gene Expression Profile. Gastroenterology 2021, 160, 2591–2594.e6. [Google Scholar] [CrossRef]
- Ouchi, R.; Togo, S.; Kimura, M.; Shinozawa, T.; Koido, M.; Koike, H.; Thompson, W.; Karns, R.A.; Mayhew, C.N.; McGrath, P.S.; et al. Modeling Steatohepatitis in Humans with Pluripotent Stem Cell-Derived Organoids. Cell Metab. 2019, 30, 374–384.e6. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Deng, P.; Tao, T.; Liu, H.; Wu, S.; Chen, W.; Qin, J. Modeling Human Nonalcoholic Fatty Liver Disease (NAFLD) with an Organoids-on-a-Chip System. ACS Biomater. Sci. Eng. 2020, 6, 5734–5743. [Google Scholar] [CrossRef] [PubMed]
- Ratziu, V.; Sanyal, A.; Harrison, S.A.; Wong, V.W.; Francque, S.; Goodman, Z.; Aithal, G.P.; Kowdley, K.V.; Seyedkazemi, S.; Fischer, L.; et al. Cenicriviroc Treatment for Adults With Nonalcoholic Steatohepatitis and Fibrosis: Final Analysis of the Phase 2b CENTAUR Study. Hepatology 2020, 72, 892–905. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Allen, A.M.; Wang, Z.; Prokop, L.J.; Murad, M.H.; Loomba, R. Fibrosis progression in nonalcoholic fatty liver vs nonalcoholic steatohepatitis: A systematic review and meta-analysis of paired-biopsy studies. Clin. Gastroenterol. Hepatol. 2015, 13, 643–654.e9; quiz e39–e40. [Google Scholar] [CrossRef] [PubMed]
- Gadd, V.L.; Skoien, R.; Powell, E.E.; Fagan, K.J.; Winterford, C.; Horsfall, L.; Irvine, K.; Clouston, A.D. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology 2014, 59, 1393–1405. [Google Scholar] [CrossRef]
- Syn, W.K.; Oo, Y.H.; Pereira, T.A.; Karaca, G.F.; Jung, Y.; Omenetti, A.; Witek, R.P.; Choi, S.S.; Guy, C.D.; Fearing, C.M.; et al. Accumulation of natural killer T cells in progressive nonalcoholic fatty liver disease. Hepatology 2010, 51, 1998–2007. [Google Scholar] [CrossRef]
- Miyao, M.; Kotani, H.; Ishida, T.; Kawai, C.; Manabe, S.; Abiru, H.; Tamaki, K. Pivotal role of liver sinusoidal endothelial cells in NAFLD/NASH progression. Lab. Investig. 2015, 95, 1130–1144. [Google Scholar] [CrossRef]
- Schwabe, R.F.; Tabas, I.; Pajvani, U.B. Mechanisms of Fibrosis Development in Nonalcoholic Steatohepatitis. Gastroenterology 2020, 158, 1913–1928. [Google Scholar] [CrossRef]
- Alisi, A.; Carsetti, R.; Nobili, V. Pathogen- or damage-associated molecular patterns during nonalcoholic fatty liver disease development. Hepatology 2011, 54, 1500–1502. [Google Scholar] [CrossRef]
- Bataller, R.; Brenner, D.A. Liver fibrosis. J. Clin. Investig. 2005, 115, 209–218. [Google Scholar] [CrossRef]
- Pellicoro, A.; Ramachandran, P.; Iredale, J.P.; Fallowfield, J.A. Liver fibrosis and repair: Immune regulation of wound healing in a solid organ. Nat. Rev. Immunol. 2014, 14, 181–194. [Google Scholar] [CrossRef]
- Loomba, R.; Friedman, S.L.; Shulman, G.I. Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 2021, 184, 2537–2564. [Google Scholar] [CrossRef] [PubMed]
- Dulai, P.S.; Singh, S.; Patel, J.; Soni, M.; Prokop, L.J.; Younossi, Z.; Sebastiani, G.; Ekstedt, M.; Hagstrom, H.; Nasr, P.; et al. Increased risk of mortality by fibrosis stage in nonalcoholic fatty liver disease: Systematic review and meta-analysis. Hepatology 2017, 65, 1557–1565. [Google Scholar] [CrossRef]
- Matteoni, C.A.; Younossi, Z.M.; Gramlich, T.; Boparai, N.; Liu, Y.C.; McCullough, A.J. Nonalcoholic fatty liver disease: A spectrum of clinical and pathological severity. Gastroenterology 1999, 116, 1413–1419. [Google Scholar] [CrossRef]
- Lackner, C.; Gogg-Kamerer, M.; Zatloukal, K.; Stumptner, C.; Brunt, E.M.; Denk, H. Ballooned hepatocytes in steatohepatitis: The value of keratin immunohistochemistry for diagnosis. J. Hepatol. 2008, 48, 821–828. [Google Scholar] [CrossRef] [PubMed]
- Kucukoglu, O.; Guldiken, N.; Chen, Y.; Usachov, V.; El-Heliebi, A.; Haybaeck, J.; Denk, H.; Trautwein, C.; Strnad, P. High-fat diet triggers Mallory-Denk body formation through misfolding and crosslinking of excess keratin 8. Hepatology 2014, 60, 169–178. [Google Scholar] [CrossRef]
- Bhattacharjee, J.; Kumar, J.M.; Arindkar, S.; Das, B.; Pramod, U.; Juyal, R.C.; Majumdar, S.S.; Nagarajan, P. Role of immunodeficient animal models in the development of fructose induced NAFLD. J. Nutr. Biochem. 2014, 25, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, Y.; Soejima, Y.; Fukusato, T. Animal models of nonalcoholic fatty liver disease/nonalcoholic steatohepatitis. World J. Gastroenterol. 2012, 18, 2300–2308. [Google Scholar] [CrossRef]
- Teufel, A.; Itzel, T.; Erhart, W.; Brosch, M.; Wang, X.Y.; Kim, Y.O.; von Schonfels, W.; Herrmann, A.; Bruckner, S.; Stickel, F.; et al. Comparison of Gene Expression Patterns Between Mouse Models of Nonalcoholic Fatty Liver Disease and Liver Tissues From Patients. Gastroenterology 2016, 151, 513–525.e0. [Google Scholar] [CrossRef]
- Ricchi, M.; Odoardi, M.R.; Carulli, L.; Anzivino, C.; Ballestri, S.; Pinetti, A.; Fantoni, L.I.; Marra, F.; Bertolotti, M.; Banni, S.; et al. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J. Gastroenterol. Hepatol. 2009, 24, 830–840. [Google Scholar] [CrossRef]
