bHLH010/089 Transcription Factors Control Pollen Wall Development via Specific Transcriptional and Metabolic Networks in Arabidopsis thaliana
Abstract
1. Introduction
2. Results
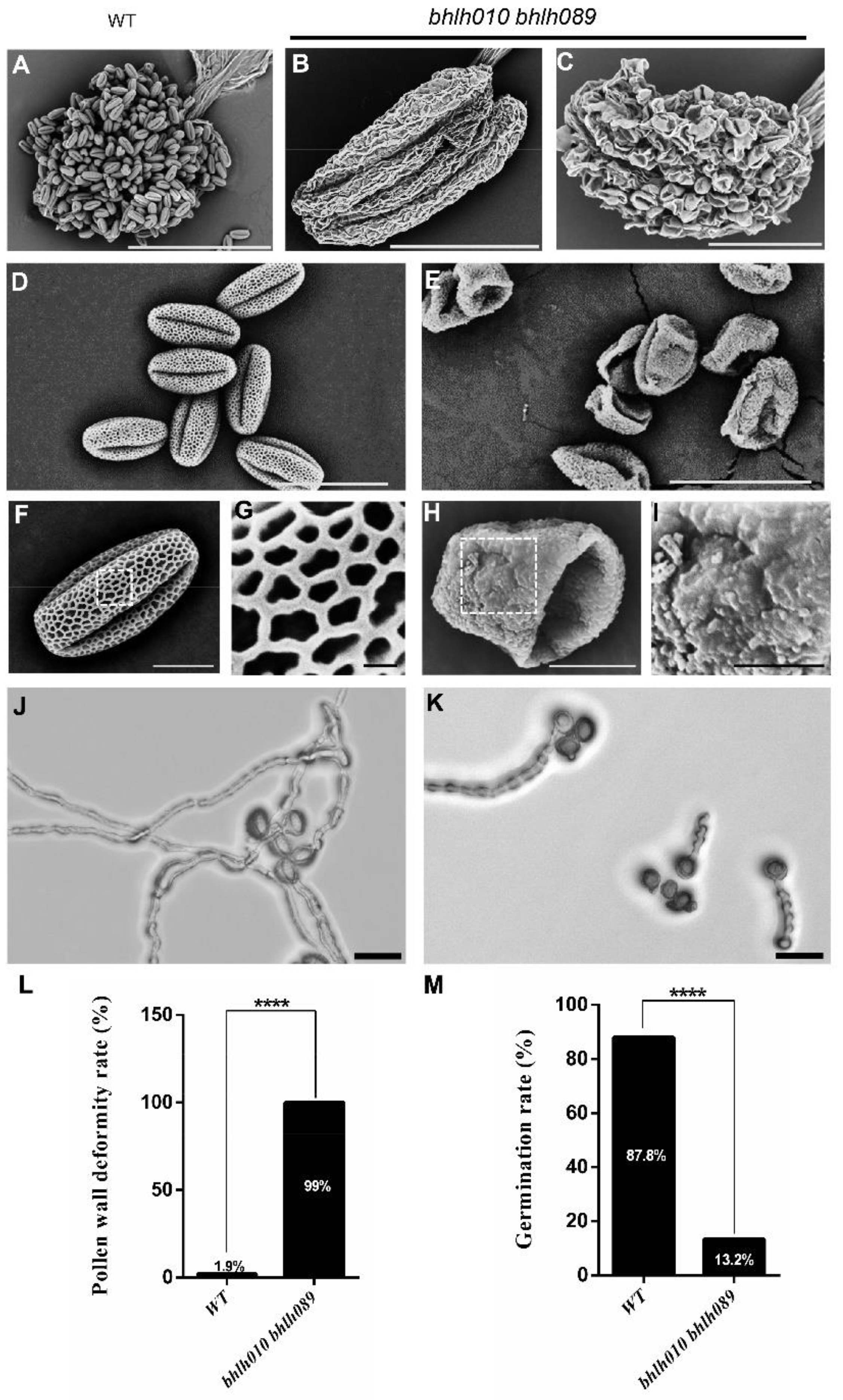
2.1. bhlh010 bhlh089 Pollen Exhibited Collapsed Structure and Severely Reduced Germination Rate
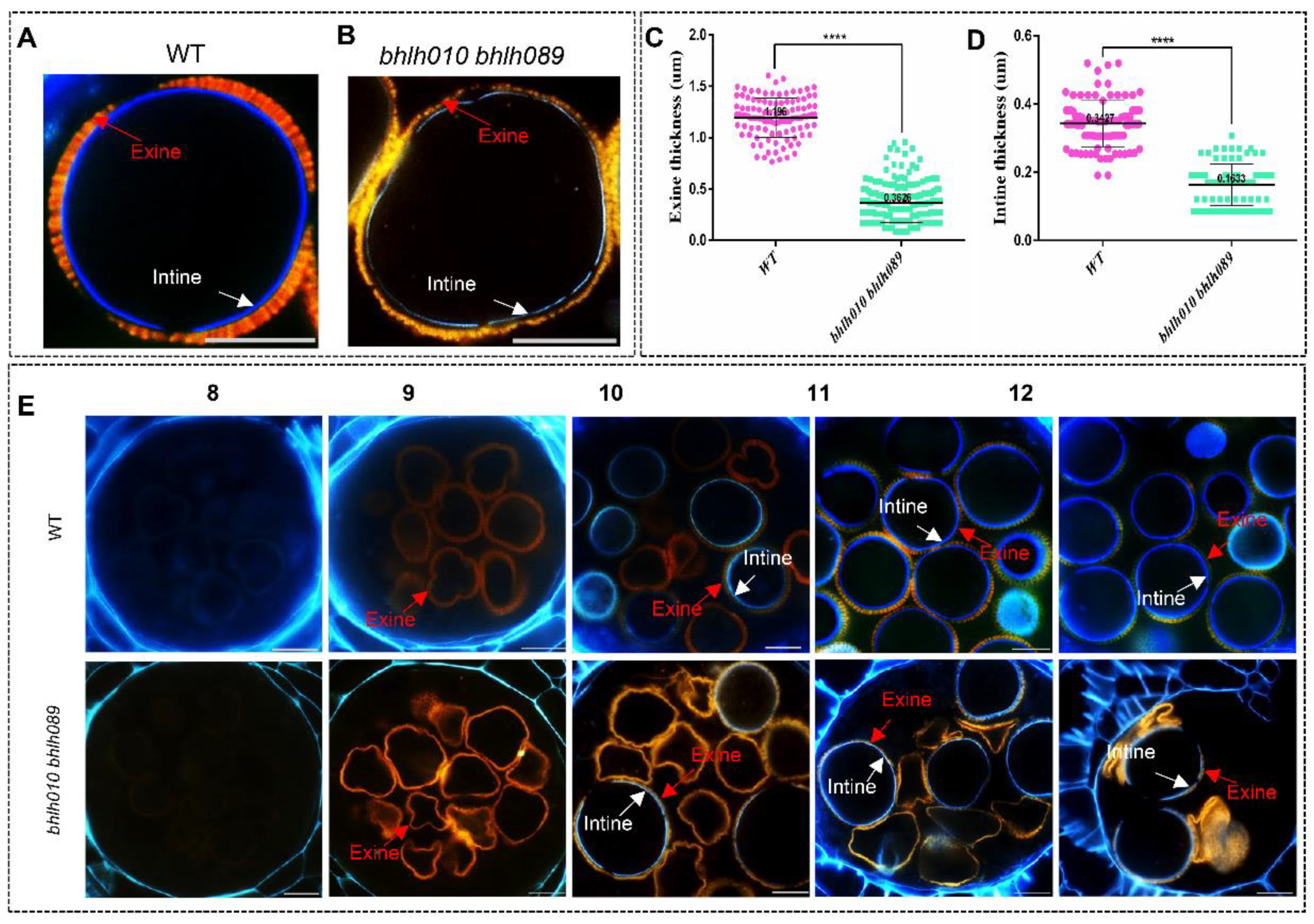
2.2. Both the Exine and Intine Developments Were Abnormal in bhlh010 bhlh089
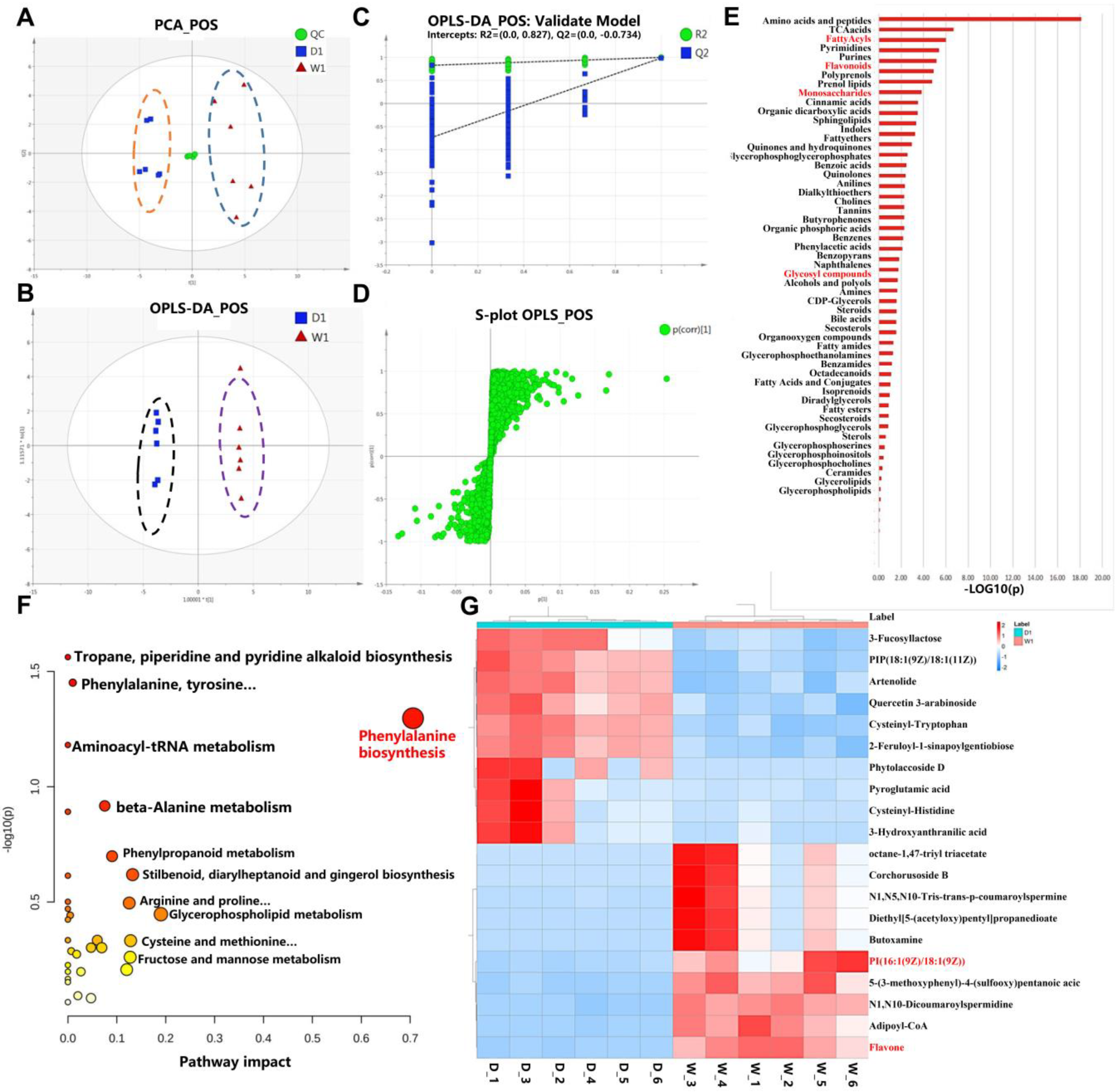
2.3. Metabolomic Comparison of WT and bhlh010 bhlh089 Inflorescences
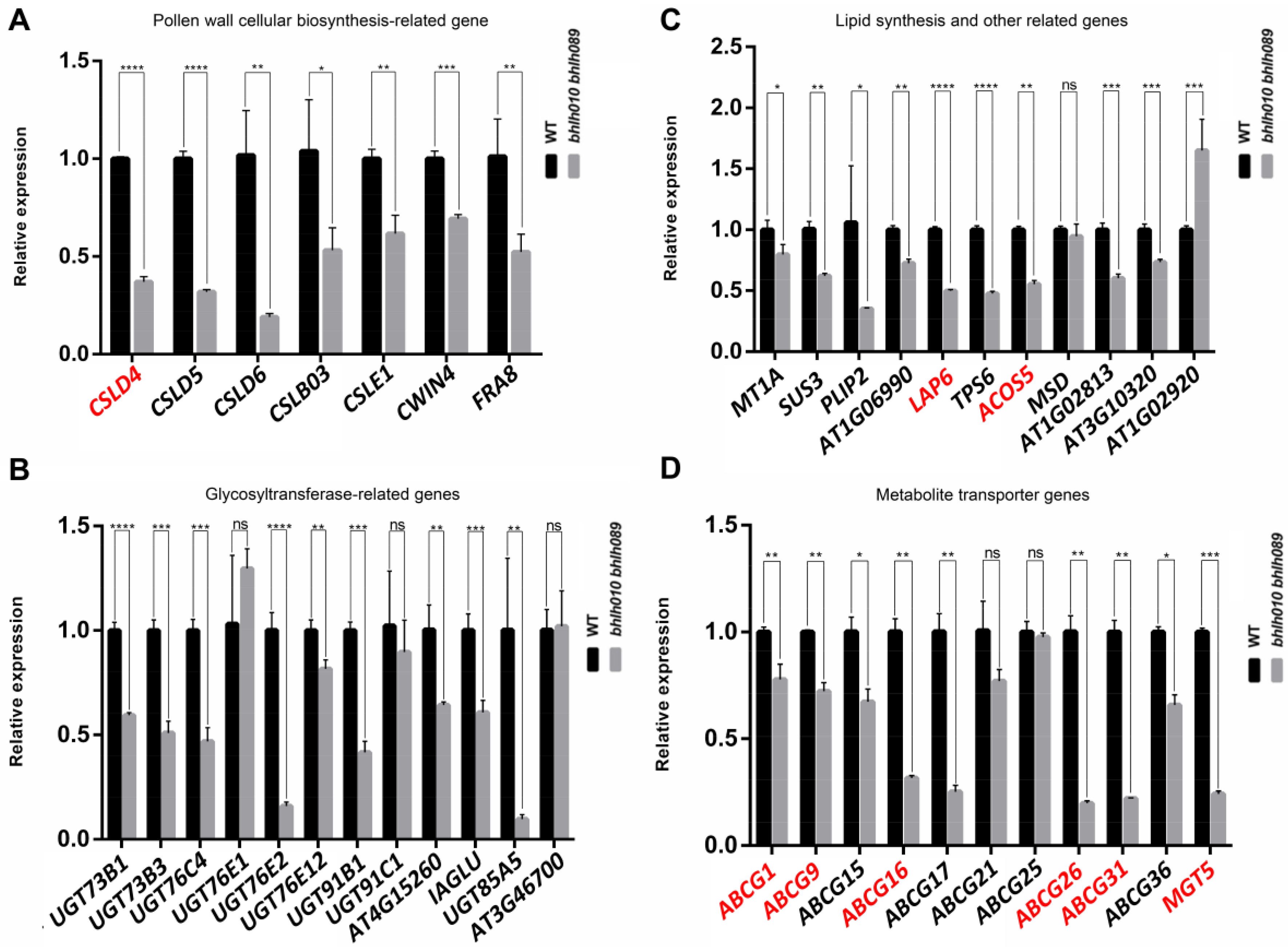
2.4. bHLH010/089 Regulate the Expression of Genes Involved in Metabolic Pathways
2.5. Screening of Genes Activated by bHLH010 or/and bHLH089
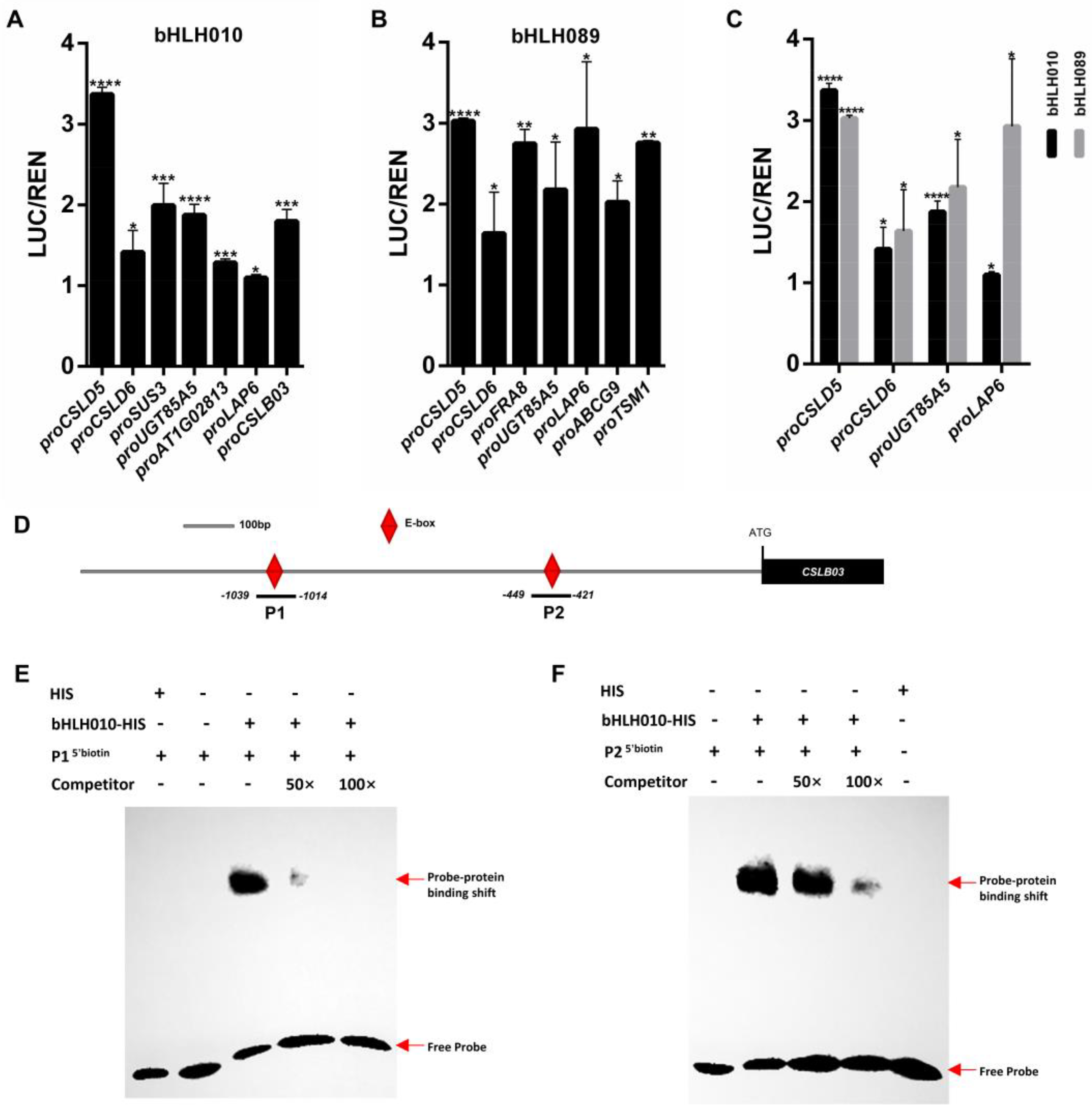
2.6. The CSLB03 Gene Is a Direct Downstream Target of bHLH010
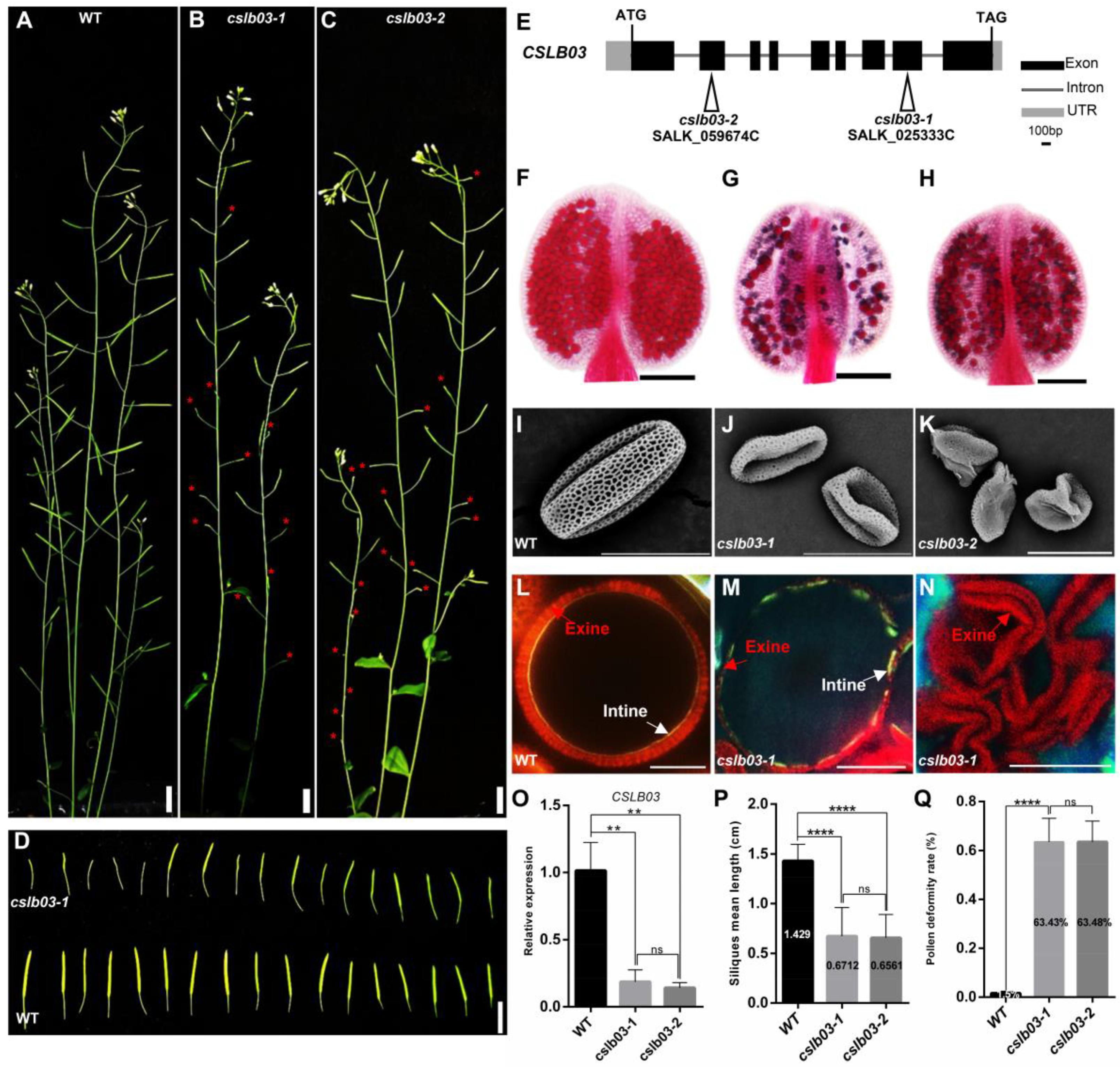
2.7. CSLB03 Gene Is Required for Pollen Wall Development
3. Discussion
3.1. bHLH010 and bHLH089 May Regulate Pollen Wall Formation through the Synthesis of Polysaccharides
3.2. bHLH010 and bHLH089 May Regulate Pollen Wall Formation through Transporters
4. Materials and Methods
4.1. Plant Material
4.2. Generation of probHLH089: β-Glucuronidase (GUS) Transgenic Lines and GUS Staining Assays
4.3. Semithin Section Examination and Pollen Wall Staining
4.4. Scanning Electron Microscopy (SEM)
4.5. Real-Time PCR
4.6. Metabolome Extraction and Detection
4.7. Differential Metabolite Analysis
4.8. Analysis of the bhlh010 bhlh089 Transcriptomic Data
4.9. Combined Metabolome and Transcriptome Analysis
4.10. Dual-Luciferase Reporter Assays
4.11. Electrophoretic Mobility Shift Assay
4.12. In Vitro Pollen Germination Analysis
4.13. Phylogenetic Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zinkl, G.M.; Zwiebel, B.I.; Grier, D.G.; Preuss, D. Pollen-stigma adhesion in Arabidopsis: A species-specific interaction mediated by lipophilic molecules in the pollen exine. Development 1999, 126, 5431–5440. [Google Scholar] [CrossRef] [PubMed]
- Xue, J.S.; Zhang, B.; Zhan, H.; Lv, Y.L.; Jia, X.L.; Wang, T.; Yang, N.Y.; Lou, Y.X.; Zhang, Z.B.; Hu, W.J.; et al. Phenylpropanoid Derivatives Are Essential Components of Sporopollenin in Vascular Plants. Mol. Plant 2020, 13, 1644–1653. [Google Scholar] [CrossRef] [PubMed]
- Heslop-Harrison, J. Wall development within the microspore tetrad of Lilium longiflorum. Can. J. Bot. 1968, 46, 1185–1192. [Google Scholar] [CrossRef]
- Blackmore, S.; Wortley, A.H.; Skvarla, J.J.; Rowley, J.R. Pollen wall development in flowering plants. New Phytol. 2007, 174, 483–498. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Toriyama, K. Genetic Regulation of Sporopollenin Synthesis and Pollen Exine Development. Annu. Rev. Plant Biol. 2011, 62, 437–460. [Google Scholar] [CrossRef] [PubMed]
- Marquez, J.; Seoane-Camba, J.A.; Suarez-Cervera, M. The role of the intine and cytoplasm in the activation and germination processes of Poaceae pollen grains. Grana 1997, 36, 328–342. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Nakamura, S.; Uheda, E.; Nakamura, N. Immunolocalization and possible roles of pectins during pollen growth and callose plug formation in angiosperms. Grana 2000, 39, 46–55. [Google Scholar] [CrossRef]
- Shi, J.; Cui, M.; Yang, L.; Kim, Y.J.; Zhang, D. Genetic and Biochemical Mechanisms of Pollen Wall Development. Trends Plant Sci. 2015, 20, 741–753. [Google Scholar] [CrossRef]
- Paxson-Sowders, D.M.; Dodrill, C.H.; Owen, H.; Makaroff, C. Dex1, a novel plant protein, is required for exine pattern formation during pollen development in Arabidopsis. Plant Physiol. 2001, 127, 1739–1749. [Google Scholar] [CrossRef]
- Ma, L.J.; Yang, Z.N.; Zhang, S. DEX1, a plasma membrane-localized protein, functions in microspore development by affecting CalS5 expression in Arabidopsis thaliana. Chin. Sci. Bull. 2013, 58, 2855–2861. [Google Scholar] [CrossRef][Green Version]
