CXCL8, CCL2, and CMV Seropositivity as New Prognostic Factors for a Severe COVID-19 Course
Abstract
1. Introduction
2. Results
2.1. Characteristics of the Study Group
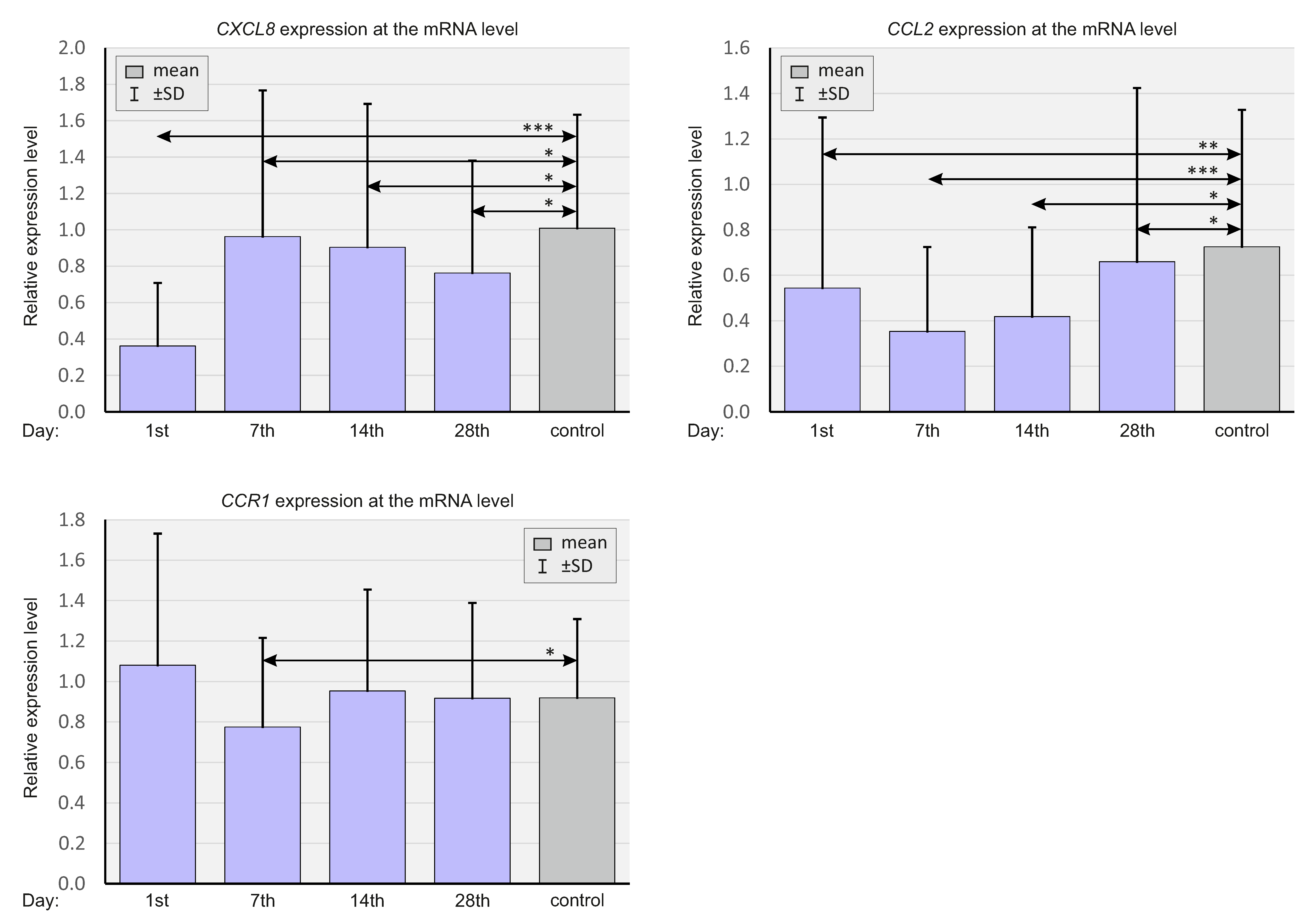
2.2. Expression of Select Chemokines at the mRNA Level
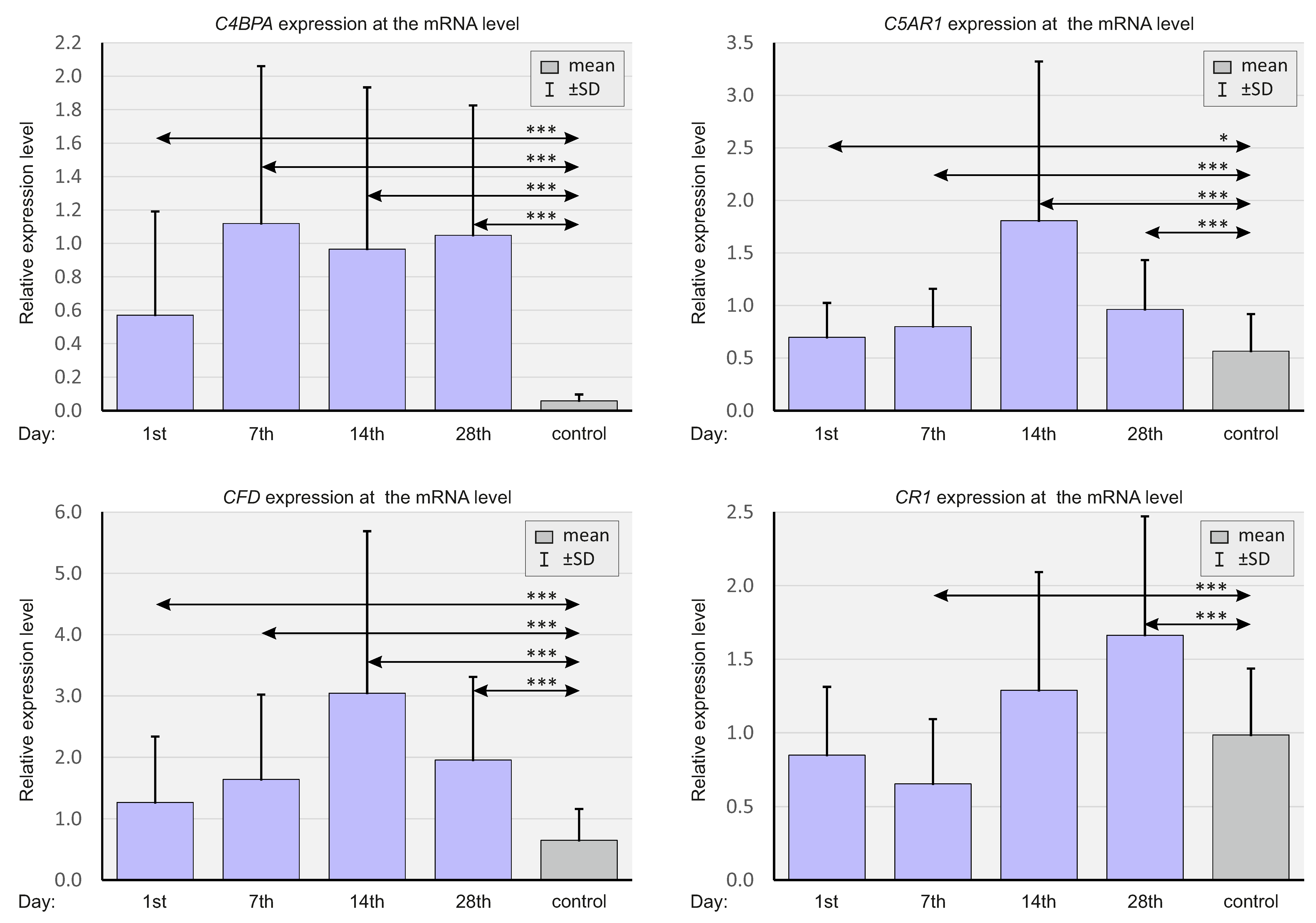
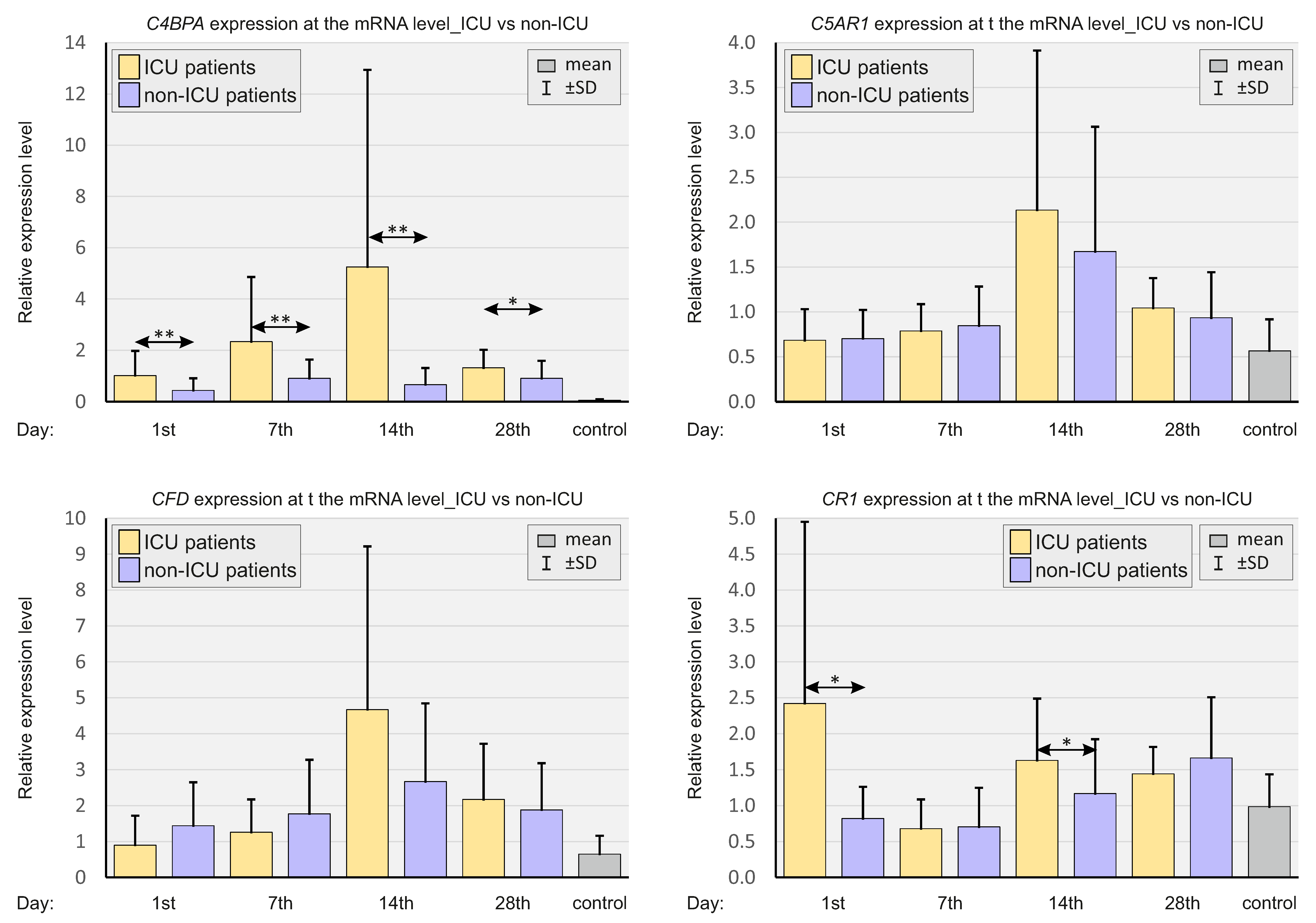
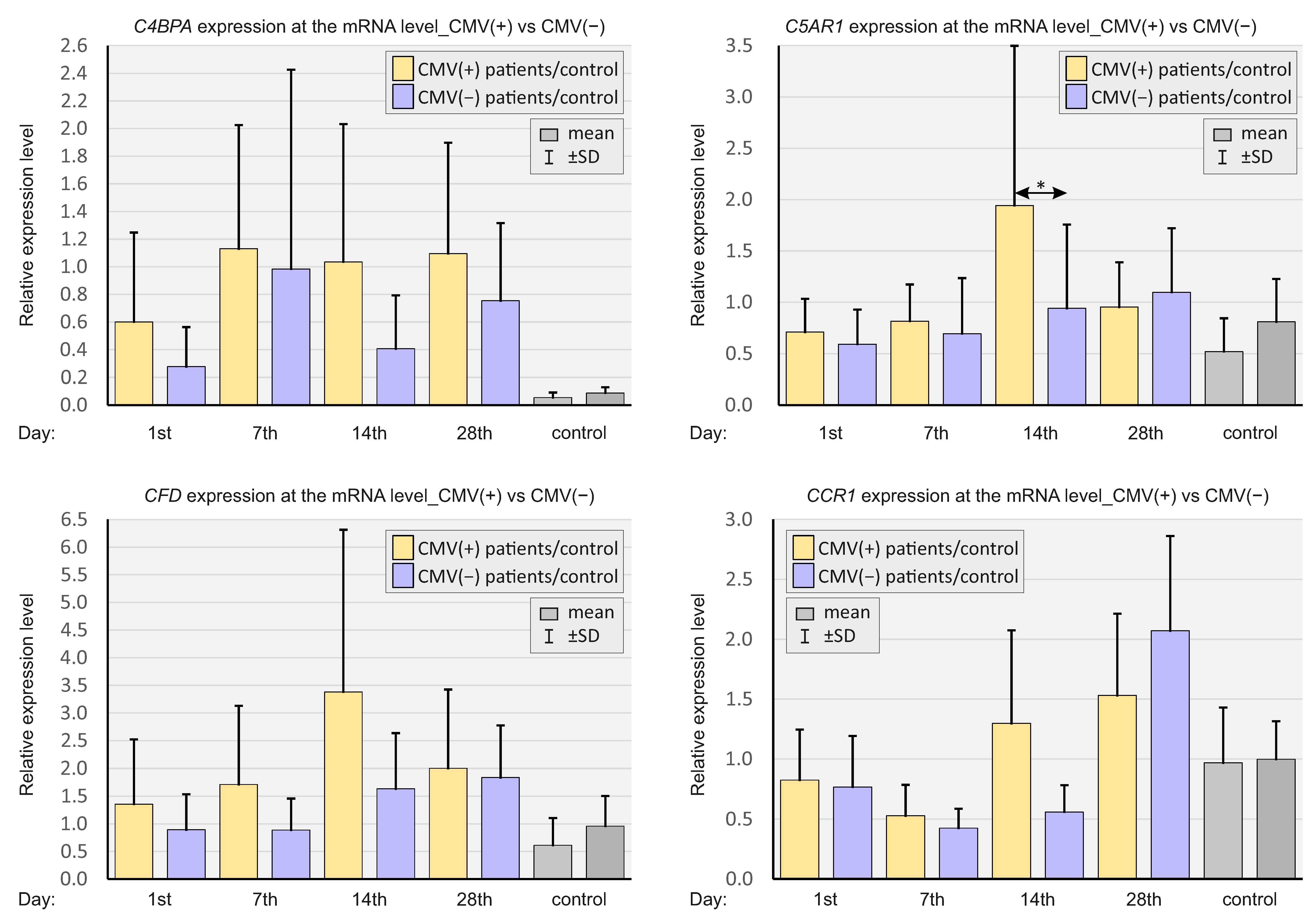
2.3. Expressions of Selected Complement System Elements at the mRNA Level
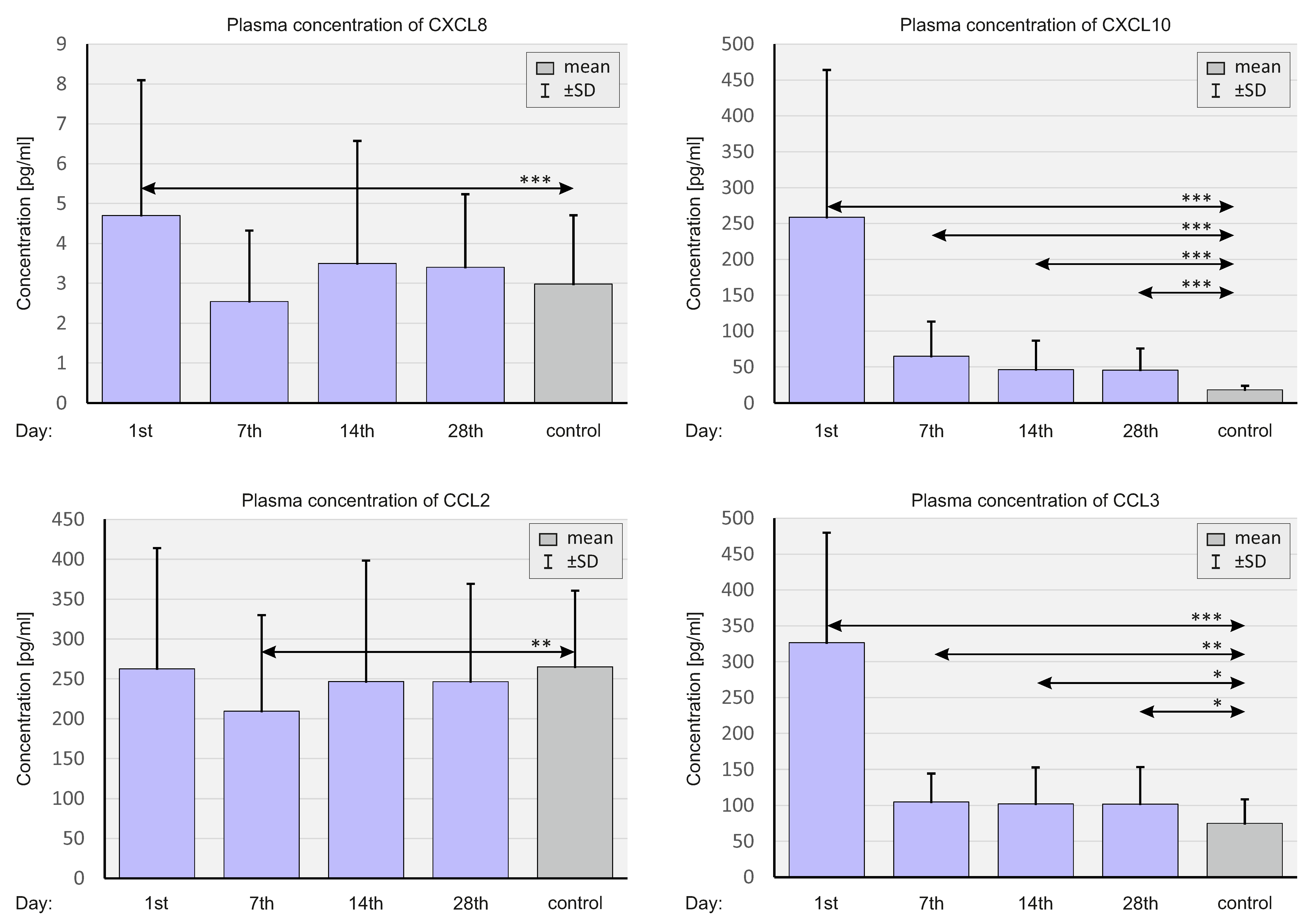
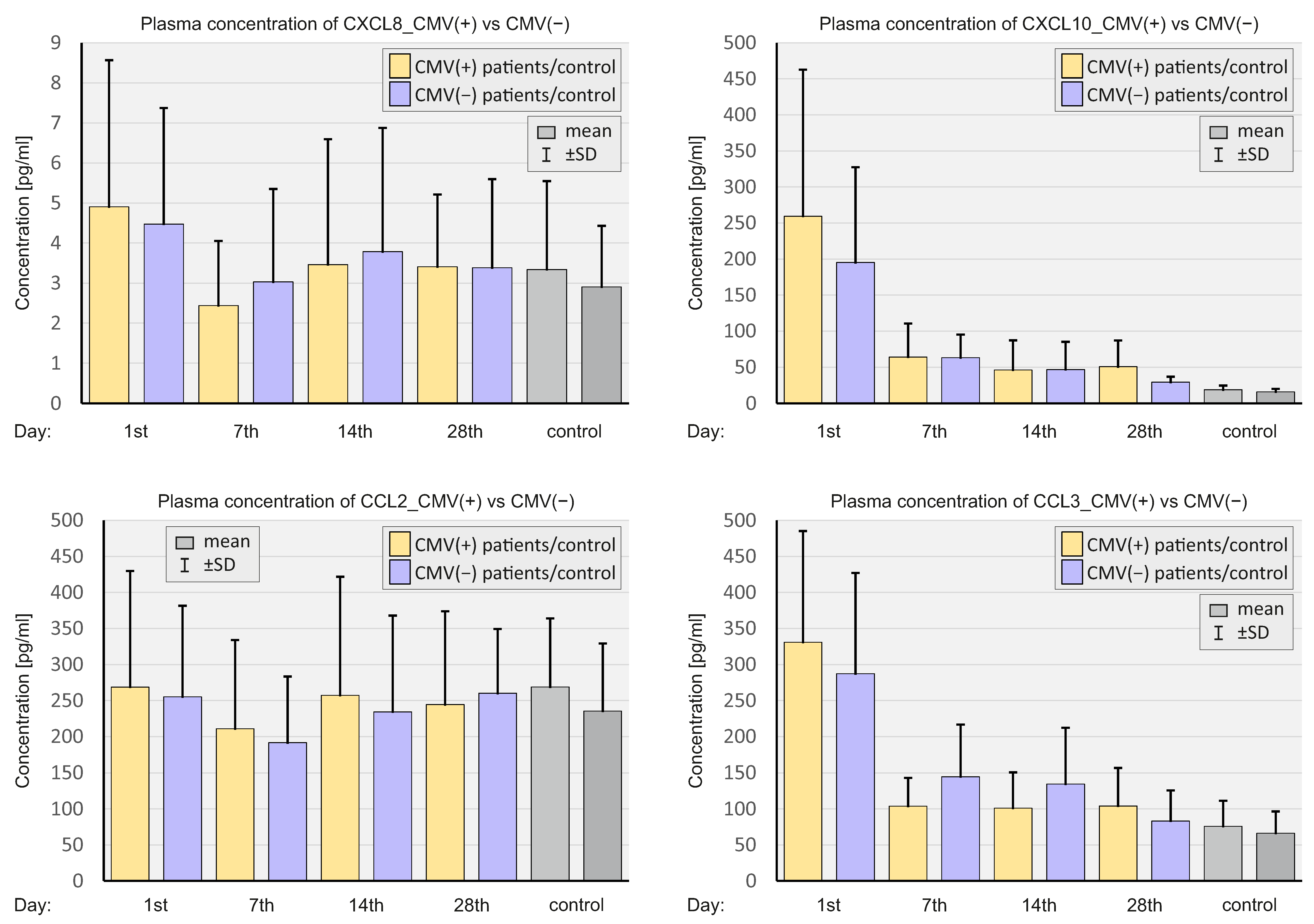
2.4. Concentration of Select Chemokines at the Protein Level
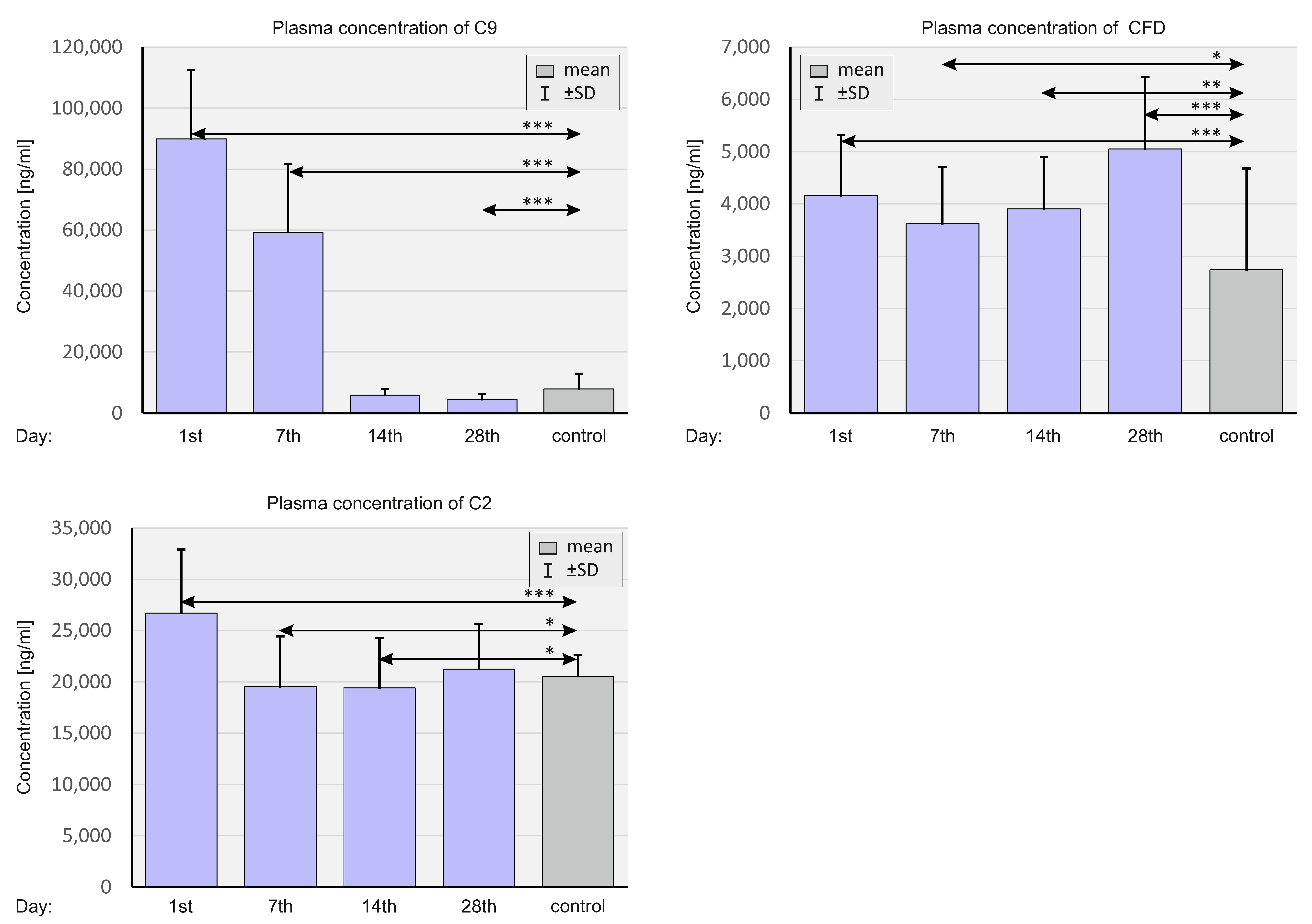
2.5. Concentration of Selected Complement System Elements at The Protein Level
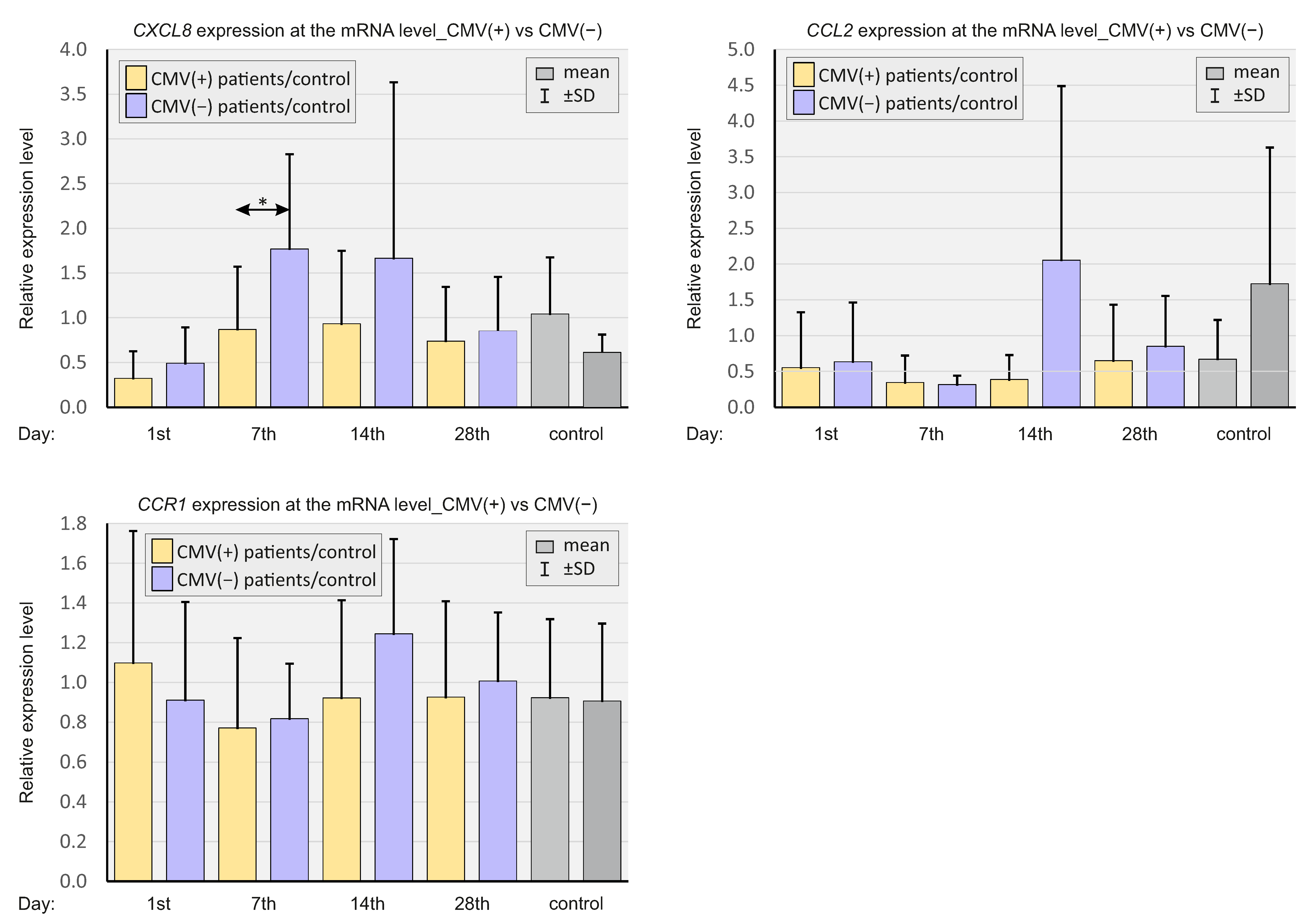
2.6. CMV Status
3. Discussion
- -
- Lower mRNA expression and higher concentrations of CXCL8 and CCL2 in the plasma correlated with more severe courses of COVID-19.
- -
- CMV-positive status correlated with a more severe COVID-19 course.
- -
- CXCL8, CCL2, and CMV seropositivity should be considered as new prognostic factors for severe COVID-19 courses.
4. Materials and Methods
4.1. Study Group
4.2. General Health Questionnaire
4.3. Material
4.3.1. Plasma Collection
4.3.2. RNA Isolation
Viral RNA Isolation
Blood Mononuclear Cell RNA Isolation
4.3.3. qRT-PCR
qRT–PCR Assays for Detecting SARS-CoV-2 RNA
qRT–PCR Assays for Evaluating Complement and Chemokine mRNA Expressions
4.3.4. Luminex Assay
4.3.5. Enzyme-Linked Immunosorbent Assay (ELISA)—Serological Assays for Specific Anti-SARS-CoV-2 IgM, IgG, IgA, and Anti-CMV IgG Antibodies Detection
4.4. Statistical Analysis
5. Study Limitations
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| BMG | Beta-2-microglobulin |
| BMI | body mass index |
| C4BPA | complement component 4 binding protein alpha |
| C5AR1 | complement component 5a receptor 1 |
| CCL3 | C-C motif chemokine ligand 3 (macrophage inflammatory protein-1α—MIP-1α) |
| CCR1 | C-C motif chemokine receptor 1 |
| CFD | Complement factor D |
| CMV | cytomegalovirus |
| CR1 | complement receptor type 1 |
| CRS | cytokine release syndrome |
| CT | computed tomography |