- Piras, I.S.; Gerhard, G.S.; DiStefano, J.K. Palmitate and Fructose Interact to Induce Human Hepatocytes to Produce Pro-Fibrotic Transcriptional Responses in Hepatic Stellate Cells Exposed to Conditioned Media. Cell Physiol. Biochem. 2020, 54, 1068–1082. [Google Scholar]
- Todoric, J.; Di Caro, G.; Reibe, S.; Henstridge, D.C.; Green, C.R.; Vrbanac, A.; Ceteci, F.; Conche, C.; McNulty, R.; Shalapour, S.; et al. Fructose stimulated de novo lipogenesis is promoted by inflammation. Nat. Metab. 2020, 2, 1034–1045. [Google Scholar] [CrossRef] [PubMed]
- Cui, W.; Chen, S.L.; Hu, K.Q. Quantification and mechanisms of oleic acid-induced steatosis in HepG2 cells. Am. J. Transl. Res. 2010, 2, 95–104. [Google Scholar] [PubMed]
- Joshi-Barve, S.; Barve, S.S.; Amancherla, K.; Gobejishvili, L.; Hill, D.; Cave, M.; Hote, P.; McClain, C.J. Palmitic acid induces production of proinflammatory cytokine interleukin-8 from hepatocytes. Hepatology 2007, 46, 823–830. [Google Scholar] [CrossRef]
- García-Ruiz, I.; Solís-Muñoz, P.; Fernández-Moreira, D.; Muñoz-Yagüe, T.; Solís-Herruzo, J.A. In vitro treatment of HepG2 cells with saturated fatty acids reproduces mitochondrial dysfunction found in nonalcoholic steatohepatitis. Dis. Model. Mech. 2015, 8, 183–191. [Google Scholar] [PubMed]
- Khalifa, O.; Al-Akl, N.S.; Errafii, K.; Arredouani, A. Exendin-4 alleviates steatosis in an in vitro cell model by lowering FABP1 and FOXA1 expression via the Wnt/-catenin signaling pathway. Sci. Rep. 2022, 12, 2226. [Google Scholar] [CrossRef] [PubMed]
- Gunn, P.J.; Green, C.J.; Pramfalk, C.; Hodson, L. In vitro cellular models of human hepatic fatty acid metabolism: Differences between Huh7 and HepG2 cell lines in human and fetal bovine culturing serum. Physiol. Rep. 2017, 5, e13532. [Google Scholar] [CrossRef]
- Chen, K.Y.; Lin, J.A.; Yao, H.Y.; Hsu, A.C.; Tai, Y.T.; Chen, J.T.; Hsieh, M.C.; Shen, T.L.; Hsu, R.Y.; Wu, H.T.; et al. Arctigenin protects against steatosis in WRL68 hepatocytes through activation of phosphoinositide 3-kinase/protein kinase B and AMP-activated protein kinase pathways. Nutr. Res. 2018, 52, 87–97. [Google Scholar] [CrossRef]
- Tolosa, L.; Gomez-Lechon, M.J.; Jimenez, N.; Hervas, D.; Jover, R.; Donato, M.T. Advantageous use of HepaRG cells for the screening and mechanistic study of drug-induced steatosis. Toxicol. Appl. Pharm. 2016, 302, 1–9. [Google Scholar] [CrossRef]
- Teng, Y.; Zhao, Z.; Tasnim, F.; Huang, X.; Yu, H. A scalable and sensitive steatosis chip with long-term perfusion of in situ differentiated HepaRG organoids. Biomaterials 2021, 275, 120904. [Google Scholar] [CrossRef]
- De Gottardi, A.; Vinciguerra, M.; Sgroi, A.; Moukil, M.; Ravier-Dall’Antonia, F.; Pazienza, V.; Pugnale, P.; Foti, M.; Hadengue, A. Microarray analyses and molecular profiling of steatosis induction in immortalized human hepatocytes. Lab. Investig. 2007, 87, 792–806. [Google Scholar] [CrossRef]
- Mahli, A.; Seitz, T.; Beckroge, T.; Freese, K.; Thasler, W.E.; Benkert, M.; Dietrich, P.; Weiskirchen, R.; Bosserhoff, A.; Hellerbrand, C. Bone Morphogenetic Protein-8B Expression is Induced in Steatotic Hepatocytes and Promotes Hepatic Steatosis and Inflammation In Vitro. Cells 2019, 8, 457. [Google Scholar] [CrossRef] [PubMed]
- Prill, S.; Caddeo, A.; Baselli, G.; Jamialahmadi, O.; Dongiovanni, P.; Rametta, R.; Kanebratt, K.P.; Pujia, A.; Pingitore, P.; Mancina, R.M.; et al. The TM6SF2 E167K genetic variant induces lipid biosynthesis and reduces apolipoprotein B secretion in human hepatic 3D spheroids. Sci. Rep. 2019, 9, 11585. [Google Scholar] [CrossRef] [PubMed]
- Parafati, M.; Bae, S.H.; Kirby, R.J.; Fitzek, M.; Iyer, P.; Engkvist, O.; Smith, D.M.; Malany, S. Pluripotent Stem Cell-Derived Hepatocytes Phenotypic Screening Reveals Small Molecules Targeting the CDK2/4-C/EBPalpha/DGAT2 Pathway Preventing ER-Stress Induced Lipid Accumulation. Int. J. Mol. Sci. 2020, 21, 9557. [Google Scholar] [CrossRef]
- Sinton, M.C.; Meseguer-Ripolles, J.; Lucendo-Villarin, B.; Wernig-Zorc, S.; Thomson, J.P.; Carter, R.N.; Lyall, M.J.; Walker, P.D.; Thakker, A.; Meehan, R.R.; et al. A human pluripotent stem cell model for the analysis of metabolic dysfunction in hepatic steatosis. iScience 2021, 24, 101931. [Google Scholar] [CrossRef]
- Aden, D.P.; Fogel, A.; Plotkin, S.; Damjanov, I.; Knowles, B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature 1979, 282, 615–616. [Google Scholar] [CrossRef]
- Mersch-Sundermann, V.; Knasmüller, S.; Wu, X.J.; Darroudi, F.; Kassie, F. Use of a human-derived liver cell line for the detection of cytoprotective, antigenotoxic and cogenotoxic agents. Toxicology 2004, 198, 329–340. [Google Scholar] [CrossRef] [PubMed]
- Moscato, S.; Ronca, F.; Campani, D.; Danti, S. Poly(vinyl alcohol)/gelatin Hydrogels Cultured with HepG2 Cells as a 3D Model of Hepatocellular Carcinoma: A Morphological Study. J. Funct. Biomater. 2015, 6, 16–32. [Google Scholar] [CrossRef]