- Ariizumi, T.; Hatakeyama, K.; Hinata, K.; Sato, S.; Kato, T.; Tabata, S.; Toriyama, K. The HKM gene, which is identical to the MS1 gene of Arabidopsis thaliana, is essential for primexine formation and exine pattern formation. Sex. Plant Reprod. 2005, 18, 1–7. [Google Scholar] [CrossRef]
- Guan, Y.F.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Zhang, H.X.; Yang, Z.N. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Hatakeyama, K.; Hinata, K.; Inatsugi, R.; Nishida, I.; Sato, S.; Kato, T.; Tabata, S.; Toriyama, K. Disruption of the novel plant protein NEF1 affects lipid accumulation in the plastids of the tapetum and exine formation of pollen, resulting in male sterility in Arabidopsis thaliana. Plant J. 2004, 39, 170–181. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Kawanabe, T.; Hatakeyama, K.; Sato, S.; Kato, T.; Tabata, S.; Toriyama, K. Ultrastructural characterization of exine development of the transient defective exine 1 mutant suggests the existence of a factor involved in constructing reticulate exine architecture from sporopollenin aggregates. Plant Cell Physiol. 2008, 49, 58–67. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.S.; Zhang, C.; Chang, Y.H.; Zhu, J.; Xu, X.F.; Shi, Z.H.; Zhang, X.L.; Xu, L.; Huang, H.; Zhang, S.; et al. No primexine and plasma membrane undulation is essential for primexine deposition and plasma membrane undulation during microsporogenesis in Arabidopsis. Plant Physiol. 2012, 158, 264–272. [Google Scholar] [CrossRef][Green Version]
- Chen, H.; Zhang, Z.; Ni, E.; Lin, J.; Peng, G.; Huang, J.; Zhu, L.; Deng, L.; Yang, F.; Luo, Q.; et al. HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol. 2020, 225, 2077–2093. [Google Scholar] [CrossRef]
- Huang, J.; Li, Z.; Biener, G.; Xiong, E.; Malik, S.; Eaton, N.; Zhao, C.Z.; Raicu, V.; Kong, H.; Zhao, D. Carbonic anhydrases function in anther cell differentiation downstream of the receptor-like kinase EMS1. Plant Cell 2017, 29, 1335–1356. [Google Scholar] [CrossRef]
- Sun, M.X.; Huang, X.Y.; Yang, J.; Guan, Y.F.; Yang, Z.N. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013, 26, 83–91. [Google Scholar] [CrossRef]
- Hu, J.; Wang, Z.; Zhang, L.; Sun, M.X. The Arabidopsis Exine Formation Defect (EFD) gene is required for primexine patterning and is critical for pollen fertility. New Phytol. 2014, 203, 140–154. [Google Scholar] [CrossRef]
- Vioque, J.; Kolattukudy, P.E. Resolution and purification of an aldehyde-generating and an alcohol-generating fatty acyl-CoA reductase from pea leaves (Pisum sativum L.). Arch. Biochem. Biophys. 1997, 340, 64–72. [Google Scholar] [CrossRef]
- Bernard, A.; Domergue, F.; Pascal, S.; Jetter, R.; Renne, C.; Faure, J.D.; Haslam, R.P.; Napier, J.A.; Lessire, R.; Joubes, J. Reconstitution of plant alkane biosynthesis in yeast demonstrates that Arabidopsis ECERIFERUM1 and ECERIFERUM3 are core components of a very-long-chain alkane synthesis complex. Plant Cell 2012, 24, 3106–3118. [Google Scholar] [CrossRef] [PubMed]
- Ariizumi, T.; Hatakeyama, K.; Hinata, K.; Sato, S.; Kato, T.; Tabata, S.; Toriyama, K. A novel male-sterile mutant of Arabidopsis thaliana, faceless pollen-1, produces pollen with a smooth surface and an acetolysis-sensitive exine. Plant Mol. Biol. 2003, 53, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.H.; Han, M.J.; Lee, D.Y.; Lee, Y.S.; Schreiber, L.; Franke, R.; Faust, A.; Yephremov, A.; Saedler, H.; Kim, Y.W.; et al. Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 2006, 18, 3015–3032. [Google Scholar] [CrossRef] [PubMed]
- de Azevedo Souza, C.; Kim, S.S.; Koch, S.; Kienow, L.; Schneider, K.; McKim, S.M.; Haughn, G.W.; Kombrink, E.; Douglas, C.J. A novel fatty Acyl-CoA Synthetase is required for pollen development and sporopollenin biosynthesis in Arabidopsis. Plant Cell 2009, 21, 507–525. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Pinot, F.; Sauveplane, V.; Werck-Reichhart, D.; Diehl, P.; Schreiber, L.; Franke, R.; Zhang, P.; Chen, L.; Gao, Y.; et al. Cytochrome P450 family member CYP704B2 catalyzes the {omega}-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 2010, 22, 173–190. [Google Scholar] [CrossRef]