| CXCL10 | C-X-C motif chemokine ligand 10 (interferon gamma-induced protein 10—IP-10) |
| DPP9 | dipeptidyl peptidase 9 |
| ELISA | enzyme-linked immunosorbent assay |
| G-CSF | granulocyte colony stimulating factor |
| GROα | growth-regulated oncogene-α (CXCL1) |
| ICU | intensive care unit |
| IL-1α | interleukin-1α |
| IL-6 | interleukin 6 |
| IL-8 | interleukin 8 (CXCL8) |
| IL-10 | interleukin 10 |
| IFN-α | interferon-α |
| ISG | interferon-stimulated gene |
| MASP-2 | mannose-binding protein-associated serine protease 2 |
| Tr5‘6wq222MBL | Mannose-binding lectin |
| MCP-1 | monocyte chemoattractant protein-1 (CCL2) |
| MCP-3 | monocyte chemotactic protein-3 (CCL7) |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| non-ICU | non-intensive care unit patients |
| RT-PCR | real-time polymerase chain reaction |
| STEMI | ST-segment elevation myocardial infarction |
| TNF-α | tumor necrosis factor-α |
| TPO | thrombopoietin |
References
- Li, Q.; Guan, X.; Wu, P.; Wang, X.; Zhou, L.; Tong, Y.; Ren, R.; Leung, K.S.M.; Lau, E.H.Y.; Wong, J.Y. Early Transmission Dynamics in Wuhan, China, of Novel Coronavirus–Infected Pneumonia. N. Engl. J. Med. 2020, 382, 1199–1207. [Google Scholar] [CrossRef] [PubMed]
- van der Valk, J.P.; Veen, J.C.I. SARS-Cov-2: The Relevance and Prevention of Aerosol Transmission. J. Occup. Environ. Med. 2021, 63, e395–e401. [Google Scholar] [CrossRef] [PubMed]
- Parasher, A. COVID-19: Current understanding of its Pathophysiology, Clinical presentation and Treatment. Postgrad. Med, J. 2020, 97, 312–320. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Min, P.; Lee, S.; Kim, S.-W. Prevalence and Duration of Acute Loss of Smell or Taste in COVID-19 Patients. J. Korean Med. Sci. 2020, 35, e174. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.-D.; Ding, M.; Dong, X.; Zhang, J.-J.; Azkur, A.K.; Azkur, D.; Gan, H.; Sun, Y.-L.; Fu, W.; Li, W.; et al. Risk factors for severe and critically ill COVID-19 patients: A review. Allergy 2021, 76, 428–455. [Google Scholar] [CrossRef]
- Tang, L.; Yin, Z.; Hu, Y.; Mei, H. Controlling Cytokine Storm Is Vital in COVID-19. Front. Immunol. 2020, 11, 570993. [Google Scholar] [CrossRef]