- Pérez-Carreras, M.; Del Hoyo, P.; Martín, M.A.; Rubio, J.C.; Martín, A.; Castellano, G.; Colina, F.; Arenas, J.; Solis-Herruzo, J.A. Defective hepatic mitochondrial respiratory chain in patients with nonalcoholic steatohepatitis. Hepatology 2003, 38, 999–1007. [Google Scholar] [CrossRef]
- Kugelmas, M.; Hill, D.B.; Vivian, B.; Marsano, L.; McClain, C.J. Cytokines and NASH: A pilot study of the effects of lifestyle modification and vitamin E. Hepatology 2003, 38, 413–419. [Google Scholar] [CrossRef]
- Bahcecioglu, I.H.; Yalniz, M.; Ataseven, H.; Ilhan, N.; Ozercan, I.H.; Seckin, D.; Sahin, K. Levels of serum hyaluronic acid, TNF-alpha and IL-8 in patients with nonalcoholic steatohepatitis. Hepatogastroenterology 2005, 52, 1549–1553. [Google Scholar]
- Jung, T.W.; Hong, H.C.; Hwang, H.J.; Yoo, H.J.; Baik, S.H.; Choi, K.M. C1q/TNF-Related Protein 9 (CTRP9) attenuates hepatic steatosis via the autophagy-mediated inhibition of endoplasmic reticulum stress. Mol. Cell Endocrinol. 2015, 417, 131–140. [Google Scholar] [CrossRef] [PubMed]
- Peterson, J.M.; Wei, Z.; Seldin, M.M.; Byerly, M.S.; Aja, S.; Wong, G.W. CTRP9 transgenic mice are protected from diet-induced obesity and metabolic dysfunction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2013, 305, R522–R533. [Google Scholar] [CrossRef] [PubMed]
- Mondello, C.; Chiodi, I. Cellular immortalization and neoplastic transformation: Simultaneous, sequential or independent? Telomeres, telomerase or karyotypic variations? Cell Cycle 2013, 12, 1804–1805. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Vinken, M. Primary hepatocyte cultures for liver disease modeling. Curr. Opin. Toxicol. 2021, 25, 1–5. [Google Scholar] [CrossRef]
- Dooley, S.; Hamzavi, J.; Ciuclan, L.; Godoy, P.; Ilkavets, I.; Ehnert, S.; Ueberham, E.; Gebhardt, R.; Kanzler, S.; Geier, A.; et al. Hepatocyte-specific Smad7 expression attenuates TGF-beta-mediated fibrogenesis and protects against liver damage. Gastroenterology 2008, 135, 642–659. [Google Scholar] [CrossRef]
- Kwon, C.; Sun, J.L.; Jeong, J.H.; Jung, T.W. Humanin attenuates palmitate-induced hepatic lipid accumulation and insulin resistance via AMPK-mediated suppression of the mTOR pathway. Biochem. Biophys. Res. Commun. 2020, 526, 539–545. [Google Scholar] [CrossRef]
- Jung, T.W.; Kang, C.; Goh, J.; Chae, S.I.; Kim, H.C.; Lee, T.J.; Abd El-Aty, A.M.; Jeong, J.H. WISP1 promotes non-alcoholic fatty liver disease and skeletal muscle insulin resistance via TLR4/JNK signaling. J. Cell Physiol. 2018, 233, 6077–6087. [Google Scholar] [CrossRef]
- Jung, T.W.; Kim, H.C.; Abd El-Aty, A.M.; Jeong, J.H. Maresin 1 attenuates NAFLD by suppression of endoplasmic reticulum stress via AMPK-SERCA2b pathway. J. Biol. Chem. 2018, 293, 3981–3988. [Google Scholar] [CrossRef]
- Teschke, R.; Uetrecht, J. Mechanism of idiosyncratic drug induced liver injury (DILI): Unresolved basic issues. Ann. Transl. Med. 2021, 9, 730. [Google Scholar] [CrossRef]
- Huch, M.; Gehart, H.; van Boxtel, R.; Hamer, K.; Blokzijl, F.; Verstegen, M.M.; Ellis, E.; van Wenum, M.; Fuchs, S.A.; de Ligt, J.; et al. Long-term culture of genome-stable bipotent stem cells from adult human liver. Cell 2015, 160, 299–312. [Google Scholar] [CrossRef]
- Broutier, L.; Andersson-Rolf, A.; Hindley, C.J.; Boj, S.F.; Clevers, H.; Koo, B.K.; Huch, M. Culture and establishment of self-renewing human and mouse adult liver and pancreas 3D organoids and their genetic manipulation. Nat. Protoc. 2016, 11, 1724–1743. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Gehart, H.; Artegiani, B.; LÖpez-Iglesias, C.; Dekkers, F.; Basak, O.; van Es, J.; Chuva de Sousa Lopes, S.M.; Begthel, H.; Korving, J.; et al. Long-Term Expansion of Functional Mouse and Human Hepatocytes as 3D Organoids. Cell 2018, 175, 1591–1606.e19. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Zhang, L.; Liu, W.; Ma, X.; Cen, J.; Sun, Z.; Wang, C.; Feng, S.; Zhang, Z.; Yue, L.; et al. In Vitro Expansion of Primary Human Hepatocytes with Efficient Liver Repopulation Capacity. Cell Stem Cell 2018, 23, 806–819.e4. [Google Scholar] [CrossRef] [PubMed]
- Fu, G.B.; Huang, W.J.; Zeng, M.; Zhou, X.; Wu, H.P.; Liu, C.C.; Wu, H.; Weng, J.; Zhang, H.D.; Cai, Y.C.; et al. Expansion and differentiation of human hepatocyte-derived liver progenitor-like cells and their use for the study of hepatotropic pathogens. Cell Res. 2019, 29, 8–22. [Google Scholar] [CrossRef] [PubMed]
- Vyas, D.; Baptista, P.M.; Brovold, M.; Moran, E.; Gaston, B.; Booth, C.; Samuel, M.; Atala, A.; Soker, S. Self-assembled liver organoids recapitulate hepatobiliary organogenesis in vitro. Hepatology 2018, 67, 750–761. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Wang, Y.; Cen, J.; Ma, X.; Cui, L.; Qiu, Z.; Zhang, Z.; Li, H.; Yang, R.Z.; Wang, C.; et al. Modelling liver cancer initiation with organoids derived from directly reprogrammed human hepatocytes. Nat. Cell Biol. 2019, 21, 1015–1026. [Google Scholar] [CrossRef]
- Artegiani, B.; van Voorthuijsen, L.; Lindeboom, R.G.H.; Seinstra, D.; Heo, I.; Tapia, P.; Lopez-Iglesias, C.; Postrach, D.; Dayton, T.; Oka, R.; et al. Probing the Tumor Suppressor Function of BAP1 in CRISPR-Engineered Human Liver Organoids. Cell Stem Cell 2019, 24, 927–943.e6. [Google Scholar] [CrossRef]
