Cellular Lactate Spectroscopy Using 1.5 Tesla Clinical Apparatus
Abstract
1. Introduction
2. Results
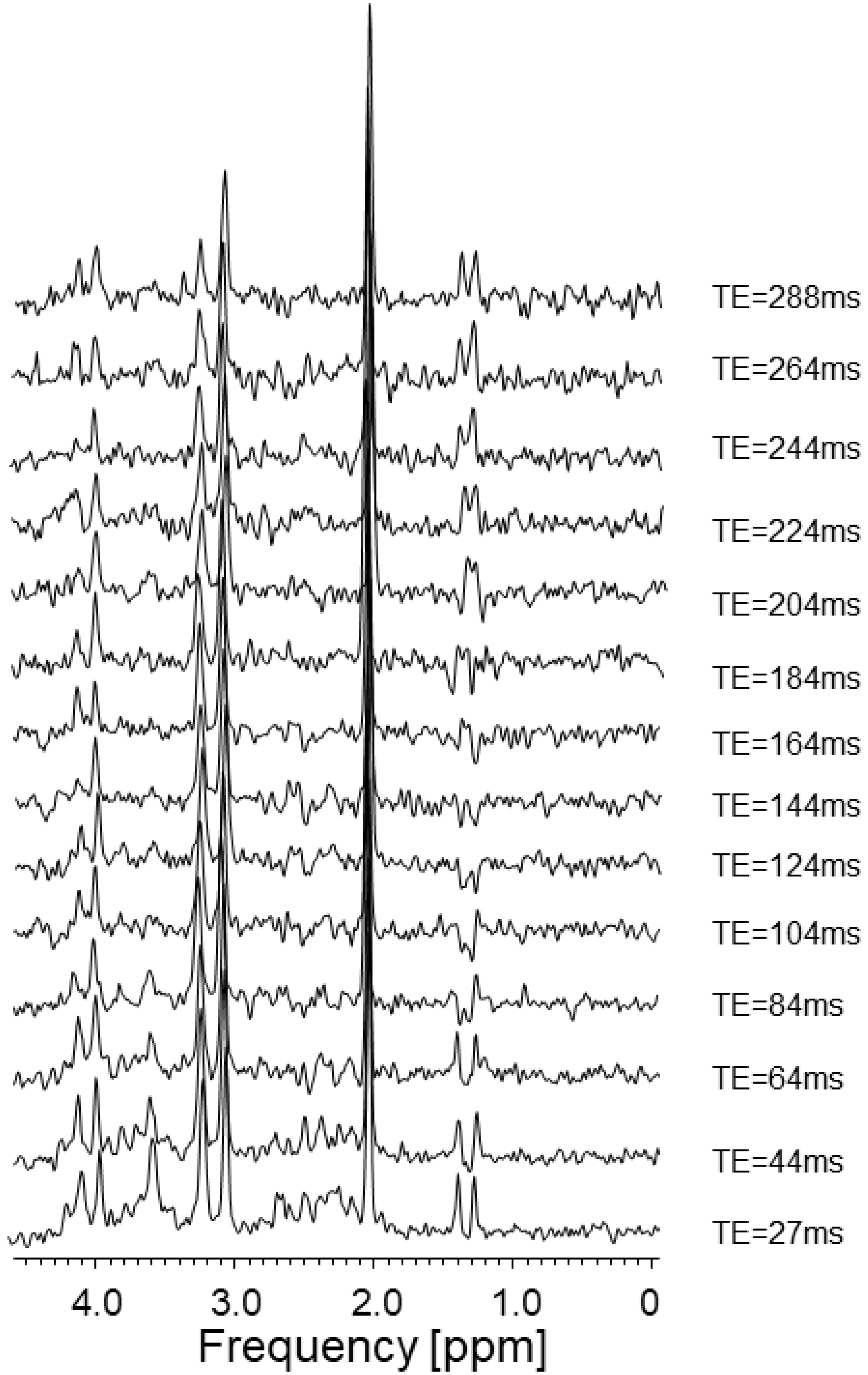
2.1. Examination of Lactate as a Standard towards Its Search in the MRS Spectrum of Cell Culture
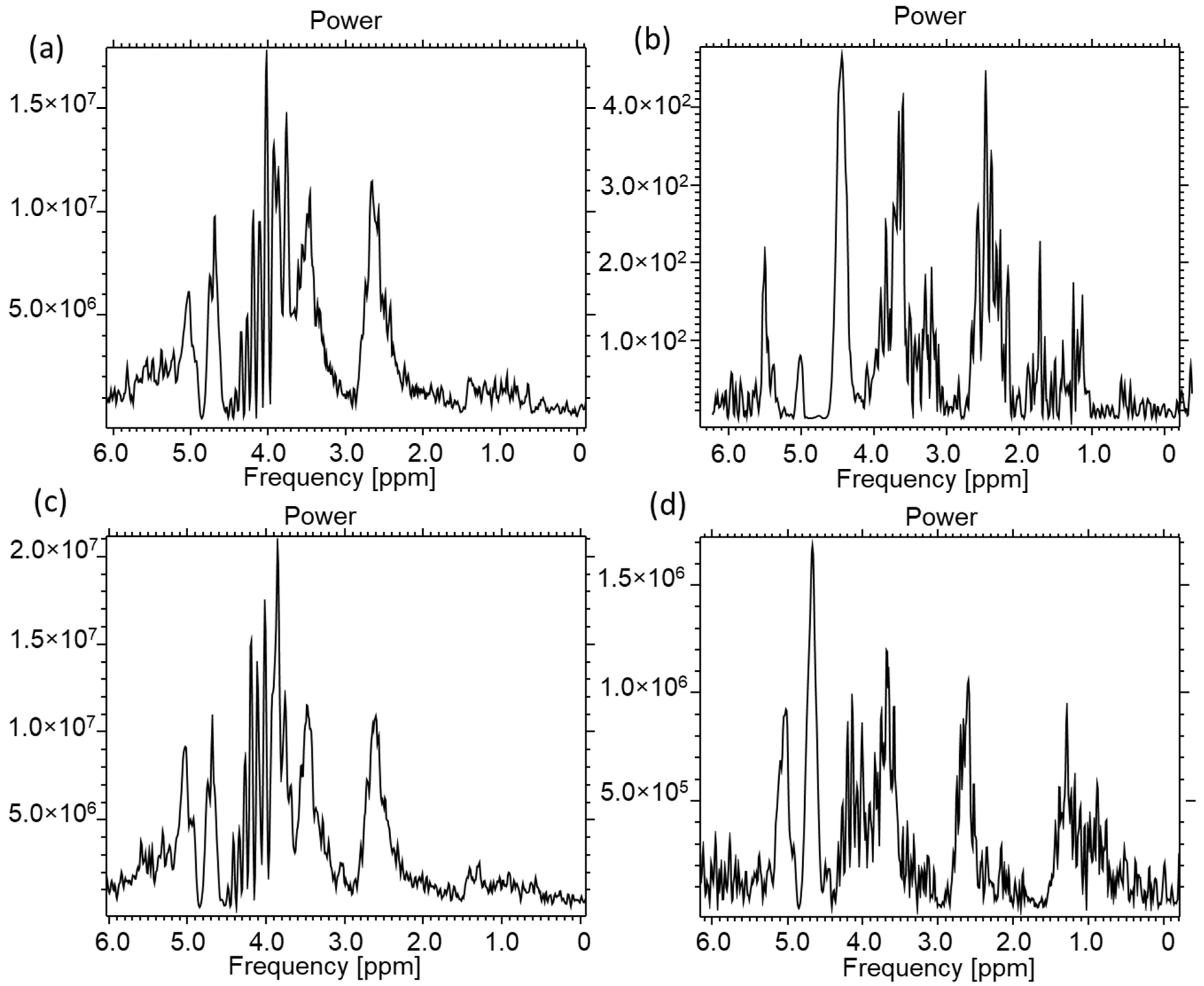
2.2. MRS Spectroscopy of RPMI-1640 Culture Fluid
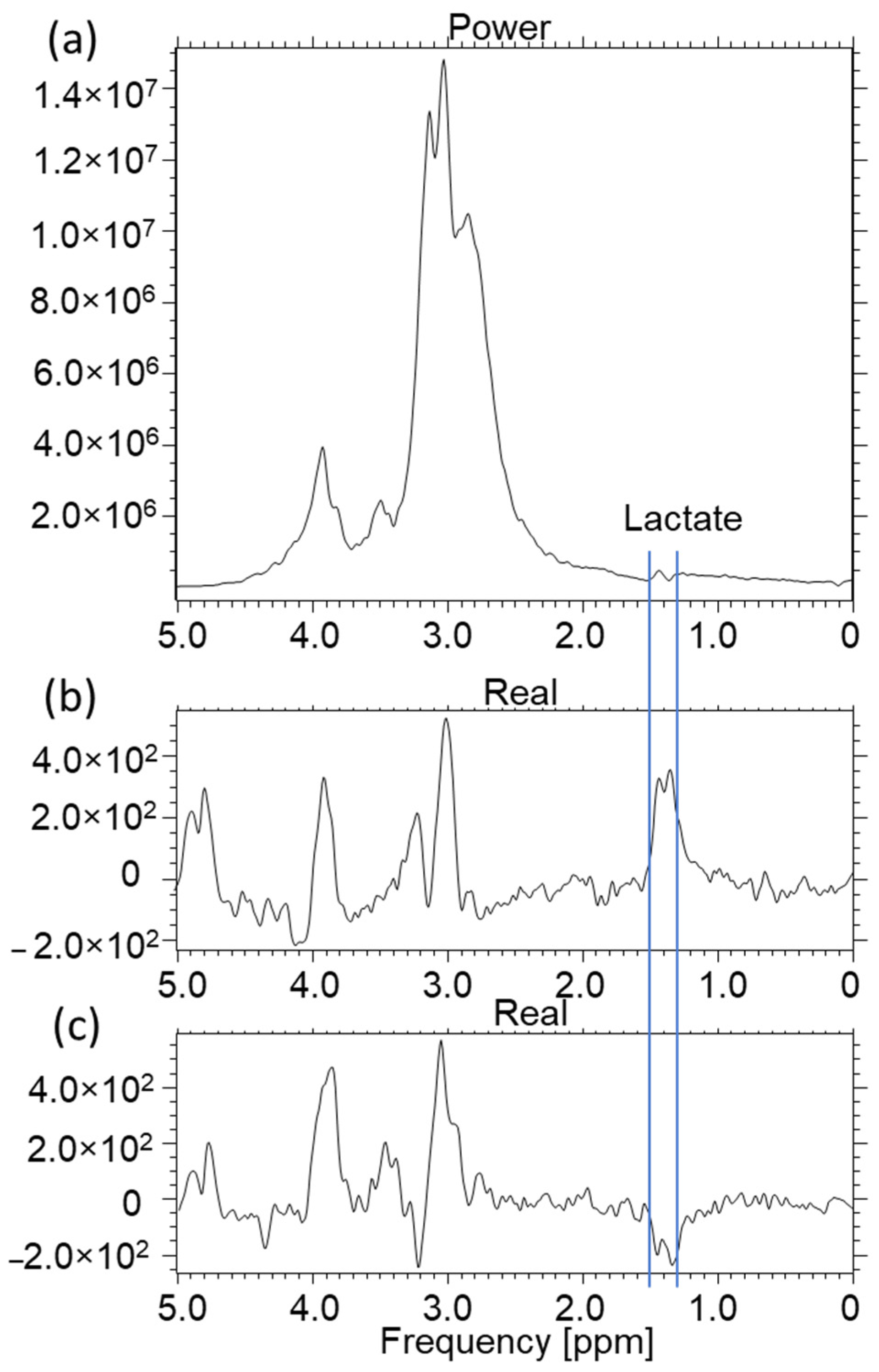
2.3. A549 and CRL2314 Cell Culture Study
3. Discussion
4. Materials and Methods
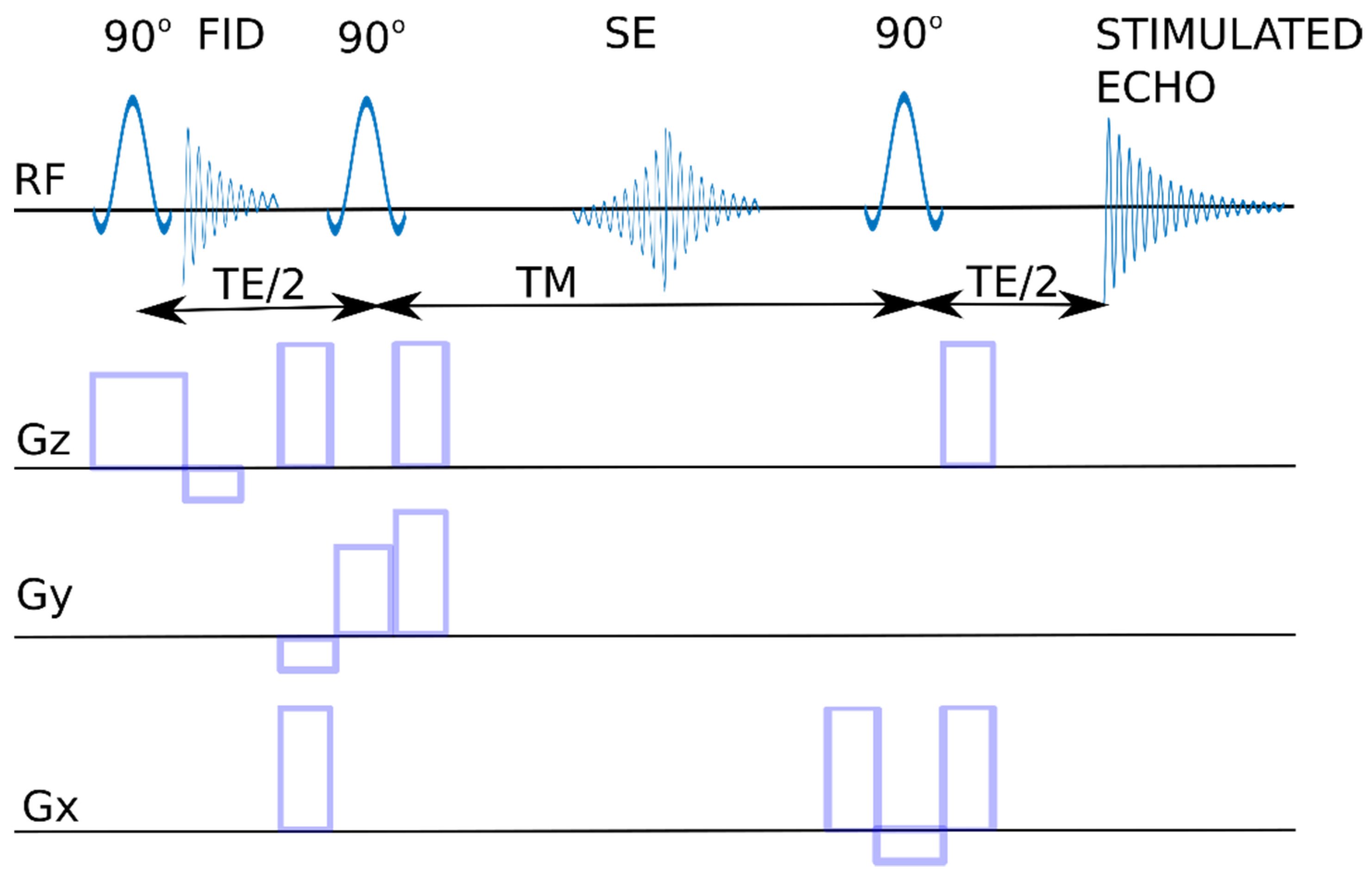
4.1. The Hardware
4.2. The Software
4.3. Samples
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| MRS | magnetic resonance spectroscopy |
| NMR | nuclear magnetic resonance |
| Glu | glutamate |
| Gln | glutamine |
| Glx | Glu + Gln |
| 1H | hydrogen |
| Cho | choline |
| PCho | phosphocholine |
| GPC | glycerophosphocholine |
| tCho | total choline |
| Cr | creatine |
| RF | radio frequency electromagnetic wave |
| RPMI 1640 | a medium for cell culture |
| MCF-7 | a breast cancer cell line |
| A549 | Lung cancer cell line |
| TE | echo time |
| TR | repetition time |
| PCB | printed circuit board |
References
- Choi, I.Y.; Andronesi, O.C.; Barker, P.; Bogner, W.; Edden, R.A.E.; Kaiser, L.G.; Lee, P.; Marjańska, M.; Terpstra, M.; de Graaf, R.A. Spectral editing in 1 H magnetic resonance spectroscopy: Experts’ consensus recommendations. NMR Biomed. 2021, 34, e4411. [Google Scholar] [CrossRef]
- Tomiyasu, M.; Harada, M. In vivo Human MR Spectroscopy Using a Clinical Scanner: Development, Applications, and Future Prospects. Magn. Reson. Med. Sci. 2022, 21, 235–252. [Google Scholar] [CrossRef]
- Kennedy, R.T. Emerging trends in in vivo neurochemical monitoring by microdialysis. Curr. Opin. Chem. Biol. 2013, 17, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Prichard, J.; Rothman, D.; Novotny, E.; Petroff, O.; Kuwabara, T.; Avison, M.; Howseman, A.; Hanstock, C.; Shulman, R. Lactate rise detected by 1H NMR in human visual cortex during physiologic stimulation. Proc. Natl. Acad. Sci. USA 1991, 88, 5829–5831. [Google Scholar] [CrossRef]
- Pohanka, M. D-Lactic Acid as a Metabolite: Toxicology, Diagnosis, and Detection. Biomed. Res. Int. 2020, 2020, 3419034. [Google Scholar] [CrossRef] [PubMed]