- Hu, B.; Huang, S.; Yin, L. The cytokine storm and COVID-19. J. Med. Virol. 2020, 93, 250–256. [Google Scholar] [CrossRef]
- Petrey, A.C.; Qeadan, F.; Middleton, E.A.; Pinchuk, I.V.; Campbell, R.A.; Beswick, E.J. Cytokine release syndrome in COVID-19: Innate immune, vascular, and platelet pathogenic factors differ in severity of disease and sex. J. Leukoc. Biol. 2020, 109, 55–66. [Google Scholar] [CrossRef]
- Wei, X.-S.; Wang, X.-R.; Zhang, J.-C.; Yang, W.-B.; Ma, W.-L.; Yang, B.-H.; Jiang, N.-C.; Gao, Z.-C.; Shi, H.-Z.; Zhou, Q.; et al. A cluster of health care workers with COVID-19 pneumonia caused by SARS-CoV-2. J. Microbiol. Immunol. Infect. 2021, 54, 54–60. [Google Scholar] [CrossRef]
- Liu, T.; Zhang, J.; Yang, Y.; Ma, H.; Li, Z.; Zhang, J.; Cheng, J.; Zhang, X.; Zhao, Y.; Xia, Z.; et al. The role of interleukin-6 in monitoring severe case of coronavirus disease 2019. EMBO Mol. Med. 2020, 12, e12421. [Google Scholar] [CrossRef]
- Abers, M.S.; Delmonte, O.M.; Ricotta, E.E.; Fintzi, J.; Fink, D.L.; de Jesus, A.A.A.; Zarember, K.A.; Alehashemi, S.; Oikonomou, V.; Desai, J.V.; et al. An immune-based biomarker signature is associated with mortality in COVID-19 patients. JCI Insight 2021, 6, e144455. [Google Scholar] [CrossRef] [PubMed]
- Sharif-Zak, M.; Abbasi-Jorjandi, M.; Asadikaram, G.; Ghoreshi, Z.-A.; Rezazadeh-Jabalbarzi, M.; Afsharipur, A.; Rashidinejad, H.; Khajepour, F.; Jafarzadeh, A.; Arefinia, N.; et al. CCR2 and DPP9 expression in the peripheral blood of COVID-19 patients: Influences of the disease severity and gender. Immunobiology 2022, 227, 152184. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Cugno, M.; Meroni, P.L.; Gualtierotti, R.; Griffini, S.; Grovetti, E.; Torri, A.; Panigada, M.; Aliberti, S.; Blasi, F.; Tedesco, F.; et al. Complement activation in patients with COVID-19: A novel therapeutic target. J. Allergy Clin. Immunol. 2020, 146, 215–217. [Google Scholar] [CrossRef] [PubMed]
- Magro, C.; Mulvey, J.J.; Berlin, D.; Nuovo, G.; Salvatore, S.; Harp, J.; Baxter-Stoltzfus, A.; Laurence, J. Complement associated microvascular injury and thrombosis in the pathogenesis of severe COVID-19 infection: A report of five cases. Transl. Res. 2020, 220, 1–13. [Google Scholar] [CrossRef]
