Integrated Multi-Omics Analysis Reveals Differential Effects of Fructo-Oligosaccharides (FOS) Supplementation on the Human Gut Ecosystem
Abstract
1. Introduction
2. Results
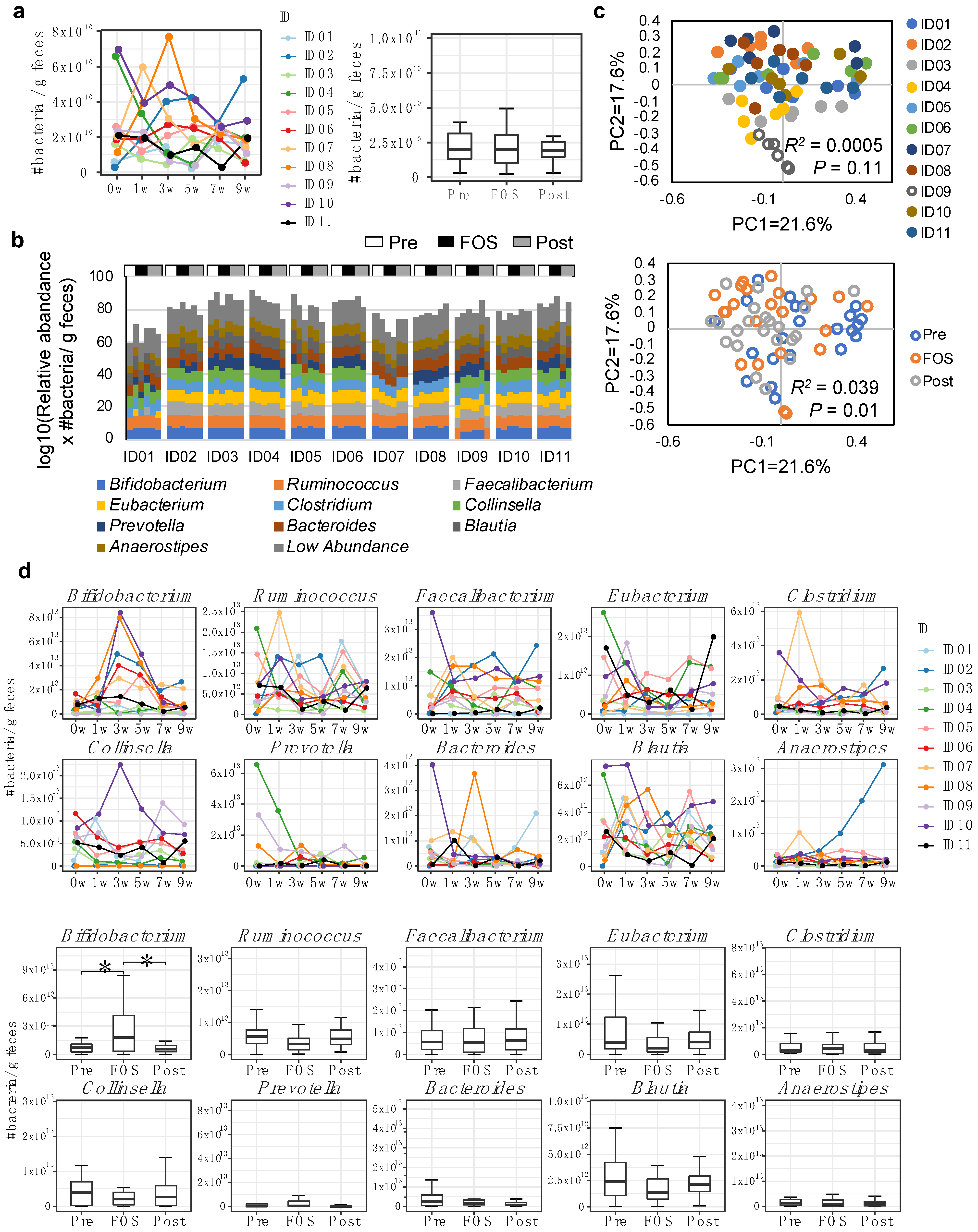
2.1. Increase of Bifidobacterium in Feces by FOS Supplementation
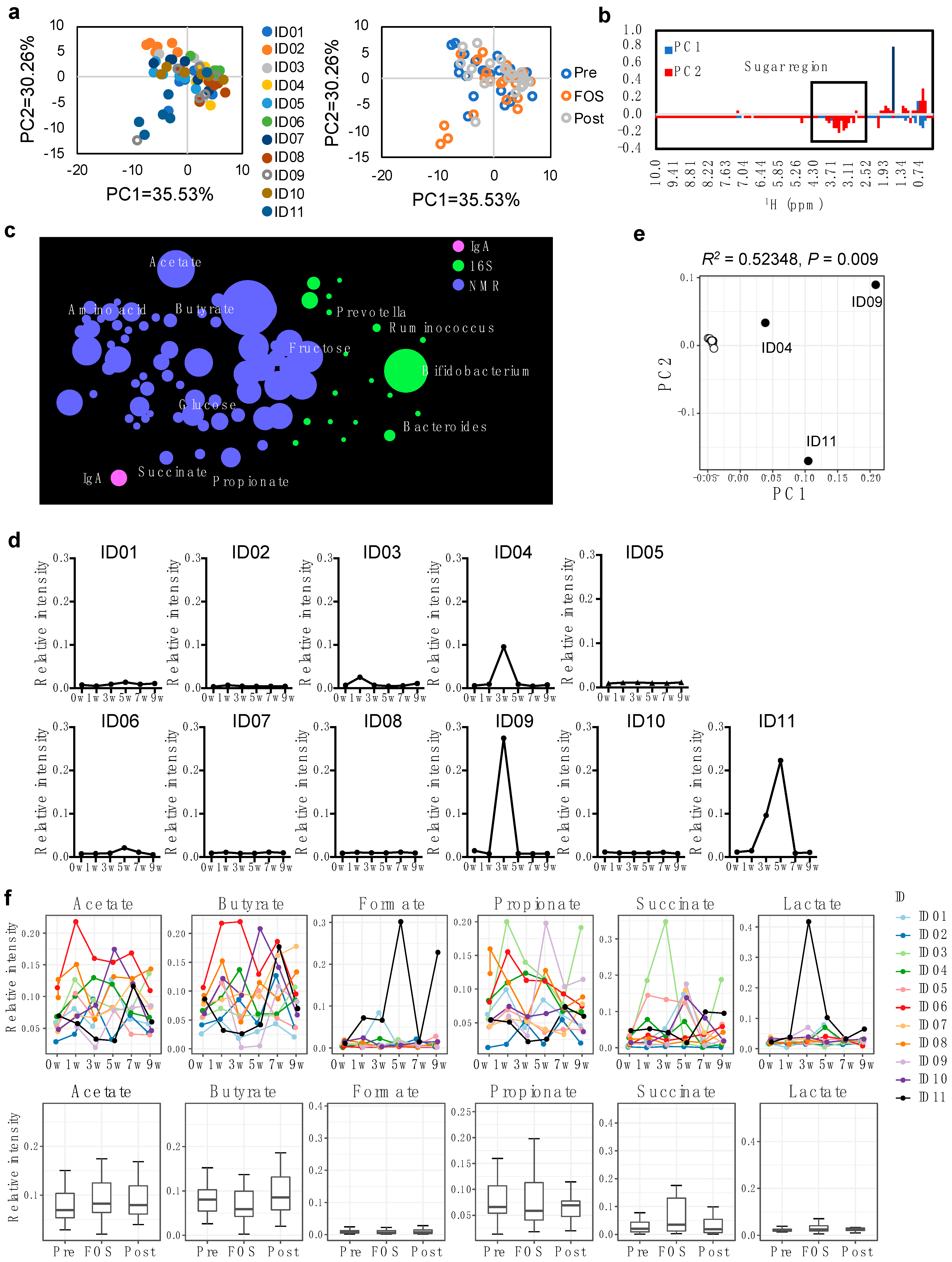
2.2. Increase of Fecal Fructose during the FOS Intake in Some Individuals
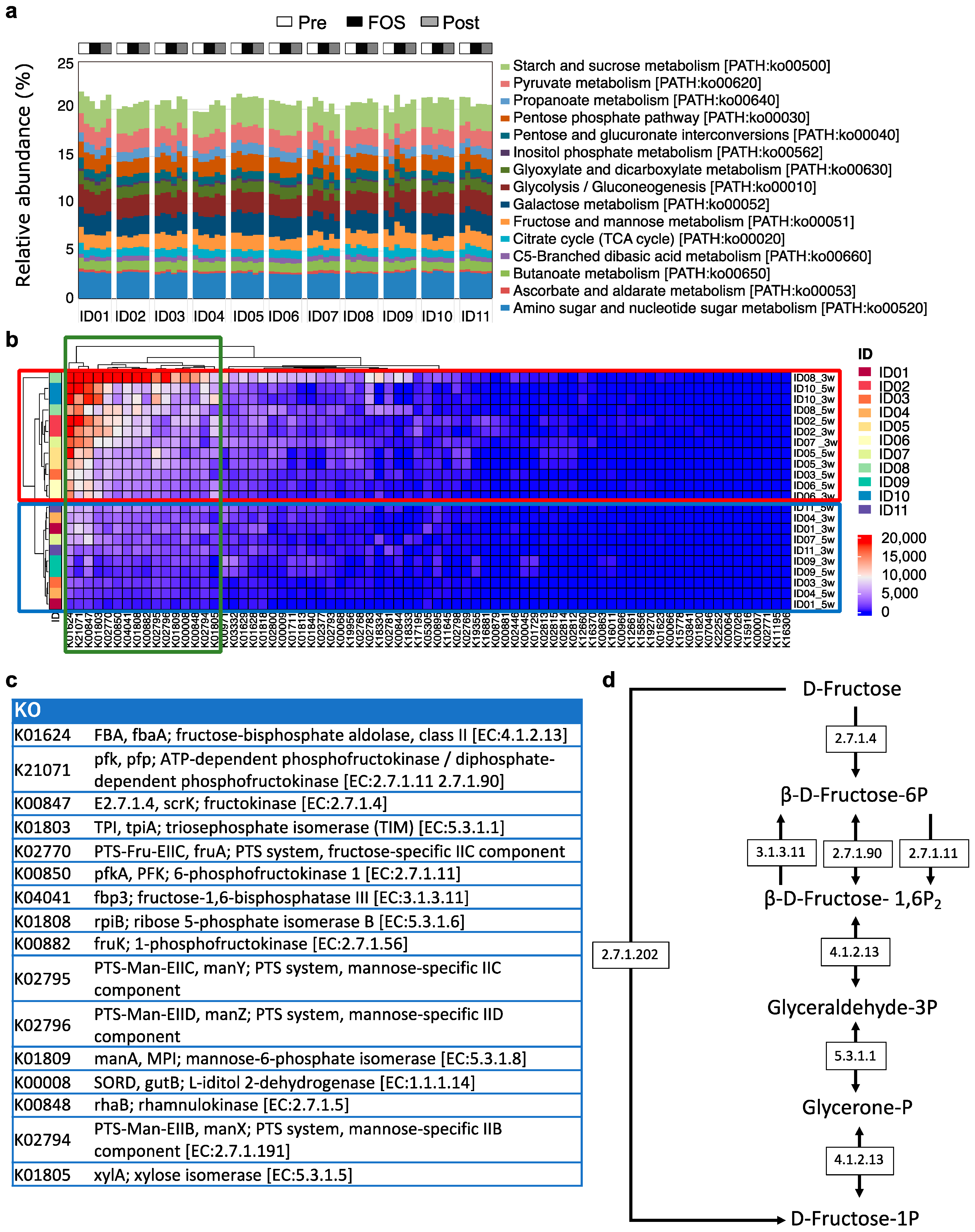
2.3. Genetic Differences in Fructose Metabolism from Metagenomic Analysis
2.4. Impact of FOS Intake on the Immune Compartment
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Study Design
4.3. FOS Supplementation
4.4. Sample Collection
4.5. Measurement of Fecal IgA
4.6. Bacterial DNA Extraction
4.7. Bacterial 16S rRNA Gene Amplicon Sequencing and Processing
4.8. Metagenomic Sequencing
4.9. Assembly of Metagenomic Sequences and Gene Prediction
4.10. Functional Assignment of Non-Redundant Genes in Human Gut Microbiomes
4.11. Quantification of the Annotated Genes in Human Gut Microbiomes
4.12. Metabolic Profiling of Human Fecal Samples
4.13. Quantification of Bacterial Number and Gene Expression Using Quantitative Polymerase Chain Reaction (qPCR)
4.14. Correlation Network Construction
4.15. Peripheral Blood Mononuclear Cell (PBMC) Isolation and Analysis
4.16. Multivariate Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rooks, M.G.; Garrett, W.S. Gut microbiota, metabolites and host immunity. Nat. Rev. Immunol. 2016, 16, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Human Microbiome Project, C. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Li, Y.; Cai, Z.; Li, S.; Zhu, J.; Zhang, F.; Liang, S.; Zhang, W.; Guan, Y.; Shen, D.; et al. A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 2012, 490, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Tremaroli, V.; Nookaew, I.; Bergstrom, G.; Behre, C.J.; Fagerberg, B.; Nielsen, J.; Backhed, F. Gut metagenome in European women with normal, impaired and diabetic glucose control. Nature 2013, 498, 99–103. [Google Scholar] [CrossRef]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef]