- Kramer, A.; Al Agha, S.; Böhm, L.; Michael, M.; Schulze-Bosse, K.; Bernhard, M. Lactat in der Notfallmedizin [Lactate in emergency medicine]. Anaesthesist 2020, 69, 826–834. [Google Scholar] [CrossRef] [PubMed]
- Ciborowski, M.; Lipska, A.; Godzien, J.; Ferrarini, A.; Korsak, J.; Radziwon, P.; Tomasiak, M.; Barbas, C. Combination of LC-MS- and GC-MS-based metabolomics to study the effect of ozonated autohemotherapy on human blood. J. Proteome Res. 2012, 11, 6231–6241. [Google Scholar] [CrossRef]
- Träber, F.; Block, W.; Lamerichs, R.; Gieseke, J.; Schild, H.H. 1H metabolite relaxation times at 3.0 Tesla: Measurements of T1 and T2 values in normal brain and determination of regional differences in transverse relaxation. J. Magn. Reson. Imaging 2004, 19, 537–545. [Google Scholar] [CrossRef]
- Ogg, R.J.; Kingsley, P.B.; Taylor, J.S. WET, a T1- and B1-insensitive water-suppression method for in vivo localized ¹H NMR spectroscopy. J. Magn. Reson. Ser. B 1994, 104, 1–10. [Google Scholar] [CrossRef]
- de Graaf, R.A.; Nicolay, K. Adiabatic water suppression using frequency selective excitation. Magn. Reson. Med. 1998, 40, 690–696. [Google Scholar] [CrossRef]
- Tkáč, I.; Gruetter, R. Methodology of H NMR Spectroscopy of the Human Brain at Very High Magnetic Fields. Appl. Magn. Reson. 2005, 29, 139–157. [Google Scholar] [CrossRef] [PubMed]
- Mescher, M.; Merkle, H.; Kirsch, J.; Garwood, M.; Gruetter, R. Simultaneous in vivo spectral editing and water suppression. NMR Biomed. 1998, 11, 266–272. [Google Scholar] [CrossRef]
- Star-Lack, J.; Nelson, S.J.; Kurhanewicz, J.; Huang, L.R.; Vigneron, D.B. Improved water and lipid suppression for 3D PRESS CSI using RF band selective inversion with gradient dephasing (BASING). Magn. Reson. Med. 1997, 38, 311–321. [Google Scholar] [CrossRef] [PubMed]
- Hollinshead, K.E.; Williams, D.S.; Tennant, D.A.; Ludwig, C. Probing Cancer Cell Metabolism Using NMR Spectroscopy. Adv. Exp. Med. Biol. 2016, 899, 89–111. [Google Scholar]
- Chun, S.I.; Mun, C.W. Cytotoxicity of TSP in 3D Agarose Gel Cultured Cell. PLoS ONE 2015, 10, e0128739. [Google Scholar]