- Weber, S.; Kehl, V.; Erber, J.; Wagner, K.I.; Jetzlsperger, A.-M.; Burrell, T.; Schober, K.; Schommers, P.; Augustin, M.; Crowell, C.S.; et al. CMV seropositivity is a potential novel risk factor for severe COVID-19 in non-geriatric patients. PLoS ONE 2022, 17, e0268530. [Google Scholar] [CrossRef]
- Legler, D.F.; Thelen, M. Chemokines: Chemistry, Biochemistry and Biological Function. CHIMIA 2016, 70, 856. [Google Scholar] [CrossRef]
- Matsushima, K. Introduction. Springer Semin. Immunopathol. 2000, 22, 321–328. [Google Scholar] [CrossRef]
- Palomino, D.C.T.; Marti, L.C. Chemokines and immunity. Einstein 2015, 13, 469–473. [Google Scholar]
- Arunachalam, P.S.; Wimmers, F.; Mok, C.K.P.; Perera, R.A.P.M.; Scott, M.; Hagan, T.; Sigal, N.; Feng, Y.; Bristow, L.; Tsang, O.T.-Y.; et al. Systems biological assessment of immunity to mild versus severe COVID-19 infection in humans. Science 2020, 369, 1210–1220. [Google Scholar] [CrossRef]
- Russo, R.C.; Garcia, C.C.; Teixeira, M.M.; A Amaral, F. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev. Clin. Immunol. 2014, 10, 593–619. [Google Scholar] [CrossRef] [PubMed]
- Shetelig, C.; Limalanathan, S.; Hoffmann, P.; Seljeflot, I.; Gran, J.M.; Eritsland, J.; Andersen, G. Association of IL-8 with Infarct Size and Clinical Outcomes in Patients with STEMI. J. Am. Coll. Cardiol. 2018, 72, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Kraft, R.; Herndon, D.N.; Finnerty, C.C.; Cox, R.A.; Song, J.; Jeschke, M.G. Predictive Value of IL-8 for Sepsis and Severe Infections After Burn Injury: A Clinical Study. Shock 2015, 43, 222–227. [Google Scholar] [CrossRef] [PubMed]
- Domingos, I.F.; Pereira-Martins, D.A.; Sobreira, M.J.V.C.; Oliveira, R.T.D.; Alagbe, A.E.; Lanaro, C.; Albuquerque, D.M.; Blotta, M.H.S.L.; Araujo, A.S.; Costa, F.F.; et al. High levels of proinflammatory cytokines IL-6 and IL-8 are associated with a poor clinical outcome in sickle cell anemia. Ann. Hematol. 2020, 99, 947–953. [Google Scholar] [CrossRef]
- Huang, Y.; Guo, H.; Zhou, Y.; Guo, J.; Wang, T.; Zhao, X.; Li, H.; Sun, Y.; Bian, X.; Fang, C. The associations between fasting plasma glucose levels and mortality of COVID-19 in patients without diabetes. Diabetes Res. Clin. Pract. 2020, 169, 108448. [Google Scholar] [CrossRef]