- Nishijima, S.; Suda, W.; Oshima, K.; Kim, S.W.; Hirose, Y.; Morita, H.; Hattori, M. The gut microbiome of healthy Japanese and its microbial and functional uniqueness. DNA Res. 2016, 23, 125–133. [Google Scholar] [CrossRef]
- Human Microbiome Project, C. A framework for human microbiome research. Nature 2012, 486, 215–221. [Google Scholar] [CrossRef]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef]
- Fan, C.; Zhang, L.; Jia, S.; Tang, X.; Fu, H.; Li, W.; Liu, C.; Zhang, H.; Cheng, Q.; Zhang, Y. Seasonal variations in the composition and functional profiles of gut microbiota reflect dietary changes in plateau pikas. Integr. Zool. 2022, 17, 379–395. [Google Scholar] [CrossRef]
- Gibson, G.R.; Hutkins, R.; Sanders, M.E.; Prescott, S.L.; Reimer, R.A.; Salminen, S.J.; Scott, K.; Stanton, C.; Swanson, K.S.; Cani, P.D.; et al. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics (ISAPP) consensus statement on the definition and scope of prebiotics. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 491–502. [Google Scholar] [CrossRef] [PubMed]
- Macfarlane, G.T.; Macfarlane, S. Fermentation in the human large intestine: Its physiologic consequences and the potential contribution of prebiotics. J. Clin. Gastroenterol. 2011, 45, S120–S127. [Google Scholar] [CrossRef] [PubMed]
- Oozeer, R.; van Limpt, K.; Ludwig, T.; Ben Amor, K.; Martin, R.; Wind, R.D.; Boehm, G.; Knol, J. Intestinal microbiology in early life: Specific prebiotics can have similar functionalities as human-milk oligosaccharides. Am. J. Clin. Nutr. 2013, 98, 561S–571S. [Google Scholar] [CrossRef] [PubMed]
- Ortega-González, M.; Ocón, B.; Romero-Calvo, I.; Anzola, A.; Guadix, E.; Zarzuelo, A.; Suárez, M.D.; Sánchez de Medina, F.; Martínez-Augustin, O. Nondigestible oligosaccharides exert nonprebiotic effects on intestinal epithelial cells enhancing the immune response via activation of TLR4-NFκB. Mol. Nutr. Food Res. 2014, 58, 384–393. [Google Scholar] [CrossRef] [PubMed]