- Targher, G.; Mantovani, A.; Byrne, C.D.; Wang, X.B.; Yan, H.D.; Sun, Q.F.; Pan, K.H.; Zheng, K.I.; Chen, Y.P.; Eslam, M.; et al. Risk of severe illness from COVID-19 in patients with metabolic dysfunction-associated fatty liver disease and increased fibrosis scores. Gut 2020, 69, 1545–1547. [Google Scholar] [CrossRef]
- Ji, D.; Qin, E.; Xu, J.; Zhang, D.; Cheng, G.; Wang, Y.; Lau, G. Non-alcoholic fatty liver diseases in patients with COVID-19: A retrospective study. J. Hepatol. 2020, 73, 451–453. [Google Scholar] [CrossRef]
- Tong, X.F.; Wang, Q.Y.; Zhao, X.Y.; Sun, Y.M.; Wu, X.N.; Yang, L.L.; Lu, Z.Z.; Ou, X.J.; Jia, J.D.; You, H. Histological assessment based on liver biopsy: The value and challenges in NASH drug development. Acta Pharm. Sin. 2022, 43, 1200–1209. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Nowak, G.; Ericzon, B.G.; Nava, S.; Jaksch, M.; Westgren, M.; Sumitran-Holgersson, S. Identification of expandable human hepatic progenitors which differentiate into mature hepatic cells in vivo. Gut 2005, 54, 972–979. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Cai, J.; Zhao, Y.; Liu, Y.; Ye, F.; Song, Z.; Qin, H.; Meng, S.; Chen, Y.; Zhou, R.; Song, X.; et al. Directed differentiation of human embryonic stem cells into functional hepatic cells. Hepatology 2007, 45, 1229–1239. [Google Scholar] [CrossRef] [PubMed]
- Si-Tayeb, K.; Noto, F.K.; Nagaoka, M.; Li, J.; Battle, M.A.; Duris, C.; North, P.E.; Dalton, S.; Duncan, S.A. Highly efficient generation of human hepatocyte-like cells from induced pluripotent stem cells. Hepatology 2010, 51, 297–305. [Google Scholar] [CrossRef] [PubMed]
- Huang, P.; He, Z.; Ji, S.; Sun, H.; Xiang, D.; Liu, C.; Hu, Y.; Wang, X.; Hui, L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 2011, 475, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Sekiya, S.; Suzuki, A. Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 2011, 475, 390–393. [Google Scholar] [CrossRef]
- Zhu, S.; Rezvani, M.; Harbell, J.; Mattis, A.N.; Wolfe, A.R.; Benet, L.Z.; Willenbring, H.; Ding, S. Mouse liver repopulation with hepatocytes generated from human fibroblasts. Nature 2014, 508, 93–97. [Google Scholar] [CrossRef]
- Ang, L.T.; Tan, A.K.Y.; Autio, M.I.; Goh, S.H.; Choo, S.H.; Lee, K.L.; Tan, J.; Pan, B.; Lee, J.J.H.; Lum, J.J.; et al. A Roadmap for Human Liver Differentiation from Pluripotent Stem Cells. Cell Rep. 2018, 22, 2190–2205. [Google Scholar] [CrossRef]
- Ma, X.; Duan, Y.; Tschudy-Seney, B.; Roll, G.; Behbahan, I.S.; Ahuja, T.P.; Tolstikov, V.; Wang, C.; McGee, J.; Khoobyari, S.; et al. Highly efficient differentiation of functional hepatocytes from human induced pluripotent stem cells. Stem Cells Transl. Med. 2013, 2, 409–419. [Google Scholar] [CrossRef]
- Wu, F.; Wu, D.; Ren, Y.; Huang, Y.; Feng, B.; Zhao, N.; Zhang, T.; Chen, X.; Chen, S.; Xu, A. Generation of hepatobiliary organoids from human induced pluripotent stem cells. J. Hepatol. 2019, 70, 1145–1158. [Google Scholar] [CrossRef] [PubMed]
- Graffmann, N.; Scherer, B.; Adjaye, J. In vitro differentiation of pluripotent stem cells into hepatocyte like cells-Basic principles and current progress. Stem Cell Res. 2022, 61, 102763. [Google Scholar] [CrossRef] [PubMed]
- Hay, D.C.; Zhao, D.; Fletcher, J.; Hewitt, Z.A.; McLean, D.; Urruticoechea-Uriguen, A.; Black, J.R.; Elcombe, C.; Ross, J.A.; Wolf, R.; et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 2008, 26, 894–902. [Google Scholar] [CrossRef] [PubMed]
- Espejel, S.; Roll, G.R.; McLaughlin, K.J.; Lee, A.Y.; Zhang, J.Y.; Laird, D.J.; Okita, K.; Yamanaka, S.; Willenbring, H. Induced pluripotent stem cell-derived hepatocytes have the functional and proliferative capabilities needed for liver regeneration in mice. J. Clin. Investig. 2010, 120, 3120–3126. [Google Scholar] [CrossRef]
- Touboul, T.; Hannan, N.R.; Corbineau, S.; Martinez, A.; Martinet, C.; Branchereau, S.; Mainot, S.; Strick-Marchand, H.; Pedersen, R.; Di Santo, J.; et al. Generation of functional hepatocytes from human embryonic stem cells under chemically defined conditions that recapitulate liver development. Hepatology 2010, 51, 1754–1765. [Google Scholar] [CrossRef]
- Mun, S.J.; Ryu, J.S.; Lee, M.O.; Son, Y.S.; Oh, S.J.; Cho, H.S.; Son, M.Y.; Kim, D.S.; Kim, S.J.; Yoo, H.J.; et al. Generation of expandable human pluripotent stem cell-derived hepatocyte-like liver organoids. J. Hepatol. 2019, 71, 970–985. [Google Scholar] [CrossRef]
- Takebe, T.; Sekine, K.; Enomura, M.; Koike, H.; Kimura, M.; Ogaeri, T.; Zhang, R.-R.; Ueno, Y.; Zheng, Y.-W.; Koike, N.; et al. Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 2013, 499, 481–484. [Google Scholar] [CrossRef]
- Shi, Y.; Inoue, H.; Takahashi, J.; Yamanaka, S. Induced pluripotent stem cell technology: Venturing into the second decade. In Principles of Tissue Engineering; Academic Press: Cambridge, MA, USA, 2020; pp. 435–443. [Google Scholar]