- Rodriguez, C.; de Prost, N.; Fourati, S.; Lamoureux, C.; Gricourt, G.; N’Debi, M.; Canoui-Poitrine, F.; Désveaux, I.; Picard, O.; Demontant, V.; et al. Viral genomic, metagenomic and human transcriptomic characterization and prediction of the clinical forms of COVID-19. PLoS Pathog. 2021, 17, e1009416. [Google Scholar] [CrossRef]
- Blot, M.; Bour, J.-B.; Quenot, J.P.; Bourredjem, A.; Nguyen, M.; Guy, J.; Monier, S.; Georges, M.; Large, A.; Dargent, A.; et al. The dysregulated innate immune response in severe COVID-19 pneumonia that could drive poorer outcome. J. Transl. Med. 2020, 18, 457. [Google Scholar] [CrossRef]
- Stikker, B.S.; Stik, G.; van Ouwerkerk, A.F.; Trap, L.; Spicuglia, S.; Hendriks, R.W.; Stadhouders, R. Severe COVID-19-associated variants linked to chemokine receptor gene control in monocytes and macrophages. Genome Biol. 2022, 23, 96. [Google Scholar] [CrossRef]
- Saraiva, M.; O’Garra, A. The regulation of IL-10 production by immune cells. Nat. Rev. Immunol. 2010, 10, 170–181. [Google Scholar] [CrossRef]
- Saraiva, M.; Vieira, P.; O’Garra, A. Biology and therapeutic potential of interleukin-10. J. Exp. Med. 2020, 217, e20190418. [Google Scholar] [CrossRef]
- Georg, P.; Astaburuaga-García, R.; Bonaguro, L.; Brumhard, S.; Michalick, L.; Lippert, L.J.; Kostevc, T.; Gäbel, C.; Schneider, M.; Streitz, M.; et al. Complement activation induces excessive T cell cytotoxicity in severe COVID-19. Cell 2022, 185, 493–512.e25. [Google Scholar] [CrossRef] [PubMed]
- Acosta-Ampudia, Y.; Monsalve, D.M.; Rojas, M.; Rodríguez, Y.; Gallo, J.E.; Salazar-Uribe, J.C.; Santander, M.J.; Cala, M.P.; Zapata, W.; Zapata, M.I.; et al. COVID-19 convalescent plasma composition and immunological effects in severe patients. J. Autoimmun. 2021, 118, 102598. [Google Scholar] [CrossRef] [PubMed]
- Espíndola, O.M.; Gomes, Y.C.P.; Brandão, C.O.; Torres, R.C.; Siqueira, M.; Soares, C.N.; Lima, M.A.S.D.; Leite, A.C.C.B.; Venturotti, C.O.; Carvalho, A.J.C.; et al. Inflammatory Cytokine Patterns Associated with Neurological Diseases in Coronavirus Disease 2019. Ann. Neurol. 2021, 89, 1041–1045. [Google Scholar] [CrossRef]