- Kruger, N.J.; Troncoso-Ponce, M.A.; Ratcliffe, R.G. 1H NMR metabolite fingerprinting and metabolomic analysis of perchloric acid extracts from plant tissues. Nat. Protoc. 2008, 3, 1001–1012. [Google Scholar] [CrossRef]
- Le Belle, J.E.; Harris, N.G.; Williams, S.R.; Bhakoo, K.K. A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR Biomed. 2002, 15, 37–44. [Google Scholar] [CrossRef]
- Albers, M.J.; Butler, T.N.; Rahwa, I.; Bao, N.; Keshari, K.R.; Swanson, M.G.; Kurhanewicz, J. Evaluation of the ERETIC method as an improved quantitative reference for 1H HR-MAS spectroscopy of prostate tissue. Magn. Reson. Med. 2009, 61, 525–532. [Google Scholar] [CrossRef]
- Gatenby, R.A.; Gillies, R.J. Why do cancers have high aerobic glycolysis? Nat. Rev. Cancer 2004, 4, 891–899. [Google Scholar] [CrossRef]
- Warburg, O. On the origin of cancer cells. Science 1956, 123, 309–314. [Google Scholar] [CrossRef]
- Elf, S.E.; Chen, J. Targeting glucose metabolism in patients with cancer. Cancer 2014, 120, 774–780. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; Cantley, L.C.; Thompson, C.B. Understanding the Warburg effect: The metabolic requirements of cell proliferation. Science 2009, 324, 1029–1033. [Google Scholar] [CrossRef] [PubMed]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef] [PubMed]
- Govindaraju, V.; Young, K.; Maudsley, A.A. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR Biomed. 2000, 13, 129–153. [Google Scholar] [CrossRef]
- Hakumäki, J.M.; Kauppinen, R.A. 1H NMR visible lipids in the life and death of cells. Trends Biochem. Sci. 2000, 25, 357–362. [Google Scholar] [CrossRef]
- Barba, I.; Cabañas, M.E.; Arús, C. The relationship between nuclear magnetic resonance-visible lipids, lipid droplets, and cell proliferation in cultured C6 cells. Cancer Res. 1999, 59, 1861–1868. [Google Scholar]
- Kaelin, W.G., Jr.; Thompson, C.B. Q&A: Cancer: Clues from cell metabolism. Nature 2010, 465, 562–564. [Google Scholar]
- Ramadan, S.; Lin, A.; Stanwell, P. Glutamate and glutamine: A review of in vivo MRS in the human brain. NMR Biomed. 2013, 26, 1630–1646. [Google Scholar] [CrossRef]