- Hosono, A.; Ozawa, A.; Kato, R.; Ohnishi, Y.; Nakanishi, Y.; Kimura, T.; Nakamura, R. Dietary fructooligosaccharides induce immunoregulation of intestinal IgA secretion by murine Peyer’s patch cells. Biosci. Biotechnol. Biochem. 2003, 67, 758–764. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Nosaka, S.; Suzuki, M.; Nagafuchi, S.; Takahashi, T.; Yajima, T.; Takenouchi-Ohkubo, N.; Iwase, T.; Moro, I. Dietary fructooligosaccharides up-regulate immunoglobulin A response and polymeric immunoglobulin receptor expression in intestines of infant mice. Clin. Exp. Immunol. 2004, 137, 52–58. [Google Scholar] [CrossRef]
- Kato, T.; Fukuda, S.; Fujiwara, A.; Suda, W.; Hattori, M.; Kikuchi, J.; Ohno, H. Multiple omics uncovers host-gut microbial mutualism during prebiotic fructooligosaccharide supplementation. DNA Res. 2014, 21, 469–480. [Google Scholar] [CrossRef]
- Sonnenburg, E.D.; Sonnenburg, J.L.; Manchester, J.K.; Hansen, E.E.; Chiang, H.C.; Gordon, J.I. A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc. Natl. Acad. Sci. USA 2006, 103, 8834–8839. [Google Scholar] [CrossRef]
- McNulty, N.P.; Wu, M.; Erickson, A.R.; Pan, C.; Erickson, B.K.; Martens, E.C.; Pudlo, N.A.; Muegge, B.D.; Henrissat, B.; Hettich, R.L.; et al. Effects of diet on resource utilization by a model human gut microbiota containing Bacteroides cellulosilyticus WH2, a symbiont with an extensive glycobiome. PLoS Biol. 2013, 11, e1001637. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef]
- Lazar, V.; Ditu, L.M.; Pircalabioru, G.G.; Picu, A.; Petcu, L.; Cucu, N.; Chifiriuc, M.C. Gut Microbiota, Host Organism, and Diet Trialogue in Diabetes and Obesity. Front. Nutr. 2019, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; Lane, J.A.; Smith, G.J.; Grimaldi, K.A.; Ross, R.P.; Stanton, C. Precision Nutrition and the Microbiome Part II: Potential Opportunities and Pathways to Commercialisation. Nutrients 2019, 11, 1468. [Google Scholar] [CrossRef] [PubMed]
- Nasidze, I.; Li, J.; Quinque, D.; Tang, K.; Stoneking, M. Global diversity in the human salivary microbiome. Genome Res. 2009, 19, 636–643. [Google Scholar] [CrossRef] [PubMed]