- Lucas, C.; Wong, P.; Klein, J.; Castro, T.B.R.; Silva, J.; Sundaram, M.; Ellingson, M.K.; Mao, T.; Oh, J.E.; Israelow, B.; et al. Longitudinal analyses reveal immunological misfiring in severe COVID-19. Nature 2020, 584, 463–469. [Google Scholar] [CrossRef]
- Hadjadj, J.; Yatim, N.; Barnabei, L.; Corneau, A.; Boussier, J.; Smith, N.; Péré, H.; Charbit, B.; Bondet, V.; Chenevier-Gobeaux, C.; et al. Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients. Science 2020, 369, 718–724. [Google Scholar] [CrossRef] [PubMed]
- Dean, M.J.; Ochoa, J.B.; Sanchez-Pino, M.D.; Zabaleta, J.; Garai, J.; Del Valle, L.; Wyczechowska, D.; Baiamonte, L.B.; Philbrook, P.; Majumder, R.; et al. Severe COVID-19 Is Characterized by an Impaired Type I Interferon Response and Elevated Levels of Arginase Producing Granulocytic Myeloid Derived Suppressor Cells. Front. Immunol. 2021, 12, 695972. [Google Scholar] [CrossRef] [PubMed]
- Carvelli, J.; Demaria, O.; Vély, F.; Batista, L.; Benmansour, N.C.; Fares, J.; Carpentier, S.; Thibult, M.-L.; Morel, A.; Remark, R.; et al. Association of COVID-19 inflammation with activation of the C5a–C5aR1 axis. Nature 2020, 588, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Skendros, P.; Mitsios, A.; Chrysanthopoulou, A.; Mastellos, D.C.; Metallidis, S.; Rafailidis, P.; Ntinopoulou, M.; Sertaridou, E.; Tsironidou, V.; Tsigalou, C.; et al. Complement and tissue factor–enriched neutrophil extracellular traps are key drivers in COVID-19 immunothrombosis. J. Clin. Investig. 2020, 130, 6151–6157. [Google Scholar] [CrossRef]
- Zhang, W.; Morris, A.B.; Peek, E.V.; Karadkhele, G.; Robertson, J.M.; Kissick, H.T.; Larsen, C.P. Faculty Opinions recommendation of CMV status drives distinct trajectories of CD4+ T cell differentiation. Front. Immunol. 2021, 12, 620386. [Google Scholar] [CrossRef]
- Higdon, L.E.; Gustafson, C.E.; Ji, X.; Sahoo, M.K.; Pinsky, B.A.; Margulies, K.B.; Maecker, H.T.; Goronzy, J.; Maltzman, J.S. Association of Premature Immune Aging and Cytomegalovirus After Solid Organ Transplant. Front. Immunol. 2021, 12, 661551. [Google Scholar] [CrossRef]