- Guan, J.; Wang, G.; Wang, J.; Zhang, Z.; Fu, Y.; Cheng, L.; Meng, G.; Lyu, Y.; Zhu, J.; Li, Y.; et al. Chemical reprogramming of human somatic cells to pluripotent stem cells. Nature 2022, 605, 325–331. [Google Scholar] [CrossRef]
- Boeckmans, J.; Buyl, K.; Natale, A.; Vandenbempt, V.; Branson, S.; De Boe, V.; Rogiers, V.; De Kock, J.; Rodrigues, R.M.; Vanhaecke, T. Elafibranor restricts lipogenic and inflammatory responses in a human skin stem cell-derived model of NASH. Pharmacol. Res. 2019, 144, 377–389. [Google Scholar] [CrossRef]
- Kazankov, K.; Jørgensen, S.M.D.; Thomsen, K.L.; Møller, H.J.; Vilstrup, H.; George, J.; Schuppan, D.; Grønbæk, H. The role of macrophages in nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 145–159. [Google Scholar] [CrossRef]
- Her, Z.; Tan, J.H.L.; Lim, Y.S.; Tan, S.Y.; Chan, X.Y.; Tan, W.W.S.; Liu, M.; Yong, K.S.M.; Lai, F.; Ceccarello, E.; et al. CD4(+) T Cells Mediate the Development of Liver Fibrosis in High Fat Diet-Induced NAFLD in Humanized Mice. Front. Immunol. 2020, 11, 580968. [Google Scholar] [CrossRef] [PubMed]
- Rau, M.; Schilling, A.K.; Meertens, J.; Hering, I.; Weiss, J.; Jurowich, C.; Kudlich, T.; Hermanns, H.M.; Bantel, H.; Beyersdorf, N.; et al. Progression from Nonalcoholic Fatty Liver to Nonalcoholic Steatohepatitis Is Marked by a Higher Frequency of Th17 Cells in the Liver and an Increased Th17/Resting Regulatory T Cell Ratio in Peripheral Blood and in the Liver. J. Immunol. 2016, 196, 97–105. [Google Scholar] [CrossRef] [PubMed]
- González-Terán, B.; Matesanz, N.; Nikolic, I.; Verdugo, M.A.; Sreeramkumar, V.; Hernández-Cosido, L.; Mora, A.; Crainiciuc, G.; Sáiz, M.L.; Bernardo, E.; et al. p38γ and p38δ reprogram liver metabolism by modulating neutrophil infiltration. EMBO J. 2016, 35, 536–552. [Google Scholar] [CrossRef]
- Malehmir, M.; Pfister, D.; Gallage, S.; Szydlowska, M.; Inverso, D.; Kotsiliti, E.; Leone, V.; Peiseler, M.; Surewaard, B.G.J.; Rath, D.; et al. Platelet GPIbα is a mediator and potential interventional target for NASH and subsequent liver cancer. Nat. Med. 2019, 25, 641–655. [Google Scholar] [CrossRef]
- Hammoutene, A.; Rautou, P.E. Role of liver sinusoidal endothelial cells in non-alcoholic fatty liver disease. J. Hepatol. 2019, 70, 1278–1291. [Google Scholar] [CrossRef]
- Xu, L.; Hui, A.Y.; Albanis, E.; Arthur, M.J.; O’Byrne, S.M.; Blaner, W.S.; Mukherjee, P.; Friedman, S.L.; Eng, F.J. Human hepatic stellate cell lines, LX-1 and LX-2: New tools for analysis of hepatic fibrosis. Gut 2005, 54, 142–151. [Google Scholar] [CrossRef] [PubMed]
- Ruehl, M.; Erben, U.; Kim, K.; Freise, C.; Dagdelen, T.; Eisele, S.; Trowitzsch-Kienast, W.; Zeitz, M.; Jia, J.; Stickel, F.; et al. Extracts of Lindera obtusiloba induce antifibrotic effects in hepatic stellate cells via suppression of a TGF-beta-mediated profibrotic gene expression pattern. J. Nutr. Biochem. 2009, 20, 597–606. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gao, J.; Zhang, D.; Zhang, J.; Ma, J.; Jiang, H. New insights into the antifibrotic effects of sorafenib on hepatic stellate cells and liver fibrosis. J. Hepatol. 2010, 53, 132–144. [Google Scholar] [CrossRef]
- Watanabe, T.; Tajima, H.; Hironori, H.; Nakagawara, H.; Ohnishi, I.; Takamura, H.; Ninomiya, I.; Kitagawa, H.; Fushida, S.; Tani, T.; et al. Sodium valproate blocks the transforming growth factor (TGF)-beta1 autocrine loop and attenuates the TGF-beta1-induced collagen synthesis in a human hepatic stellate cell line. Int. J. Mol. Med. 2011, 28, 919–925. [Google Scholar]
- Wang, Z.Y.; Burlak, C.; Klaunig, J.E.; Kamendulis, L.M. Development of a cytokine-producing immortalized murine Kupffer cell line. Cytokine 2014, 70, 165–172. [Google Scholar] [CrossRef]
- Ogawa, M.; Ogawa, S.; Bear, C.E.; Ahmadi, S.; Chin, S.; Li, B.; Grompe, M.; Keller, G.; Kamath, B.M.; Ghanekar, A. Directed differentiation of cholangiocytes from human pluripotent stem cells. Nat. Biotechnol. 2015, 33, 853–861. [Google Scholar] [CrossRef] [PubMed]
- Sampaziotis, F.; de Brito, M.C.; Madrigal, P.; Bertero, A.; Saeb-Parsy, K.; Soares, F.A.C.; Schrumpf, E.; Melum, E.; Karlsen, T.H.; Bradley, J.A.; et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat. Biotechnol. 2015, 33, 845–852. [Google Scholar] [CrossRef] [PubMed]
- Kochan, K.; Kus, E.; Szafraniec, E.; Wislocka, A.; Chlopicki, S.; Baranska, M. Changes induced by non-alcoholic fatty liver disease in liver sinusoidal endothelial cells and hepatocytes: Spectroscopic imaging of single live cells at the subcellular level. Analyst 2017, 142, 3948–3958. [Google Scholar] [CrossRef] [PubMed]
- Coll, M.; Perea, L.; Boon, R.; Leite, S.B.; Vallverdu, J.; Mannaerts, I.; Smout, A.; El Taghdouini, A.; Blaya, D.; Rodrigo-Torres, D.; et al. Generation of Hepatic Stellate Cells from Human Pluripotent Stem Cells Enables In Vitro Modeling of Liver Fibrosis. Cell Stem Cell 2018, 23, 101–113.e7. [Google Scholar] [CrossRef] [PubMed]