- Chen, W.; Yu, X.H.; Zhang, K.; Shi, J.; De Oliveira, S.; Schreiber, L.; Shanklin, J.; Zhang, D. Male Sterile2 encodes a plastid-localized fatty acyl carrier protein reductase required for pollen exine development in Arabidopsis. Plant Physiol. 2011, 157, 842–853. [Google Scholar] [CrossRef]
- Ni, E.; Zhou, L.; Li, J.; Jiang, D.; Wang, Z.; Zheng, S.; Qi, H.; Zhou, Y.; Wang, C.; Xiao, S.; et al. OsCER1 Plays a Pivotal Role in Very-Long-Chain Alkane Biosynthesis and Affects Plastid Development and Programmed Cell Death of Tapetum in Rice (Oryza sativa L.). Front. Plant Sci. 2018, 9, 1217–1226. [Google Scholar] [CrossRef]
- Xiong, S.X.; Lu, J.Y.; Lou, Y.; Teng, X.D.; Gu, J.N.; Zhang, C.; Shi, Q.S.; Yang, Z.N.; Zhu, J. The transcription factors MS188 and AMS form a complex to activate the expression of CYP703A2 for sporopollenin biosynthesis in Arabidopsis thaliana. Plant J. 2016, 88, 936–946. [Google Scholar] [CrossRef]
- Xu, F.; Zheng, L.; Yang, Z.; Zhang, S. Arabidopsis ECERIFERUM3 (CER3) Functions to Maintain Hydration for Pollen–Stigma Recognition During Fertilization. J. Plant Biol. 2020, 63, 347–359. [Google Scholar] [CrossRef]
- Wang, K.; Guo, Z.L.; Zhou, W.T.; Zhang, C.; Zhang, Z.Y.; Lou, Y.; Xiong, S.X.; Yao, X.Z.; Fan, J.J.; Zhu, J.; et al. The Regulation of Sporopollenin Biosynthesis Genes for Rapid Pollen Wall Formation. Plant Physiol. 2018, 178, 283–294. [Google Scholar] [CrossRef]
- Xie, H.H.; Chen, L.; Xu, F.Q.; Guo, W.S.; Wang, S.; Yang, Z.N.; Zhang, S. ACOS5 is Required for Primexine Formation and Exine Pattern Formation During Microsporogenesis in Arabidopsis. J. Plant Biol. 2017, 60, 404–412. [Google Scholar] [CrossRef]
- Kim, S.S.; Grienenberger, E.; Lallemand, B.; Colpitts, C.C.; Kim, S.Y.; Souza, C.d.A.; Geoffroy, P.; Heintz, D.; Krahn, D.; Kaiser, M.; et al. LAP6/POLYKETIDE SYNTHASE A and LAP5/POLYKETIDE SYNTHASE B Encode Hydroxyalkyl α-Pyrone Synthases Required for Pollen Development and Sporopollenin Biosynthesis in Arabidopsis thaliana. Plant Cell 2010, 22, 4045–4066. [Google Scholar] [CrossRef] [PubMed]
- Pighin, J.A.; Zheng, H.; Balakshin, L.J.; Goodman, I.P.; Western, T.L.; Jetter, R.; Kunst, L.; Samuels, A.L. Plant cuticular lipid export requires an ABC transporter. Science 2004, 306, 702–704. [Google Scholar] [CrossRef] [PubMed]
- Bird, D.; Beisson, F.; Brigham, A.; Shin, J.; Greer, S.; Jetter, R.; Kunst, L.; Wu, X.; Yephremov, A.; Samuels, L. Characterization of Arabidopsis ABCG11/WBC11, an ATP binding cassette (ABC) transporter that is required for cuticular lipid secretion. Plant J. 2007, 52, 485–498. [Google Scholar] [CrossRef]
- Choi, H.; Jin, J.Y.; Choi, S.; Hwang, J.U.; Kim, Y.Y.; Suh, M.C.; Lee, Y. An ABCG/WBC-type ABC transporter is essential for transport of sporopollenin precursors for exine formation in developing pollen. Plant J. 2011, 65, 181–193. [Google Scholar] [CrossRef]
- Qin, P.; Tu, B.; Wang, Y.; Deng, L.; Quilichini, T.D.; Li, T.; Wang, H.; Ma, B.; Li, S. ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol. 2013, 54, 138–154. [Google Scholar] [CrossRef]
- Kuromori, T.; Ito, T.; Sugimoto, E.; Shinozaki, K. Arabidopsis mutant of AtABCG26, an ABC transporter gene, is defective in pollen maturation. J. Plant Physiol. 2011, 168, 2001–2005. [Google Scholar] [CrossRef]
- Chang, Z.; Chen, Z.; Yan, W.; Xie, G.; Lu, J.; Wang, N.; Lu, Q.; Yao, N.; Yang, G.; Xia, J.; et al. An ABC transporter, OsABCG26, is required for anther cuticle and pollen exine formation and pollen-pistil interactions in rice. Plant Sci. 2016, 253, 21–30. [Google Scholar] [CrossRef]