- Takeshita, T.; Matsuo, K.; Furuta, M.; Shibata, Y.; Fukami, K.; Shimazaki, Y.; Akifusa, S.; Han, D.H.; Kim, H.D.; Yokoyama, T.; et al. Distinct composition of the oral indigenous microbiota in South Korean and Japanese adults. Sci. Rep. 2014, 4, 6990. [Google Scholar] [CrossRef] [PubMed]
- Gifford, S.M.; Sharma, S.; Rinta-Kanto, J.M.; Moran, M.A. Quantitative analysis of a deeply sequenced marine microbial metatoranscriptome. ISME J. 2011, 5, 461–472. [Google Scholar] [CrossRef] [PubMed]
- Vandeputte, D.; Kathagen, G.; D’Hoe, K.; Vieira-Silva, S.; Valles-Colomer, M.; Sabino, J.; Wang, J.; Tito, R.Y.; De Commer, L.; Darzi, Y.; et al. Quantitative microbiome profiling links gut community variation to microbial load. Nature 2017, 551, 507–511. [Google Scholar] [CrossRef]
- Rao, C.; Coyte, K.Z.; Bainter, W.; Geha, R.S.; Martin, C.R.; Rakoff-Nahoum, S. Multi-kingdom ecological drivers of microbiota assembly in preterm infants. Nature 2021, 591, 633–638. [Google Scholar] [CrossRef]
- Kowalska-Duplaga, K.; Gosiewski, T.; Kapusta, P.; Sroka-Oleksiak, A.; Wedrychowicz, A.; Pieczarkowski, S.; Ludwig-Slomczynska, A.H.; Wolkow, P.P.; Fyderek, K. Differences in the intestinal microbiome of healthy children and patients with newly diagnosed Crohn’s disease. Sci. Rep. 2019, 9, 18880. [Google Scholar] [CrossRef]
- Quince, C.; Ijaz, U.Z.; Loman, N.; Eren, A.M.; Saulnier, D.; Russell, J.; Haig, S.J.; Calus, S.T.; Quick, J.; Barclay, A.; et al. Extensive Modulation of the Fecal Metagenome in Children With Crohn’s Disease During Exclusive Enteral Nutrition. Am. J. Gastroenterol. 2015, 110, 1718–1729. [Google Scholar] [CrossRef]
- Manichanh, C.; Rigottier-Gois, L.; Bonnaud, E.; Gloux, K.; Pelletier, E.; Frangeul, L.; Nalin, R.; Jarrin, C.; Chardon, P.; Marteau, P.; et al. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut 2006, 55, 205–211. [Google Scholar] [CrossRef]