- Gage, B.K.; Liu, J.C.; Innes, B.T.; MacParland, S.A.; McGilvray, I.D.; Bader, G.D.; Keller, G.M. Generation of Functional Liver Sinusoidal Endothelial Cells from Human Pluripotent Stem-Cell-Derived Venous Angioblasts. Cell Stem Cell 2020, 27, 254–269.e9. [Google Scholar] [CrossRef]
- Verhulst, S.; van Os, E.A.; De Smet, V.; Eysackers, N.; Mannaerts, I.; van Grunsven, L.A. Gene Signatures Detect Damaged Liver Sinusoidal Endothelial Cells in Chronic Liver Diseases. Front. Med. 2021, 8, 750044. [Google Scholar] [CrossRef]
- De Smedt, J.; van Os, E.A.; Talon, I.; Ghosh, S.; Toprakhisar, B.; Furtado Madeiro Da Costa, R.; Zaunz, S.; Vazquez, M.A.; Boon, R.; Baatsen, P.; et al. PU.1 drives specification of pluripotent stem cell-derived endothelial cells to LSEC-like cells. Cell Death Dis. 2021, 12, 84. [Google Scholar] [CrossRef]
- Takata, N.; Ishii, K.A.; Takayama, H.; Nagashimada, M.; Kamoshita, K.; Tanaka, T.; Kikuchi, A.; Takeshita, Y.; Matsumoto, Y.; Ota, T.; et al. LECT2 as a hepatokine links liver steatosis to inflammation via activating tissue macrophages in NASH. Sci. Rep. 2021, 11, 555. [Google Scholar] [CrossRef]
- Rajab, N.; Angel, P.W.; Deng, Y.; Gu, J.; Jameson, V.; Kurowska-Stolarska, M.; Milling, S.; Pacheco, C.M.; Rutar, M.; Laslett, A.L.; et al. An integrated analysis of human myeloid cells identifies gaps in in vitro models of in vivo biology. Stem Cell Rep. 2021, 16, 1629–1643. [Google Scholar] [CrossRef]
- Lam, D.; Dan, Y.Y.; Chan, Y.S.; Ng, H.H. Emerging liver organoid platforms and technologies. Cell Regen. 2021, 10, 27. [Google Scholar] [CrossRef]
- Burt, A.D.; Ferrell, L.D.; Hübscher, S.G. MacSween’s Pathology of the Liver-Elsevier, 7th ed.; Elsevier: Amsterdam, The Netherlands, 2017. [Google Scholar]
- Segovia-Miranda, F.; Morales-Navarrete, H.; Kucken, M.; Moser, V.; Seifert, S.; Repnik, U.; Rost, F.; Brosch, M.; Hendricks, A.; Hinz, S.; et al. Three-dimensional spatially resolved geometrical and functional models of human liver tissue reveal new aspects of NAFLD progression. Nat. Med. 2019, 25, 1885–1893. [Google Scholar] [CrossRef]
- Strobel, S.; Kostadinova, R.; Fiaschetti-Egli, K.; Rupp, J.; Bieri, M.; Pawlowska, A.; Busler, D.; Hofstetter, T.; Sanchez, K.; Grepper, S.; et al. A 3D primary human cell-based in vitro model of non-alcoholic steatohepatitis for efficacy testing of clinical drug candidates. Sci. Rep. 2021, 11, 22765. [Google Scholar] [CrossRef] [PubMed]
- Sendi, H.; Mead, I.; Wan, M.; Mehrab-Mohseni, M.; Koch, K.; Atala, A.; Bonkovsky, H.L.; Bishop, C.E. miR-122 inhibition in a human liver organoid model leads to liver inflammation, necrosis, steatofibrosis and dysregulated insulin signaling. PLoS ONE 2018, 13, e0200847. [Google Scholar] [CrossRef] [PubMed]
- Pingitore, P.; Sasidharan, K.; Ekstrand, M.; Prill, S.; Linden, D.; Romeo, S. Human Multilineage 3D Spheroids as a Model of Liver Steatosis and Fibrosis. Int. J. Mol. Sci. 2019, 20, 1629. [Google Scholar] [CrossRef] [PubMed]
- Duriez, M.; Jacquet, A.; Hoet, L.; Roche, S.; Bock, M.D.; Rocher, C.; Haussy, G.; Vigé, X.; Bocskei, Z.; Slavnic, T.; et al. A 3D Human Liver Model of Nonalcoholic Steatohepatitis. J. Clin. Transl. Hepatol. 2020, 8, 359–370. [Google Scholar] [CrossRef]
- Gori, M.; Simonelli, M.C.; Giannitelli, S.M.; Businaro, L.; Trombetta, M.; Rainer, A. Investigating Nonalcoholic Fatty Liver Disease in a Liver-on-a-Chip Microfluidic Device. PLoS ONE 2016, 11, e0159729. [Google Scholar] [CrossRef]
- Kostrzewski, T.; Cornforth, T.; Snow, S.A.; Ouro-Gnao, L.; Rowe, C.; Large, E.M.; Hughes, D.J. Three-dimensional perfused human in vitro model of non-alcoholic fatty liver disease. World J. Gastroenterol. 2017, 23, 204–215. [Google Scholar] [CrossRef]
- Du, K.; Li, S.; Li, C.; Li, P.; Miao, C.; Luo, T.; Qiu, B.; Ding, W. Modeling nonalcoholic fatty liver disease on a liver lobule chip with dual blood supply. Acta Biomater. 2021, 134, 228–239. [Google Scholar] [CrossRef]
- Cho, H.J.; Kim, H.J.; Lee, K.; Lasli, S.; Ung, A.; Hoffman, T.; Nasiri, R.; Bandaru, P.; Ahadian, S.; Dokmeci, M.R.; et al. Bioengineered Multicellular Liver Microtissues for Modeling Advanced Hepatic Fibrosis Driven Through Non-Alcoholic Fatty Liver Disease. Small 2021, 17, e2007425. [Google Scholar] [CrossRef]
- Davidson, M.D.; Kukla, D.A.; Khetani, S.R. Microengineered cultures containing human hepatic stellate cells and hepatocytes for drug development. Integr. Biol. 2017, 9, 662–677. [Google Scholar] [CrossRef]
- Feaver, R.E.; Cole, B.K.; Lawson, M.J.; Hoang, S.A.; Marukian, S.; Blackman, B.R.; Figler, R.A.; Sanyal, A.J.; Wamhoff, B.R.; Dash, A. Development of an in vitro human liver system for interrogating nonalcoholic steatohepatitis. JCI Insight 2016, 1, e90954. [Google Scholar] [CrossRef] [PubMed]
- Ijssennagger, N.; Janssen, A.W.F.; Milona, A.; Ramos Pittol, J.M.; Hollman, D.A.A.; Mokry, M.; Betzel, B.; Berends, F.J.; Janssen, I.M.; van Mil, S.W.C.; et al. Gene expression profiling in human precision cut liver slices in response to the FXR agonist obeticholic acid. J. Hepatol. 2016, 64, 1158–1166. [Google Scholar] [CrossRef] [PubMed]