- Thompson, R.B.; Allen, P.S. Sources of variability in the response of coupled spins to the PRESS sequence and their potential impact on metabolite quantification. Magn. Reson. Med. 1999, 41, 1162–1169. [Google Scholar] [CrossRef]
- Schubert, F.; Gallinat, J.; Seifert, F.; Rinneberg, H. Glutamate concentrations in human brain using single voxel proton magnetic resonance spectroscopy at 3 Tesla. NeuroImage 2004, 21, 1762–1771. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef] [PubMed]
- Cheng, M.; Rizwan, A.; Jiang, L.; Bhujwalla, Z.M.; Glunde, K. Molecular Effects of Doxorubicin on Choline Metabolism in Breast Cancer. Neoplasia 2017, 19, 617–627. [Google Scholar] [CrossRef] [PubMed]
- Danishad, K.K.; Sharma, U.; Sah, R.G.; Seenu, V.; Parshad, R.; Jagannathan, N.R. Assessment of therapeutic response of locally advanced breast cancer (LABC) patients undergoing neoadjuvant chemotherapy (NACT) monitored using sequential magnetic resonance spectroscopic imaging (MRSI). NMR Biomed. 2010, 23, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Payne, G.S.; Leach, M.O. Applications of magnetic resonance spectroscopy in radiotherapy treatment planning. Br. J. Radiol. 2006, 79, S16–S26. [Google Scholar] [CrossRef]
- Sonkar, K.; Ayyappan, V.; Tressler, C.M.; Adelaja, O.; Cai, R.; Cheng, M.; Glunde, K. Focus on the glycerophosphocholine pathway in choline phospholipid metabolism of cancer. NMR Biomed. 2019, 32, e4112. [Google Scholar] [CrossRef]
- Glunde, K.; Bhujwalla, Z.M.; Ronen, S.M. Choline metabolism in malignant transformation. Nat. Rev. Cancer 2011, 11, 835–848. [Google Scholar] [CrossRef]
- Grande, S.; Giovannini, C.; Guidoni, L.; Luciani, A.M.; Palma, A.; Rosi, A.; Sapora, O.; Viti, V. 1H MRS signals from gluta-thione may act as predictive markers of apoptosis in irradiated tumour cells. Radiat. Prot. Dosim. 2006, 122, 205–206. [Google Scholar] [CrossRef]
- Ward, C.S.; Eriksson, P.; Izquierdo-Garcia, J.L.; Brandes, A.H.; Ronen, S.M. HDAC inhibition induces increased choline uptake and elevated phosphocholine levels in MCF7 breast cancer cells. PLoS ONE 2013, 8, e62610. [Google Scholar] [CrossRef]
- Lodi, A.; Woods, S.M.; Ronen, S.M. MR-detectable metabolic consequences of mitogen-activated protein kinase kinase (MEK) inhibition. NMR Biomed. 2014, 27, 700–708. [Google Scholar] [CrossRef]