- Bray, G.A.; Nielsen, S.J.; Popkin, B.M. Consumption of high-fructose corn syrup in beverages may play a role in the epidemic of obesity. Am. J. Clin. Nutr. 2004, 79, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Joh, H.-K.; Lee, D.H.; Hur, J.; Nimptsch, K.; Chang, Y.; Joung, H.; Zhang, X.; Rezende, L.F.M.; Lee, J.E.; Ng, K.; et al. Simple sugar and sugar-sweetened beverage intake during adolescence and risk of colorectal cancer precursors. Gastroenterology 2021, 161, 128–142. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Manolescu, A.; Ritzel, M.; Yao, S.; Slugoski, M.; Young, J.D.; Chen, X.-Z.; Cheeseman, C.I. Cloning and functional characterization of the human GLUT7 isoform SLC2A7 from the small intestine. Am. J. Physiol. Gastrointest. Liver Physiol. 2004, 287, G236–G242. [Google Scholar] [CrossRef] [PubMed]
- Cheeseman, C. GLUT7: A new intestinal facilitated hexose transporter. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E238–E241. [Google Scholar] [CrossRef]
- Mao, B.; Li, D.; Zhao, J.; Liu, X.; Gu, Z.; Chen, Y.Q.; Zhang, H.; Chen, W. In vitro fermentation of fructooligosaccharides with human gut bacteria. Food Funct. 2015, 6, 947–954. [Google Scholar] [CrossRef]
- Belenguer, A.; Duncan, S.H.; Calder, A.G.; Holtrop, G.; Louis, P.; Lobley, G.E.; Flint, H.J. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl. Environ. Microbiol. 2006, 72, 3593–3599. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef]
- Adamberg, S.; Tomson, K.; Vija, H.; Puurand, M.; Kabanova, N.; Visnapuu, T.; Jogi, E.; Alamae, T.; Adamberg, K. Degradation of Fructans and Production of Propionic Acid by Bacteroides thetaiotaomicron are Enhanced by the Shortage of Amino Acids. Front. Nutr. 2014, 1, 21. [Google Scholar] [CrossRef]
- Komura, M.; Fukuta, T.; Genda, T.; Hino, S.; Aoe, S.; Kawagishi, H.; Morita, T. A short-term ingestion of fructo-oligosaccharides increases immunoglobulin A and mucin concentrations in the rat cecum, but the effects are attenuated with the prolonged ingestion. Biosci. Biotechnol. Biochem. 2014, 78, 1592–1602. [Google Scholar] [CrossRef]
- Kim, S.W.; Suda, W.; Kim, S.; Oshima, K.; Fukuda, S.; Ohno, H.; Morita, H.; Hattori, M. Robustness of gut microbiota of healthy adults in response to probiotic intervention revealed by high-throughput pyrosequencing. DNA Res. 2013, 20, 241–253. [Google Scholar] [CrossRef]