- Collin de l’Hortet, A.; Takeishi, K.; Guzman-Lepe, J.; Morita, K.; Achreja, A.; Popovic, B.; Wang, Y.; Handa, K.; Mittal, A.; Meurs, N.; et al. Generation of Human Fatty Livers Using Custom-Engineered Induced Pluripotent Stem Cells with Modifiable SIRT1 Metabolism. Cell Metab. 2019, 30, 385–401.e9. [Google Scholar] [CrossRef] [PubMed]
- Hofer, M.; Lutolf, M.P. Engineering organoids. Nat. Rev. Mater. 2021, 6, 402–420. [Google Scholar] [CrossRef] [PubMed]
- Underhill, G.H.; Khetani, S.R. Bioengineered Liver Models for Drug Testing and Cell Differentiation Studies. Cell Mol. Gastroenterol. Hepatol. 2018, 5, 426–439.e1. [Google Scholar] [CrossRef]
- Kukla, D.A.; Khetani, S.R. Bioengineered Liver Models for Investigating Disease Pathogenesis and Regenerative Medicine. Semin. Liver Dis. 2021, 41, 368–392. [Google Scholar] [CrossRef]
- Bernal, P.N.; Bouwmeester, M.; Madrid-Wolff, J.; Falandt, M.; Florczak, S.; Rodriguez, N.G.; Li, Y.; Größbacher, G.; Samsom, R.A.; van Wolferen, M.; et al. Volumetric Bioprinting of Organoids and Optically Tuned Hydrogels to Build Liver-Like Metabolic Biofactories. Adv. Mater. 2022, 34, e2110054. [Google Scholar] [CrossRef]
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772. [Google Scholar] [CrossRef]
- Zhang, B.; Korolj, A.; Lai, B.F.L.; Radisic, M. Advances in organ-on-a-chip engineering. Nat. Rev. Mater. 2018, 3, 257–278. [Google Scholar] [CrossRef]
- Orbach, S.M.; Less, R.R.; Kothari, A.; Rajagopalan, P. In Vitro Intestinal and Liver Models for Toxicity Testing. ACS Biomater. Sci. Eng. 2017, 3, 1898–1910. [Google Scholar] [CrossRef]
- Nawroth, J.C.; Petropolis, D.B.; Manatakis, D.V.; Maulana, T.I.; Burchett, G.; Schlunder, K.; Witt, A.; Shukla, A.; Kodella, K.; Ronxhi, J.; et al. Modeling alcohol-associated liver disease in a human Liver-Chip. Cell Rep. 2021, 36, 109393. [Google Scholar] [CrossRef] [PubMed]
- Freag, M.S.; Namgung, B.; Reyna Fernandez, M.E.; Gherardi, E.; Sengupta, S.; Jang, H.L. Human Nonalcoholic Steatohepatitis on a Chip. Hepatol. Commun. 2021, 5, 217–233. [Google Scholar] [CrossRef] [PubMed]
- Khetani, S.R.; Bhatia, S.N. Microscale culture of human liver cells for drug development. Nat. Biotechnol. 2008, 26, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Palma, E.; Doornebal, E.J.; Chokshi, S. Precision-cut liver slices: A versatile tool to advance liver research. Hepatol. Int. 2019, 13, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Neuberger, J.; Patel, J.; Caldwell, H.; Davies, S.; Hebditch, V.; Hollywood, C.; Hubscher, S.; Karkhanis, S.; Lester, W.; Roslund, N.; et al. Guidelines on the use of liver biopsy in clinical practice from the British Society of Gastroenterology, the Royal College of Radiologists and the Royal College of Pathology. Gut 2020, 69, 1382–1403. [Google Scholar] [CrossRef] [PubMed]
- Hussein, K.H.; Park, K.M.; Ghim, J.H.; Yang, S.R.; Woo, H.M. Three dimensional culture of HepG2 liver cells on a rat decellularized liver matrix for pharmacological studies. J. Biomed. Mater. Res. B Appl. Biomater. 2016, 104, 263–273. [Google Scholar] [CrossRef] [PubMed]
- Mazza, G.; Rombouts, K.; Rennie Hall, A.; Urbani, L.; Vinh Luong, T.; Al-Akkad, W.; Longato, L.; Brown, D.; Maghsoudlou, P.; Dhillon, A.P.; et al. Decellularized human liver as a natural 3D-scaffold for liver bioengineering and transplantation. Sci. Rep. 2015, 5, 13079. [Google Scholar] [CrossRef]
- Huby, T.; Gautier, E.L. Immune cell-mediated features of non-alcoholic steatohepatitis. Nat. Rev. Immunol. 2022, 22, 429–443. [Google Scholar] [CrossRef]
- Cai, J.; Zhang, X.J.; Li, H. The Role of Innate Immune Cells in Nonalcoholic Steatohepatitis. Hepatology 2019, 70, 1026–1037. [Google Scholar] [CrossRef]
- Morgantini, C.; Jager, J.; Li, X.; Levi, L.; Azzimato, V.; Sulen, A.; Barreby, E.; Xu, C.; Tencerova, M.; Näslund, E.; et al. Liver macrophages regulate systemic metabolism through non-inflammatory factors. Nat. Metab. 2019, 1, 445–459. [Google Scholar] [CrossRef]
- MacParland, S.A.; Liu, J.C.; Ma, X.Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.; Dobie, R.; Wilson-Kanamori, J.R.; Dora, E.F.; Henderson, B.E.P.; Luu, N.T.; Portman, J.R.; Matchett, K.P.; Brice, M.; Marwick, J.A.; et al. Resolving the fibrotic niche of human liver cirrhosis at single-cell level. Nature 2019, 575, 512–518. [Google Scholar] [CrossRef] [PubMed]
- Sutti, S.; Albano, E. Adaptive immunity: An emerging player in the progression of NAFLD. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 81–92. [Google Scholar] [CrossRef] [PubMed]
- Bruzzì, S.; Sutti, S.; Giudici, G.; Burlone, M.E.; Ramavath, N.N.; Toscani, A.; Bozzola, C.; Schneider, P.; Morello, E.; Parola, M.; et al. B2-Lymphocyte responses to oxidative stress-derived antigens contribute to the evolution of nonalcoholic fatty liver disease (NAFLD). Free Radic. Biol. Med. 2018, 124, 249–259. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, J.; Kirby, M.; Softic, S.; Miles, L.; Salazar-Gonzalez, R.M.; Shivakumar, P.; Kohli, R. Hepatic Natural Killer T-cell and CD8+ T-cell Signatures in Mice with Nonalcoholic Steatohepatitis. Hepatol. Commun. 2017, 1, 299–310. [Google Scholar] [CrossRef]