- Rosi, A.; Grande, S.; Luciani, A.M.; Palma, A.; Giovannini, C.; Guidoni, L.; Sapora, O.; Viti, V. Role of glutathione in apopto-sis induced by radiation as determined by 1H MR spectra of cultured tumor cells. Radiat. Res. 2007, 167, 268–282. [Google Scholar] [CrossRef]
- Brandes, A.H.; Ward, C.S.; Ronen, S.M. 17-allyamino-17-demethoxygeldanamycin treatment results in a magnetic resonance spectroscopy-detectable elevation in choline-containing metabolites associated with increased expression of choline trans-porter SLC44A1 and phospholipase A2. Breast Cancer Res. 2010, 12, R84. [Google Scholar] [CrossRef] [PubMed]
- Jensen, L.R.; Huuse, E.M.; Bathen, T.F.; Goa, P.E.; Bofin, A.M.; Pedersen, T.B.; Lundgren, S.; Gribbestad, I.S. Assessment of early docetaxel response in an experimental model of human breast cancer using DCE-MRI, ex vivo HR MAS, and in vivo 1H MRS. NMR Biomed. 2010, 23, 56–65. [Google Scholar] [CrossRef] [PubMed]
- Wijnen, J.P.; Jiang, L.; Greenwood, T.R.; van der Kemp, W.J.; Klomp, D.W.; Glunde, K. 1H/31P polarization transfer at 9.4 Tesla for improved specificity of detecting phosphomonoesters and phosphodiesters in breast tumor models. PLoS ONE 2014, 9, e102256. [Google Scholar] [CrossRef]
- Le Moyec, L.; Tatoud, R.; Eugène, M.; Gauvillé, C.; Primot, I.; Charlemagne, D.; Calvo, F. Cell and membrane lipid analysis by proton magnetic resonance spectroscopy in five breast cancer cell lines. Br. J. Cancer 1992, 66, 623–628. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Le Moyec, L.; Kawakami, M.; Leray, G.; Larue, V.; Briane, D.; Hantz, E.; De Certaines, J. Magnetic resonance spectroscopy and fluorescence microscopy to investigate mobile lipids in sensitive, resistant and reverting K562 cells and their membranes. Anticancer Res. 2000, 20, 4513–4518. [Google Scholar] [PubMed]
- Mannechez, A.; Reungpatthanaphong, P.; de Certaines, J.D.; Leray, G.; Le Moyec, L. Proton NMR visible mobile lipid signals in sensitive and multidrug-resistant K562 cells are modulated by rafts. Cancer Cell Int. 2005, 5, 2. [Google Scholar] [CrossRef][Green Version]