- Morita, H.; Kuwahara, T.; Ohshima, K.; Sasamoto, H.; Itoh, K.; Hattori, M.; Hayashi, T.; Takami, H. An improved DNA isolation method for metagenomic analysis of the microbial flora of the human intestine. Microbes Environ. 2007, 22, 214–222. [Google Scholar] [CrossRef]
- Said, H.S.; Suda, W.; Nakagome, S.; Chinen, H.; Oshima, K.; Kim, S.; Kimura, R.; Iraha, A.; Ishida, H.; Fujita, J.; et al. Dysbiosis of salivary microbiota in inflammatory bowel disease and its association with oral immunological biomarkers. DNA Res. 2014, 21, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Takayasu, L.; Suda, W.; Takanashi, K.; Iioka, E.; Kurokawa, R.; Shindo, C.; Hattori, Y.; Yamashita, N.; Nishijima, S.; Oshima, K.; et al. Circadian oscillations of microbial and functional composition in the human salivary microbiome. DNA Res. 2017, 24, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Good, I.J.; Toulmin, G.H. The number of new species, and the increase in population coverage, when a sample is increased. Biometrika 1956, 43, 45–63. [Google Scholar] [CrossRef]
- Lozupone, C.; Hamady, M.; Knight, R. UniFrac -an online tool for comparing microbial community diversity in a phylogenetic context. BMC Bioinform. 2006, 7, 371. [Google Scholar] [CrossRef]
- Li, J.; Jia, H.; Cai, X.; Zhong, H.; Feng, Q.; Sunagawa, S.; Arumugam, M.; Kultima, J.R.; Prifti, E.; Nielsen, T.; et al. An integrated catalog of reference genes in the human gut microbiome. Nat. Biotechnol. 2014, 32, 834–841. [Google Scholar] [CrossRef]
- Misawa, T.; Date, Y.; Kikuchi, J. Human metabolic, mineral, and microbiota fluctuations across daily nutritional intake visualized by a data-driven approach. J. Proteome Res. 2015, 14, 1526–1534. [Google Scholar] [CrossRef]
- Motegi, H.; Tsuboi, Y.; Saga, A.; Kagami, T.; Inoue, M.; Toki, H.; Minowa, O.; Noda, T.; Kikuchi, J. Identification of Reliable Components in Multivariate Curve Resolution-Alternating Least Squares (MCR-ALS): A data-driven approach across metabolic processes. Sci. Rep. 2015, 5, 15710. [Google Scholar] [CrossRef]