- Quilichini, T.D.; Samuels, A.L.; Douglas, C.J. ABCG26-mediated polyketide trafficking and hydroxycinnamoyl spermidines contribute to pollen wall exine formation in Arabidopsis. Plant Cell 2014, 26, 4483–4498. [Google Scholar] [CrossRef]
- Ukitsu, H.; Kuromori, T.; Toyooka, K.; Goto, Y.; Matsuoka, K.; Sakuradani, E.; Shimizu, S.; Kamiya, A.; Imura, Y.; Yuguchi, M.; et al. Cytological and biochemical analysis of COF1, an Arabidopsis mutant of an ABC transporter gene. Plant Cell Physiol. 2007, 48, 1524–1533. [Google Scholar] [CrossRef]
- Dou, X.Y.; Yang, K.Z.; Zhang, Y.; Wang, W.; Liu, X.L.; Chen, L.Q.; Zhang, X.Q.; Ye, D. WBC27, an adenosine tri-phosphate-binding cassette protein, controls pollen wall formation and patterning in Arabidopsis. J. Integr. Plant Biol. 2011, 53, 74–88. [Google Scholar] [CrossRef] [PubMed]
- Quilichini, T.D.; Friedmann, M.C.; Samuels, A.L.; Douglas, C.J. ATP-binding cassette transporter G26 is required for male fertility and pollen exine formation in Arabidopsis. Plant Physiol. 2010, 154, 678–690. [Google Scholar] [CrossRef] [PubMed]
- Schnurr, J.A.; Storey, K.K.; Jung, H.J.; Somers, D.A.; Gronwald, J.W. UDP-sugar pyrophosphorylase is essential for pollen development in Arabidopsis. Planta 2006, 224, 520–532. [Google Scholar] [CrossRef] [PubMed]
- Drakakaki, G.; Zabotina, O.; Delgado, I.; Robert, S.; Keegstra, K.; Raikhel, N. Arabidopsis reversibly glycosylated polypeptides 1 and 2 are essential for pollen development. Plant Physiol. 2006, 142, 1480–1492. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Yu, M.; Geng, L.L.; Zhao, J. The fasciclin-like arabinogalactan protein gene, FLA3, is involved in microspore development of Arabidopsis. Plant J. 2010, 64, 482–497. [Google Scholar] [CrossRef] [PubMed]
- Han, M.J.; Jung, K.H.; Yi, G.; Lee, D.Y.; An, G. Rice Immature Pollen 1 (RIP1) is a regulator of late pollen development. Plant Cell Physiol. 2006, 47, 1457–1472. [Google Scholar] [CrossRef]
- Moon, S.; Kim, S.R.; Zhao, G.; Yi, J.; Yoo, Y.; Jin, P.; Lee, S.W.; Jung, K.H.; Zhang, D.; An, G. Rice glycosyltransferase1 encodes a glycosyltransferase essential for pollen wall formation. Plant Physiol. 2013, 161, 663–675. [Google Scholar] [CrossRef]
- Persson, S.; Paredez, A.; Carroll, A.; Palsdottir, H.; Doblin, M.; Poindexter, P.; Khitrov, N.; Auer, M.; Somerville, C.R. Genetic evidence for three unique components in primary cell-wall cellulose synthase complexes in Arabidopsis. Proc. Natl. Acad. Sci. USA 2007, 104, 15566–15571. [Google Scholar] [CrossRef]
- Kotake, T.; Hojo, S.; Yamaguchi, D.; Aohara, T.; Konishi, T.; Tsumuraya, Y. Properties and physiological functions of UDP-sugar pyrophosphorylase in Arabidopsis. Biosci. Biotechnol. Biochem. 2007, 71, 761–771. [Google Scholar] [CrossRef]
- Sumiyoshi, M.; Inamura, T.; Nakamura, A.; Aohara, T.; Ishii, T.; Satoh, S.; Iwai, H. UDP-arabinopyranose mutase 3 is required for pollen wall morphogenesis in rice (Oryza sativa). Plant Cell Physiol. 2015, 56, 232–241. [Google Scholar] [CrossRef]
- Yang, K.; Zhou, X.; Wang, Y.; Feng, H.; Ren, X.; Liu, H.; Liu, W. Carbohydrate metabolism and gene regulation during anther development in an androdioecious tree, Tapiscia sinensis. Ann. Bot. 2017, 120, 967–977. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.; Khare, D.; Kang, J.; Hwang, J.U.; Liang, W.; Martinoia, E.; Zhang, D.; Kang, B.; Lee, Y. Postmeiotic development of pollen surface layers requires two Arabidopsis ABCG-type transporters. Plant Cell Rep. 2016, 35, 1863–1873. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Moon, S.; Lee, Y.S.; Zhu, L.; Liang, W.; Zhang, D.; Jung, K.H.; An, G. Defective Tapetum Cell Death 1 (DTC1) Regulates ROS Levels by Binding to Metallothionein during Tapetum Degeneration. Plant Physiol. 2016, 170, 1611–1623. [Google Scholar] [CrossRef]
- Yi, J.; Kim, S.R.; Lee, D.Y.; Moon, S.; Lee, Y.S.; Jung, K.H.; Hwang, I.; An, G. The rice gene DEFECTIVE TAPETUM and MEIOCYTES 1 (DTM1) is required for early tapetum development and meiosis. Plant J. 2012, 70, 256–270. [Google Scholar] [CrossRef]