- Grande, S.; Luciani, A.M.; Rosi, A.; Palma, A.; Giovannini, C.; Sapora, O.; Guidoni, L.; Viti, V. Metabolism of glutathione in tumour cells as evidenced by 1H MRS. FEBS Lett. 2007, 581, 637–643. [Google Scholar] [CrossRef]
- Haase, A.; Frahm, J.; Hänicke, W.; Matthaei, D. ¹H NMR chemical shift selective (CHESS) imaging. Phys. Med. Biol. 1985, 30, 341–344. [Google Scholar] [CrossRef]







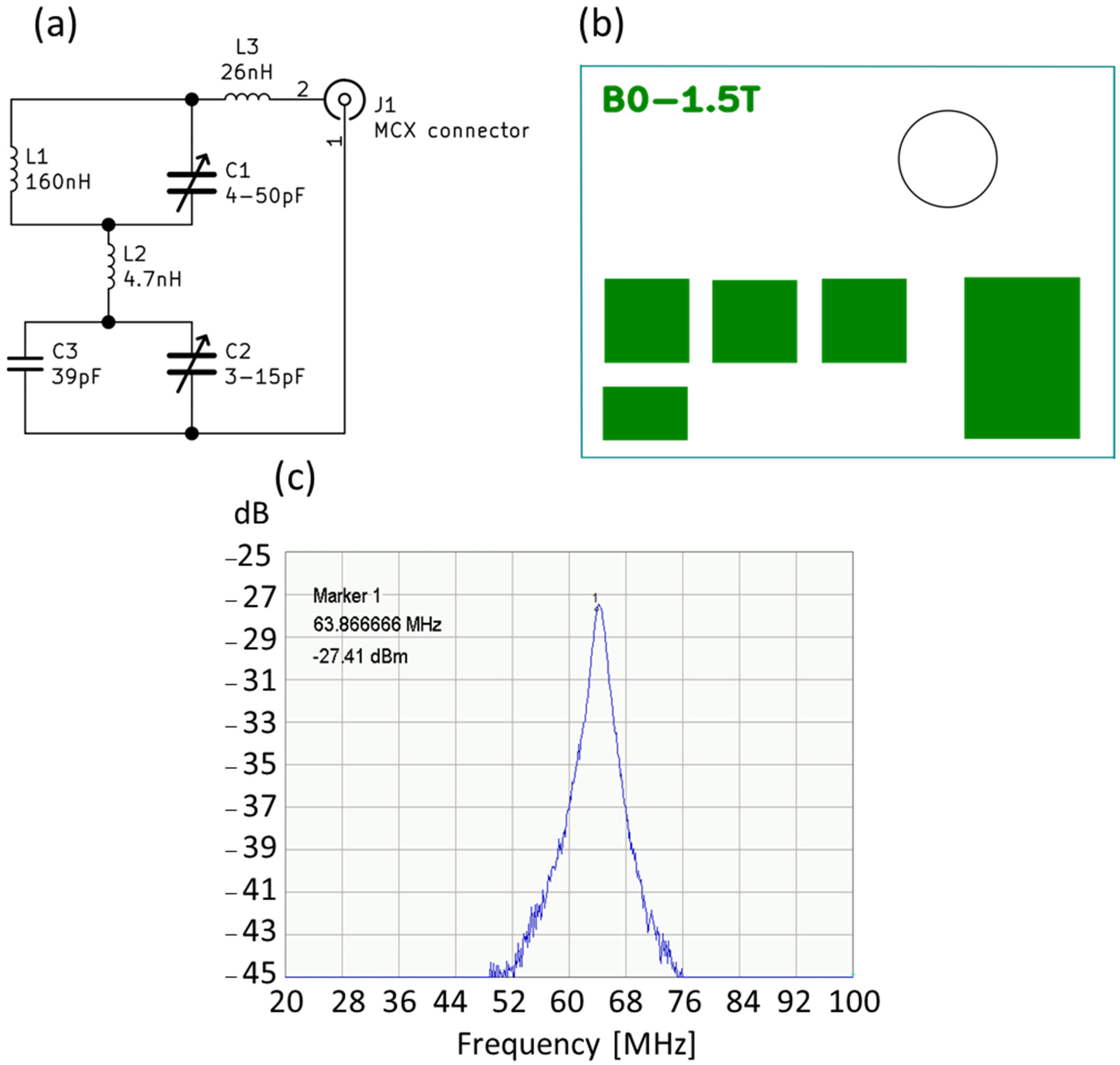
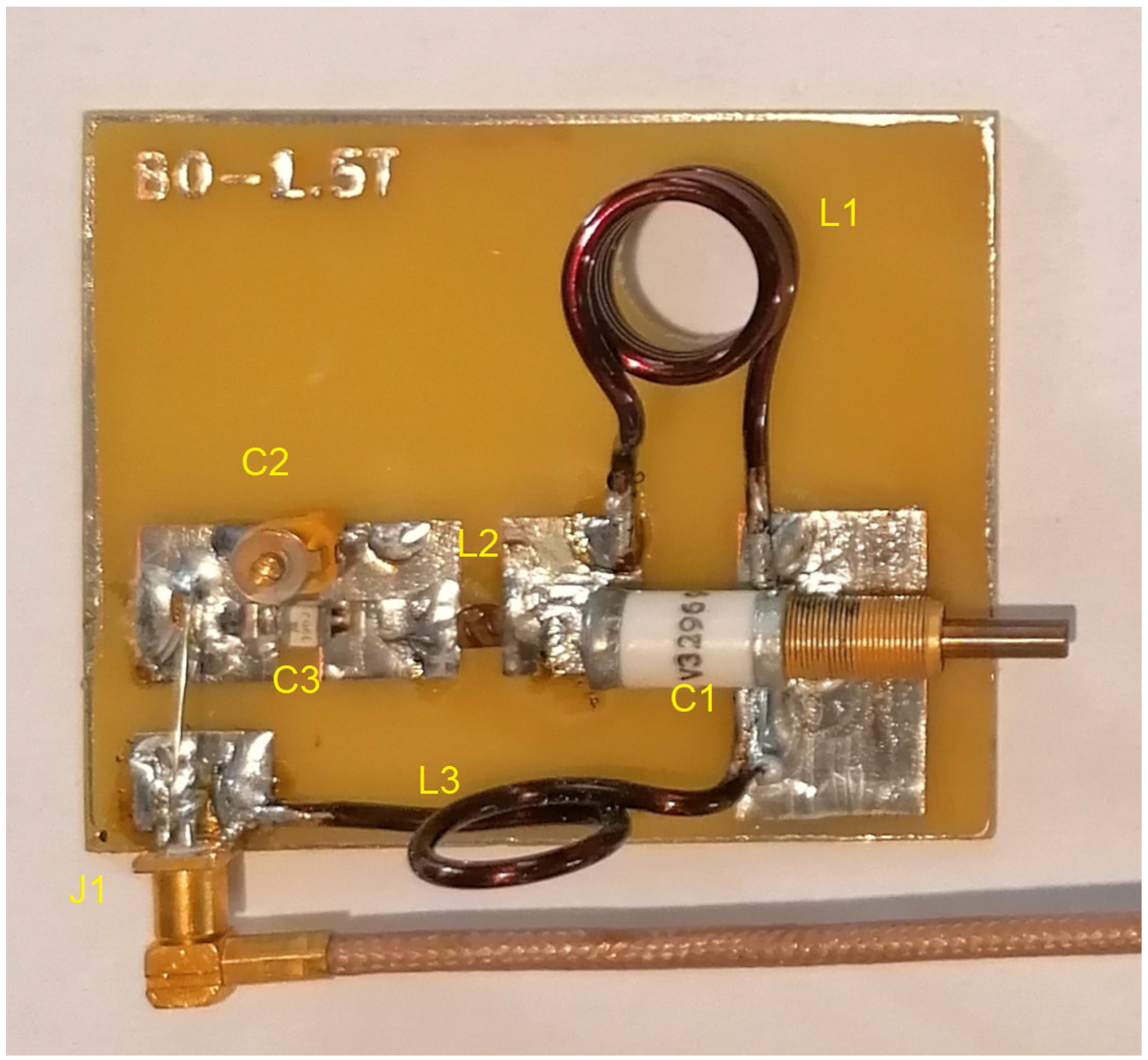
| Component | Concentration [g/L] |
|---|---|
| Inorganic salts | |
| Ca(NO3)2 × 4H2O | 0.1 |
| MgSO4 (anhyd) | 0.04884 |
| KCl | 0.4 |
| NaCl | 6.0 |
| Na2HPO4 (Anhyd) | 0.8 |
| Amino acids | |
| L-Arginine (free base) | 0.2 |
| L-Asparagine (anhyd) | 0.05 |
| L-Aspartic Acid | 0.02 |
| L-Cystine × 2HCl | 0.0652 |
| L-Glutamic Acid | 0.02 |
| L-Glutamine | 0.3 |
| Glycine | 0.01 |
| L-Histidine (free base) | 0.015 |
| Hydroxy-L-Proline | 0.02 |
| L-Isoleucine | 0.05 |
| L-Leucine | 0.05 |
| L-Lysine × HCl | 0.04 |
| L-Methionine | 0.015 |
| L-Phenylalanine | 0.015 |
| L-Proline | 0.02 |
| L-Serine | 0.03 |
| L-Threonine | 0.02 |
| L-Tryptophan | 0.005 |
| L-Tyrosine × 2Na × 2H2O | 0.02883 |
| L-Valine | 0.02 |
| Vitamins | |
| D-Biotin | 0.0002 |
| Choline Chloride | 0.003 |
| Folic Acid | 0.001 |
| myo-Inositol | 0.035 |
| Niacinamide | 0.001 |
| p-Amino Benzoic Acid | 0.001 |
| D-Pantothenic Acid × ½Ca | 0.00025 |
| Pyridoxine·HCl | 0.001 |
| Riboflavin | 0.0002 |
| Thiamine × HCl | 0.001 |
| Vitamin B-12 | 0.000005 |
| Other | |
| D-Glucose | 2.0 |
| Glutathione (reduced) | 0.001 |
| Phenol Red × Na | 0.0053 |
| NaHCO3 | 2.0 |
| MR System/Research System | Test Cells | Type of Cells | Source | Medium | Number of Cells | Analyzed Biomarker and/or Its Function |
|---|---|---|---|---|---|---|
| 400.14 MHz, Avance (Bruker, AG, Darmstadt, Germany) | MCF-7 | breast cancer | (Grande, 2006) [37] | RPMI 1640 | ~5 × 106 | GSH metabolism (γ-glutamylcysteinylglycine tripeptide, glutathione) |
| 400.14 MHz, Avance (Bruker, AG, Darmstadt, Germany) | TG98 | malignant glioma | (Grande, 2006) [37] | RPMI 1640 | ~5 × 106 | GSH metabolism (γ-glutamylcysteinylglycine tripeptide, glutathione) |