- Misawa, T.; Wei, F.; Kikuchi, J. Application of two-dimensional nuclear magnetic resonance for signal enhancement by spectral integration using a large data set of metabolic mixtures. Anal. Chem. 2016, 88, 6130–6134. [Google Scholar] [CrossRef]
- Kikuchi, J.; Tsuboi, Y.; Komatsu, K.; Gomi, M.; Chikayama, E.; Date, Y. SpinCouple: Development of a web tool for analyzing metabolite mixtures via two-dimensional J-resolved NMR database. Anal. Chem. 2016, 88, 659–665. [Google Scholar] [CrossRef]
- Wishart, D.S.; Jewison, T.; Guo, A.C.; Wilson, M.; Knox, C.; Liu, Y.; Djoumbou, Y.; Mandal, R.; Aziat, F.; Dong, E.; et al. HMDB 3.0-The Human Metabolome Database in 2013. Nucleic Acids Res. 2013, 41, D801–D807. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kato, T.; Kagawa, M.; Suda, W.; Tsuboi, Y.; Inoue-Suzuki, S.; Kikuchi, J.; Hattori, M.; Ohta, T.; Ohno, H. Integrated Multi-Omics Analysis Reveals Differential Effects of Fructo-Oligosaccharides (FOS) Supplementation on the Human Gut Ecosystem. Int. J. Mol. Sci. 2022, 23, 11728. https://doi.org/10.3390/ijms231911728
Kato T, Kagawa M, Suda W, Tsuboi Y, Inoue-Suzuki S, Kikuchi J, Hattori M, Ohta T, Ohno H. Integrated Multi-Omics Analysis Reveals Differential Effects of Fructo-Oligosaccharides (FOS) Supplementation on the Human Gut Ecosystem. International Journal of Molecular Sciences. 2022; 23(19):11728. https://doi.org/10.3390/ijms231911728
Chicago/Turabian StyleKato, Tamotsu, Masaharu Kagawa, Wataru Suda, Yuuri Tsuboi, Sayo Inoue-Suzuki, Jun Kikuchi, Masahira Hattori, Toshiko Ohta, and Hiroshi Ohno. 2022. "Integrated Multi-Omics Analysis Reveals Differential Effects of Fructo-Oligosaccharides (FOS) Supplementation on the Human Gut Ecosystem" International Journal of Molecular Sciences 23, no. 19: 11728. https://doi.org/10.3390/ijms231911728
APA StyleKato, T., Kagawa, M., Suda, W., Tsuboi, Y., Inoue-Suzuki, S., Kikuchi, J., Hattori, M., Ohta, T., & Ohno, H. (2022). Integrated Multi-Omics Analysis Reveals Differential Effects of Fructo-Oligosaccharides (FOS) Supplementation on the Human Gut Ecosystem. International Journal of Molecular Sciences, 23(19), 11728. https://doi.org/10.3390/ijms231911728