- Nugent, J.M.; Byrne, T.; McCormack, G.; Quiwa, M.; Stafford, E. Progressive programmed cell death inwards across the anther wall in male sterile flowers of the gynodioecious plant Plantago lanceolata. Planta 2019, 249, 913–923. [Google Scholar] [CrossRef] [PubMed]
- Flores-Rentería, L.; Orozco-Arroyo, G.; Cruz-García, F.; García-Campusano, F.; Alfaro, I.; Vázquez-Santana, S. Programmed cell death promotes male sterility in the functional dioecious Opuntia stenopetala (Cactaceae). Ann. Bot. 2013, 112, 789–800. [Google Scholar] [CrossRef]
- Çetinbaş-Genç, A.; Vardar, F. Programmed cell death in plant reproductive development. In Recent Advances in Plant Science; Nova Science Publishers: Hauppauge, NY, USA, 2020; pp. 341–361. [Google Scholar]
- Hernández-Cruz, R.; Barrón-Pacheco, F.; Sánchez, D.; Arias, S.; Vázquez-Santana, S. Functional dioecy in Echinocereus: Ontogenetic patterns, programmed cell death, and evolutionary significance. Int. J. Plant Sci. 2018, 179, 257–274. [Google Scholar] [CrossRef]
- Wu, H.-m.; Cheung, A.Y. Programmed cell death in plant reproduction. Plant Mol. Biol. 2000, 44, 267–281. [Google Scholar] [CrossRef]
- Yang, X.; Liang, W.; Chen, M.; Zhang, D.; Zhao, X.; Shi, J. Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility. Planta 2017, 246, 105–122. [Google Scholar] [CrossRef]
- Vardar, F.; Ünal, M. Ultrastructural aspects and programmed cell death in the tapetal cells of Lathyrus undulatus Boiss. Acta Biol. Hung. 2012, 63, 52–66. [Google Scholar] [CrossRef]
- Solís, M.T.; Chakrabarti, N.; Corredor, E.; Cortés-Eslava, J.; Rodríguez-Serrano, M.; Biggiogera, M.; Risueño, M.C.; Testillano, P.S. Epigenetic changes accompany developmental programmed cell death in tapetum cells. Plant Cell Physiol. 2014, 55, 16–29. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Zhou, Z.; Tang, S.; Zhang, Z.; Xia, S.; Qin, M.; Li, B.; Wen, J.; Yi, B.; Shen, J.; et al. Ectopic expression of BnaC.CP20.1 results in premature tapetal programmed cell death in Arabidopsis. Plant Cell Physiol. 2016, 57, 1972–1984. [Google Scholar] [CrossRef] [PubMed]
- Van Hautegem, T.; Waters, A.J.; Goodrich, J.; Nowack, M.K. Only in dying, life: Programmed cell death during plant development. Trends Plant Sci. 2015, 20, 102–113. [Google Scholar] [CrossRef]
- Vizcay-Barrena, G.; Wilson, Z.A. Altered tapetal PCD and pollen wall development in the Arabidopsis ms1 mutant. J. Exp. Bot. 2006, 57, 2709–2717. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Xiang, X.; Yang, Z.; Yu, P.; Wen, X.; Wang, H.; Abbas, A.; Khan, R.M.; Zhang, Y.; Cheng, S.; et al. OsGPAT3 plays a critical role in anther wall programmed cell death and pollen development in rice. Int. J. Mol. Sci. 2018, 19, 4017. [Google Scholar] [CrossRef] [PubMed]
- Phan, H.A.; Iacuone, S.; Li, S.F.; Parish, R.W. The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 2011, 23, 2209–2224. [Google Scholar] [CrossRef] [PubMed]
- Wilson, Z.A.; Morroll, S.M.; Dawson, J.; Swarup, R.; Tighe, P.J. The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 2001, 28, 27–39. [Google Scholar] [CrossRef]
- Lu, J.-Y.; Xiong, S.-X.; Yin, W.; Teng, X.-D.; Lou, Y.; Zhu, J.; Zhang, C.; Gu, J.-N.; Wilson, Z.A.; Yang, Z.-N.; et al. MS1, a direct target of MS188, regulates the expression of key sporophytic pollen coat protein genes in Arabidopsis. J. Exp. Bot. 2020, 71, 4877–4889. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Shen, Y.; Zhang, Z.; Jia, Q.; Xu, M.; Zhang, T.; Fang, H.; Yu, X.; Li, L.; Liu, D.; et al. A GPAT1 Mutation in Arabidopsis Enhances Plant Height but Impairs Seed Oil Biosynthesis. Int. J. Mol. Sci. 2021, 22, 785. [Google Scholar] [CrossRef]
- Feng, B.; Lu, D.; Ma, X.; Peng, Y.; Sun, Y.; Ning, G.; Ma, H. Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 2012, 72, 612–624. [Google Scholar] [CrossRef]