| 600 MHz, Varian | MCF-7 | breast cancer | (Ward, 2013) [38] | DMEM | 5 × 106 | Phosphocholine (PC) |
| 500 MHz, 600 MHz (Varian/Agilent Palo Alto, CA, USA) | PC3 | prostate cancer | (Ward, 2013) [38] | DMEM low in glucose | - | Glucose → lactate; choline-containing metabolites: (tCho, consisting of choline, phosphocholine (PC) and glycerophospho choline (GPC)) |
| 500 MHz, 600 MHz (Varian/Agilent Palo Alto, CA, USA) | MCF-7 | breast cancer | (Lodi, 2014) [39] | DMEM low in glucose | - | Glucose → lactate; choline-containing metabolites: (tCho, consisting of choline, phosphocholine (PC) and glycerophospho choline (GPC)) |
| 500 MHz, 600 MHz (Varian/Agilent Palo Alto, CA, USA) | A375 | melanoma cells | (Lodi, 2014) [39] | DMEM high in glucose | - | Glucose → lactate; choline-containing metabolites: (tCho, consisting of choline, phosphocholine (PC) and glycerophospho choline (GPC)) |
| 400.14 MHz, Bruker, AG, Darmstadt, Germany | MCF-7 | breast cancer | (Rosi, 2007) [40] | RPMI 1640 | 20 × 106 | glutamate (Glu), glutamine (Gln), glutathione (GSH) |
| 400.14 MHz, Bruker, AG, Darmstadt, Germany | HeLa | cervical cancer | (Rosi, 2007) [40] | EMEM | 20 × 106 | glutamate (Glu), glutamine (Gln), glutathione (GSH) |
| 500 MHz INOVA (Varian, Santa Clara, CA, USA) | MCF-7 | breast cancer | (Brandes, 2010) [41] | Eagle | phosphocholine (PC) | |
| 7 T Bruker BioSpec Avance 70/20 | MCF-7 | breast cancer | (Jensen, 2010) [42] | Earle | 1 × 107 | Choline (Cho) |
| 9.4 T Bruker Horizontal Bore Small Animal Scanner (Bruker BioSpin Corp.) | MCF-7 | breast cancer | (Wijnen, 2014) [43] | HBSS | 2 × 106 | metabolites phospholipids, phosphoethanolamine (PC), phosphoethanolamine (PE), glycerophosphocholine (GPC) and glycerophosphoethanolamine (GPE) |
| 9.4 T Bruker Horizontal Bore Small Animal Scanner (Bruker BioSpin Corp.) | MDA-MB-231 | breast cancer (highly aggressive) | (Wijnen, 2014) [43] | HBSS | 2 × 106 | metabolites phospholipids, phosphoethanolamine (PC), phosphoethanolamine (PE), glycerophosphocholine (GPC) and glycerophosphoethanolamine (GPE) |
| 400 MHz Bruker; AM400 WB | MCF-7 | breast cancer | (Le Moyec, 1992) [44] | Eagle (DMEM, Gibco), 10% fetal calf serum | - | lipids |