- Musso, G.; Gambino, R.; Cassader, M.; Paschetta, E.; Sircana, A. Specialized Proresolving Mediators: Enhancing Nonalcoholic Steatohepatitis and Fibrosis Resolution. Trends Pharmacol. Sci. 2018, 39, 387–401. [Google Scholar] [CrossRef]
- Kim, A.D.; Kim, S.E.; Leszczynska, A.; Kaufmann, B.; Reca, A.; Kim, D.J.; Feldstein, A.E. Dual role of neutrophils in modulating liver injury and fibrosis during development and resolution of diet-induced murine steatohepatitis. Sci. Rep. 2021, 11, 24194. [Google Scholar] [CrossRef]
- Ampuero, J.; Romero-Gomez, M. Stratification of patients in NASH clinical trials: A pitfall for trial success. JHEP Rep. 2020, 2, 100148. [Google Scholar] [CrossRef]
- Hebbard, L.; George, J. Animal models of nonalcoholic fatty liver disease. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 35–44. [Google Scholar] [CrossRef]
- Im, Y.R.; Hunter, H.; de Gracia Hahn, D.; Duret, A.; Cheah, Q.; Dong, J.; Fairey, M.; Hjalmarsson, C.; Li, A.; Lim, H.K.; et al. A Systematic Review of Animal Models of NAFLD Finds High-Fat, High-Fructose Diets Most Closely Resemble Human NAFLD. Hepatology 2021, 74, 1884–1901. [Google Scholar] [CrossRef]
- Burt, A.D.; Lackner, C.; Tiniakos, D.G. Diagnosis and Assessment of NAFLD: Definitions and Histopathological Classification. Semin. Liver Dis. 2015, 35, 207–220. [Google Scholar] [PubMed]
- Brunt, E.M.; Janney, C.G.; Di Bisceglie, A.M.; Neuschwander-Tetri, B.A.; Bacon, B.R. Nonalcoholic steatohepatitis: A proposal for grading and staging the histological lesions. Am. J. Gastroenterol. 1999, 94, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
- Kleiner, D.E.; Brunt, E.M.; Van Natta, M.; Behling, C.; Contos, M.J.; Cummings, O.W.; Ferrell, L.D.; Liu, Y.C.; Torbenson, M.S.; Unalp-Arida, A.; et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005, 41, 1313–1321. [Google Scholar] [CrossRef] [PubMed]
- U.S. Food & Drug Administration. Noncirrhotic Nonalcoholic Steatohepatitis with Liver Fibrosis: Developing Drugs for Treatment Guidance for Industry (Draft Guidance). 2018. Available online: https://www.fda.gov/regulatory-information/search-fda-guidance-documents/noncirrhotic-nonalcoholic-steatohepatitis-liver-fibrosis-developing-drugs-treatment (accessed on 1 August 2022).
- Terada, T.; Nakanuma, Y.; Hoso, M.; Saito, K.; Sasaki, M.; Nonomura, A. Fatty macroregenerative nodule in non-steatotic liver cirrhosis. A morphologic study. Virchows Arch. A Pathol. Anat. Histopathol. 1989, 415, 131–136. [Google Scholar] [CrossRef] [PubMed]
- van der Poorten, D.; Samer, C.F.; Ramezani-Moghadam, M.; Coulter, S.; Kacevska, M.; Schrijnders, D.; Wu, L.E.; McLeod, D.; Bugianesi, E.; Komuta, M.; et al. Hepatic fat loss in advanced nonalcoholic steatohepatitis: Are alterations in serum adiponectin the cause? Hepatology 2013, 57, 2180–2188. [Google Scholar] [CrossRef] [PubMed]
- Kessoku, T.; Kobayashi, T.; Tanaka, K.; Yamamoto, A.; Takahashi, K.; Iwaki, M.; Ozaki, A.; Kasai, Y.; Nogami, A.; Honda, Y.; et al. The Role of Leaky Gut in Nonalcoholic Fatty Liver Disease: A Novel Therapeutic Target. Int. J. Mol. Sci. 2021, 22, 8161. [Google Scholar] [CrossRef]
- Loria, P.; Carulli, L.; Bertolotti, M.; Lonardo, A. Endocrine and liver interaction: The role of endocrine pathways in NASH. Nat. Rev. Gastroenterol. Hepatol. 2009, 6, 236–247. [Google Scholar] [CrossRef]
- Zhang, X.; Ji, X.; Wang, Q.; Li, J.Z. New insight into inter-organ crosstalk contributing to the pathogenesis of non-alcoholic fatty liver disease (NAFLD). Protein Cell 2018, 9, 164–177. [Google Scholar] [CrossRef]
- Byrne, C.D.; Targher, G. NAFLD as a driver of chronic kidney disease. J. Hepatol. 2020, 72, 785–801. [Google Scholar] [CrossRef]
| Cell Type | Cell Lines | Major Features |
|---|---|---|
| Immortalized Cell lines | HepG2 |
|
| Huh7 | ||
| WRL68 |
| |
| HepaRG | ||
| Tissue-derived primary cells | Primary Hepatocyte (PHH) |
|
| Bipotent ductal stem cells |
| |
| Pluripotent stem cells (PSC)-derived primary cells | Hepatocyte-like cells (HLC) |
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, S.-X.; Yan, J.-S.; Chan, Y.-S. Advancements in MAFLD Modeling with Human Cell and Organoid Models. Int. J. Mol. Sci. 2022, 23, 11850. https://doi.org/10.3390/ijms231911850
Wang S-X, Yan J-S, Chan Y-S. Advancements in MAFLD Modeling with Human Cell and Organoid Models. International Journal of Molecular Sciences. 2022; 23(19):11850. https://doi.org/10.3390/ijms231911850
Chicago/Turabian StyleWang, Shi-Xiang, Ji-Song Yan, and Yun-Shen Chan. 2022. "Advancements in MAFLD Modeling with Human Cell and Organoid Models" International Journal of Molecular Sciences 23, no. 19: 11850. https://doi.org/10.3390/ijms231911850
APA StyleWang, S.-X., Yan, J.-S., & Chan, Y.-S. (2022). Advancements in MAFLD Modeling with Human Cell and Organoid Models. International Journal of Molecular Sciences, 23(19), 11850. https://doi.org/10.3390/ijms231911850