- Alanio, C.; Verma, A.; Mathew, D.; Gouma, S.; Liang, G.; Dunn, T.; Oldridge, D.A.; Weaver, J.; Kuri-Cervantes, L.; Pampena, M.B.; et al. Cytomegalovirus Latent Infection is Associated with an Increased Risk of COVID-19-Related Hospitalization. J. Infect. Dis. 2022, 226, 463–473. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Karasu, E.; Huber-Lang, M. Complement After Trauma: Suturing Innate and Adaptive Immunity. Front. Immunol. 2018, 9, 2050. [Google Scholar] [CrossRef] [PubMed]
- Declercq, J.; Bosteels, C.; Van Damme, K.; De Leeuw, E.; Maes, B.; Vandecauter, A.; Vermeersch, S.; Delporte, A.; Demeyere, B.; Vuylsteke, M.; et al. Zilucoplan in patients with acute hypoxic respiratory failure due to COVID-19 (ZILU-COV): A structured summary of a study protocol for a randomised controlled trial. Trials 2020, 21, 934. [Google Scholar] [CrossRef] [PubMed]




| Parameter | SARS-CoV-2 Negative Controls (n = 80) | SARS-CoV-2 Positive Patients (n = 210) | p | |
|---|---|---|---|---|
| Age (mean ± SD) | 56.27 ± 5.56 | 57.78 ± 14.16 | 0.052 | |
| Sex (male/female) | 5/75 | 114/96 | <0.001 | |
| Body mass index (mean ± SD) | 25.85 ± 4.74 | 29.17 ± 5.52 | <0.001 | |
| Parameter | SARS-CoV-2 positive patients non-ICU (n = 160) | SARS-CoV-2 positive patients ICU (n = 50) | ||
| Percent of patient population (%) | 76.19 | 23.81 | ||
| Age (mean ± SD) | 56.26 ± 14.53 | 62.64 ± 11.79 | <0.01 | |
| Sex (male/female) | 85/74 | 28/22 | 1 | |
| Body mass index (mean ± SD) | 28.87 ± 5.55 | 30.44 ± 5.42 | 0.04 | |
| % of patients with a given parameter | Medical history | |||
| Hypertension | 44.38 | 54 | 0.25 | |
| Diabetes | 20.63 | 20 | 1 | |
| Ischemic heart disease | 7.5 | 12 | 0.38 | |
| Hypercholesterolemia | 14.38 | 14 | 1 | |
| Liver disease | 0.63 | 2 | 0.42 | |
| Respiratory system disease | 8.13 | 14 | 0.27 | |
| Rheumatic disease | 9.38 | 10 | 1 | |
| Cancer | 10.63 | 10 | 1 | |
| Other diseases | 33.13 | 50 | 0.04 | |
| Tobacco use (previous/now) | 43.13/5.63 | 30/0 | <0.001 | |
| Current medications | ||||
| Drugs taken on a permanent basis | 63.75 | 46 | 0.03 | |
| NSAIDs | 12.5 | 16 | 0.49 | |
| Statins | 15 | 22 | 0.28 | |
| Antihypertensive drugs | 47.5 | 46 | 0.87 | |