- Cui, J.; You, C.; Zhu, E.; Huang, Q.; Ma, H.; Chang, F. Feedback Regulation of DYT1 by Interactions with Downstream bHLH Factors Promotes DYT1 Nuclear Localization and Anther Development. Plant Cell 2016, 28, 1078–1093. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, H.; Shi, Q.L.; Jiang, H.; Chen, H.; Zhong, X.L.; Gao, J.F.; Cui, Y.L.; Yang, Z.-N. The Arabidopsis bHLH transcription factor DYT1 is essential for anther development by regulating callose dissolution. J. Shanghai Jiao Tong Univ. 2009, 38, 174–182. [Google Scholar]
- Ferjentsikova, I.; Wilson, Z. AMS function during late pollen meiosis and subsequent pollen wall formation. Dev. Biol. 2011, 356, 194. [Google Scholar] [CrossRef][Green Version]
- Zhu, E.; You, C.; Wang, S.; Cui, J.; Niu, B.; Wang, Y.; Qi, J.; Ma, H.; Chang, F. The DYT1-interacting proteins bHLH010, bHLH089 and bHLH091 are redundantly required for Arabidopsis anther development and transcriptome. Plant J. 2015, 83, 976–990. [Google Scholar] [CrossRef]
- Gu, J.N.; Zhu, J.; Yu, Y.; Teng, X.D.; Lou, Y.; Xu, X.F.; Liu, J.L.; Yang, Z.N. DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 2014, 80, 1005–1013. [Google Scholar] [CrossRef]
- Weterings, P.M.S.A.Q.B.K.; Hsu, K.N.M.Y.-C.; Beals, P.Y.L.M.T.T.T.P.; Goldberg, R.B. Anther developmental defects in Arabidopsis thaliana male-sterile mutants. Sex. Plant Reprod. 1999, 11, 297–322. [Google Scholar]
- Koltunow, A.M.; Truettner, J.; Cox, K.H.; Wallroth, M.; Goldberg, R.B. Different Temporal and Spatial Gene Expression Patterns Occur during Anther Development. Plant Cell 1990, 2, 1201–1224. [Google Scholar] [CrossRef]
- Fu, Y.; Li, M.; Zhang, S.; Yang, Q.; Zhu, E.; You, C.; Qi, J.; Ma, H.; Chang, F. Analyses of functional conservation and divergence reveal requirement of bHLH010/089/091 for pollen development at elevated temperature in Arabidopsis. J. Genet. Genom. 2020, 47, 477–492. [Google Scholar] [CrossRef]
- Wang, W.; Wang, L.; Chen, C.; Xiong, G.; Tan, X.Y.; Yang, K.Z.; Wang, Z.C.; Zhou, Y.; Ye, D.; Chen, L.Q. Arabidopsis CSLD1 and CSLD4 are required for cellulose deposition and normal growth of pollen tubes. J. Exp. Bot. 2011, 62, 5161–5177. [Google Scholar] [CrossRef]
- Dobritsa, A.; Lei, Z.; Nishikawa, S.-I.; Urbanczyk-Wochniak, E.; Huhman, D.; Preuss, D.; Sumner, L. LAP5 and LAP6 Encode Anther-Specific Proteins with Similarity to Chalcone Synthase Essential for Pollen Exine Development in Arabidopsis. Plant Physiol. 2010, 153, 937–955. [Google Scholar] [CrossRef]
- Langlois-Meurinne, M.; Gachon, C.M.; Saindrenan, P. Pathogen-responsive expression of glycosyltransferase genes UGT73B3 and UGT73B5 is necessary for resistance to Pseudomonas syringae pv tomato in Arabidopsis. Plant Physiol. 2005, 139, 1890–1901. [Google Scholar] [CrossRef] [PubMed]
- Haroth, S.; Feussner, K.; Kelly, A.A.; Zienkiewicz, K.; Shaikhqasem, A.; Herrfurth, C.; Feussner, I. The glycosyltransferase UGT76E1 significantly contributes to 12-O-glucopyranosyl-jasmonic acid formation in wounded Arabidopsis thaliana leaves. J. Biol. Chem. 2019, 294, 9858–9872. [Google Scholar] [CrossRef] [PubMed]
- Ruhlmann, J.M.; Kram, B.W.; Carter, C.J. CELL WALL INVERTASE 4 is required for nectar production in Arabidopsis. J. Exp. Bot. 2010, 61, 395–404. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Foster, R.; Ma, S.; Liao, S.J.; Bliss, S.; Kartika, D.; Wang, L.; Wu, L.; Eamens, A.L.; Ruan, Y.L. Identification of transcription factors controlling cell wall invertase gene expression for reproductive development via bioinformatic and transgenic analyses. Plant J. 2021, 106, 1058–1074. [Google Scholar] [CrossRef] [PubMed]
- Gu, F.; Bringmann, M.; Combs, J.R.; Yang, J.; Bergmann, D.C.; Nielsen, E. Arabidopsis CSLD5 Functions in Cell Plate Formation in a Cell Cycle-Dependent Manner. Plant Cell 2016, 28, 1722–1737. [Google Scholar] [CrossRef]