| 400 MHz Bruker; AM400 WB | T47D | breast cancer | (Le Moyec, 1992) [44] | Eagle (DMEM, Gibco), 10% fetal calf serum | - | lipids |
| 400 MHz Bruker; AM400 WB | ZR-75-1 | breast cancer epithelial cell | (Le Moyec, 1992) [44] | Eagle (DMEM, Gibco), 10% fetal calf serum | - | lipids |
| 400 MHz Bruker; AM400 WB | SKBR3 | breast cancer | (Le Moyec, 1992) [44] | Eagle (DMEM, Gibco), 10% fetal calf serum | - | lipids |
| 400 MHz Bruker; AM400 WB | MDA-MB231 | breast cancer (highly aggressive) | (Le Moyec, 1992) [44] | Eagle (DMEM, Gibco), 10% fetal calf serum | - | lipids |
| - | K562 | myeloid leukemia | (Le Moyec, 2000) [45] | RPMI 1640 | - | lipids |
| 500 MHz, Variaan; Unity Inova | K562 | myeloid leukemia | (Mannechez, 2005) [46] | RPMI | 1 × 107 | lipids |
| 400.14 MHz Bruker, Avance | MCF-7 | breast cancer | (Grande, 2007) [47] | RPMI 1640 | glutathione (GSH) | |
| 400.14 MHz Bruker, Avance | HeLa | cervical cancer | (Grande, 2007) [47] | EMEM | 20 × 106 | glutathione (GSH) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Truszkiewicz, A.; Bartusik-Aebisher, D.; Zalejska-Fiolka, J.; Kawczyk-Krupka, A.; Aebisher, D. Cellular Lactate Spectroscopy Using 1.5 Tesla Clinical Apparatus. Int. J. Mol. Sci. 2022, 23, 11355. https://doi.org/10.3390/ijms231911355
Truszkiewicz A, Bartusik-Aebisher D, Zalejska-Fiolka J, Kawczyk-Krupka A, Aebisher D. Cellular Lactate Spectroscopy Using 1.5 Tesla Clinical Apparatus. International Journal of Molecular Sciences. 2022; 23(19):11355. https://doi.org/10.3390/ijms231911355
Chicago/Turabian StyleTruszkiewicz, Adrian, Dorota Bartusik-Aebisher, Jolanta Zalejska-Fiolka, Aleksandra Kawczyk-Krupka, and David Aebisher. 2022. "Cellular Lactate Spectroscopy Using 1.5 Tesla Clinical Apparatus" International Journal of Molecular Sciences 23, no. 19: 11355. https://doi.org/10.3390/ijms231911355
APA StyleTruszkiewicz, A., Bartusik-Aebisher, D., Zalejska-Fiolka, J., Kawczyk-Krupka, A., & Aebisher, D. (2022). Cellular Lactate Spectroscopy Using 1.5 Tesla Clinical Apparatus. International Journal of Molecular Sciences, 23(19), 11355. https://doi.org/10.3390/ijms231911355