- Yin, L.; Verhertbruggen, Y.; Oikawa, A.; Manisseri, C.; Knierim, B.; Prak, L.; Jensen, J.K.; Knox, J.P.; Auer, M.; Willats, W.G.; et al. The cooperative activities of CSLD2, CSLD3, and CSLD5 are required for normal Arabidopsis development. Mol. Plant 2011, 4, 1024–1037. [Google Scholar] [CrossRef]
- Hong, Z.L.; Zhang, Z.M.; Olson, J.M.; Verma, D.P.S. A novel UDP-glucose transferase is part of the callose synthase complex and interacts with phragmoplastin at the forming cell plate. Plant Cell 2001, 13, 769–779. [Google Scholar] [CrossRef]
- Liu, L.; Zhang, F.; Li, G.; Wang, G. Qualitative and Quantitative NAD(+) Metabolomics Lead to Discovery of Multiple Functional Nicotinate N-Glycosyltransferase in Arabidopsis. Front. Plant Sci. 2019, 10, 1164–1173. [Google Scholar] [CrossRef]
- Messner, B.; Thulke, O.; Schaffner, A.R. Arabidopsis glucosyltransferases with activities toward both endogenous and xenobiotic substrates. Planta 2003, 217, 138–146. [Google Scholar] [CrossRef]
- Wang, K.; Guo, Q.; Froehlich, J.E.; Hersh, H.L.; Zienkiewicz, A.; Howe, G.A.; Benning, C. Two Abscisic Acid-Responsive Plastid Lipase Genes Involved in Jasmonic Acid Biosynthesis in Arabidopsis thaliana. Plant Cell 2018, 30, 1006–1022. [Google Scholar] [CrossRef]
- Lai, C.P.; Huang, L.M.; Chen, L.O.; Chan, M.T.; Shaw, J.F. Genome-wide analysis of GDSL-type esterases/lipases in Arabidopsis. Plant Mol. Biol. 2017, 95, 181–197. [Google Scholar] [CrossRef] [PubMed]
- Cabib, E.; Leloir, L.F. The biosynthesis of trehalose phosphate. J. Biol. Chem. 1958, 231, 259–275. [Google Scholar] [CrossRef]
- Heslop-Harrison, J. Structural and functional variation in pollen intines. Pollen Spores 1991, 222, 331–343. [Google Scholar]
- Castells, T.; Seoane-Camba, J.A.; Suaez-Cervera, M. Intine wall modifications during germination of Zygophyllum fabago (Zygophyllaceae) pollen grains. Can. J. Bot. 2003, 81, 1269–1279. [Google Scholar] [CrossRef]
- Suárez-Cervera, M.; Arcalís, E.; Le Thomas, A.; Seoane-Camba, J. Pectin distribution pattern in the apertural intine of Euphorbia peplus L. (Euphorbiaceae) pollen. Sex. Plant Reprod. 2002, 14, 291–298. [Google Scholar] [CrossRef]
- Dardelle, F.; Lehner, A.; Ramdani, Y.; Bardor, M.; Lerouge, P.; Driouich, A.; Mollet, J.C. Biochemical and immunocytological characterizations of Arabidopsis pollen tube cell wall. Plant Physiol. 2010, 153, 1563–1576. [Google Scholar] [CrossRef]
- Yin, Y.; Johns, M.A.; Cao, H.; Rupani, M. A survey of plant and algal genomes and transcriptomes reveals new insights into the evolution and function of the cellulose synthase superfamily. BMC Genom. 2014, 15, 260. [Google Scholar] [CrossRef]
- Bernal, A.J.; Yoo, C.M.; Mutwil, M.; Jensen, J.K.; Hou, G.; Blaukopf, C.; Sorensen, I.; Blancaflor, E.B.; Scheller, H.V.; Willats, W.G. Functional analysis of the cellulose synthase-like genes CSLD1, CSLD2, and CSLD4 in tip-growing Arabidopsis cells. Plant Physiol. 2008, 148, 1238–1253. [Google Scholar] [CrossRef]
- Konishi, T.; Ohmiya, Y.; Hayashi, T. Evidence that sucrose loaded into the phloem of a poplar leaf is used directly by sucrose synthase associated with various beta-glucan synthases in the stem. Plant Physiol. 2004, 134, 1146–1152. [Google Scholar] [CrossRef][Green Version]
- Owens, D.K.; McIntosh, C.A. Identification, recombinant expression, and biochemical characterization of a flavonol 3-O-glucosyltransferase clone from Citrus paradisi. Phytochemistry 2009, 70, 1382–1391. [Google Scholar] [CrossRef]
- Tomkins, J.; Peterson, D.; Yang, T.-J.; Main, D.; Lee, L.; Henry, R.; Wing, R. Development of genomic resources for grape: BAC library construction preliminary STC analysis and identification of clones associated with flavonoid and stilbene biosynthesis. In Centre for Plant Conservation Genetics Papers; FAO: Rome, Italy, 2022. [Google Scholar]
- Ramon, M.; De Smet, I.; Vandesteene, L.; Naudts, M.; Leyman, B.; Van Dijck, P.; Rolland, F.; Beeckman, T.; Thevelein, J.M. Extensive expression regulation and lack of heterologous enzymatic activity of the Class II trehalose metabolism proteins from Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1015–1032. [Google Scholar] [CrossRef] [PubMed]
- Chary, S.N.; Hicks, G.R.; Choi, Y.G.; Carter, D.; Raikhel, N.V. Trehalose-6-phosphate synthase/phosphatase regulates cell shape and plant architecture in Arabidopsis. Plant Physiol. 2008, 146, 97–107. [Google Scholar] [CrossRef] [PubMed]
- Decottignies, A.; Goffeau, A. Complete inventory of the yeast ABC proteins. Nat. Genet. 1997, 15, 137–145. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhao, L.; Chen, P.; Cai, H.; Hou, Z.; Jin, X.; Aslam, M.; Chai, M.; Lai, L.; He, Q.; et al. ATP binding cassette transporters ABCG1 and ABCG16 affect reproductive development via auxin signalling in Arabidopsis. Plant J. 2020, 102, 1172–1186. [Google Scholar] [CrossRef]
- Choi, H.; Ohyama, K.; Kim, Y.Y.; Jin, J.Y.; Lee, S.B.; Yamaoka, Y.; Muranaka, T.; Suh, M.C.; Fujioka, S.; Lee, Y. The role of Arabidopsis ABCG9 and ABCG31 ATP binding cassette transporters in pollen fitness and the deposition of steryl glycosides on the pollen coat. Plant Cell 2014, 26, 310–324. [Google Scholar] [CrossRef]
- Campe, R.; Langenbach, C.; Leissing, F.; Popescu, G.V.; Popescu, S.C.; Goellner, K.; Beckers, G.J.; Conrath, U. ABC transporter PEN3/PDR8/ABCG36 interacts with calmodulin that, like PEN3, is required for Arabidopsis nonhost resistance. New Phytol. 2016, 209, 294–306. [Google Scholar] [CrossRef]
- Niu, B.X.; He, F.R.; He, M.; Ren, D.; Chen, L.T.; Liu, Y.G. The ATP-binding cassette transporter OsABCG15 is required for anther development and pollen fertility in rice. J. Integr. Plant Biol. 2013, 55, 710–720. [Google Scholar] [CrossRef]
- Li, L.; Tutone, A.F.; Drummond, R.S.; Gardner, R.C.; Luan, S. A novel family of magnesium transport genes in Arabidopsis. Plant Cell 2001, 13, 2761–2775. [Google Scholar] [CrossRef]
- Xu, X.F.; Wang, B.; Lou, Y.; Han, W.J.; Lu, J.Y.; Li, D.D.; Li, L.G.; Zhu, J.; Yang, Z.N. Magnesium Transporter 5 plays an important role in Mg transport for male gametophyte development in Arabidopsis. Plant J. 2015, 84, 925–936. [Google Scholar] [CrossRef]
- Li, L.G.; Sokolov, L.N.; Yang, Y.H.; Li, D.P.; Ting, J.; Pandy, G.K.; Luan, S. A mitochondrial magnesium transporter functions in Arabidopsis pollen development. Mol. Plant 2008, 1, 675–685. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef] [PubMed]
- Rozewicki, J.; Li, S.; Amada, K.M.; Standley, D.M.; Katoh, K. MAFFT-DASH: Integrated protein sequence and structural alignment. Nucleic Acids Res. 2019, 47, W5–W10. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lai, Z.; Wang, J.; Peng, S.-Q.; Chang, F. bHLH010/089 Transcription Factors Control Pollen Wall Development via Specific Transcriptional and Metabolic Networks in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 11683. https://doi.org/10.3390/ijms231911683
Lai Z, Wang J, Peng S-Q, Chang F. bHLH010/089 Transcription Factors Control Pollen Wall Development via Specific Transcriptional and Metabolic Networks in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(19):11683. https://doi.org/10.3390/ijms231911683
Chicago/Turabian StyleLai, Zesen, Jianzheng Wang, Shi-Qing Peng, and Fang Chang. 2022. "bHLH010/089 Transcription Factors Control Pollen Wall Development via Specific Transcriptional and Metabolic Networks in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 19: 11683. https://doi.org/10.3390/ijms231911683
APA StyleLai, Z., Wang, J., Peng, S.-Q., & Chang, F. (2022). bHLH010/089 Transcription Factors Control Pollen Wall Development via Specific Transcriptional and Metabolic Networks in Arabidopsis thaliana. International Journal of Molecular Sciences, 23(19), 11683. https://doi.org/10.3390/ijms231911683