| Anticoagulants | 7.5 | 6 | 1 | |
| Cardiac drugs | 7.5 | 14 | 0.17 | |
| Anti-asthmatic drugs | 8.13 | 10 | 0.77 | |
| Other drugs | 41.25 | 38 | 0.74 | |
| Parameter | Negative Controls (n = 80) | |
|---|---|---|
| % of patients with a given parameter | Medical history | |
| Hypertension | 14.08 | |
| Diabetes | 2.82 | |
| Ischemic heart disease | 0 | |
| Hypercholesterolemia | 8.45 | |
| Liver disease | 0 | |
| Respiratory system disease | 0 | |
| Rheumatic disease | 11.27 | |
| Cancer | 4.41 | |
| Other diseases | 0 | |
| Tobacco use (previous/now) | 40.26/33.80 | |
| Current medications | ||
| Drugs taken on a permanent basis | 21.27 | |
| NSAIDs | 8.45 | |
| Statins | 5.63 | |
| Antihypertensive drugs | 15.49 | |
| Anticoagulants | 6.63 | |
| Cardiac drugs | 0 | |
| Anti-asthmatic drugs | 2.82 | |
| Other drugs | 16.90 | |
| Time of Sample Collection | Day 1 | Day 7 | Day 14 | Day 28 | |
|---|---|---|---|---|---|
| Groups | CMV (+) (n = 185) | ||||
| Number of individuals within the group | n = 181 | n = 122 | n = 69 | n = 58 | |
| Chemokine expression | Gene relative expression level | ||||
| mean ± SD (IQR) | CXCL8 | 0.32 ± 0.30 (0.21) | 0.87 ± 0.70 (0.65) *** | 0.93 ± 0.82 (0.67) *** | 0.74 ± 0.61 (0.59) ** |
| CCL2 | 0.55 ± 0.77 (0.23) | 0.35 ± 0.38 (0.21) | 0.39 ± 0.34 (0.28) | 0.65 ± 0.78 (0.37) | |
| CCR1 | 1.09 ± 0.66 (0.98) | 0.77 ± 0.45 (0.69) * | 0.92 ± 0,49 (0.84) | 0.93 ± 0.48 (0.81) | |
| Chemokine concentration | Protein concentration (pg/mL) | ||||
| Mean ± SD (IQR) | CXCL8 | 4.90 ± 3.66 (4.07) | 2.44 ± 1.61 (2.04) * | 3.46 ± 3.13 (1.96) | 3.41 ± 1.8 (3.11) |
| CXCL10 | 259.36 ± 203.24 (234) | 64.11 ± 46.51 (51.76) *** | 46.06 ± 41.20 (29.12) *** | 50.59 ± 36.30 (41.82) *** | |
| CCL2 | 268.67 ± 160.97 (232) | 210.87 ± 123.17 (182.5) ** | 257.36 ± 164.32 (222.5) | 244.45 ± 129.27 (221) | |
| CCL3 | 331.06 ± 154.05 (346) | 103.89 ± 39.08 (92.80) *** | 101.05 ± 49.59 (92.79) *** | 104.11 ± 52.29 (80.01) *** | |
| Complement expression | Gene relative expression level | ||||
| Mean ± SD (IQR) | C4BPA | 0.60 ± 0.65 (0.37) | 1.13 ± 0.89 (0.99) *** | 1.04 ± 0,99 (0.62) * | 1.10 ± 0.80 (0.84) ** |
| C5AR1 | 0.71 ± 0.32 (0.70) | 0.82 ± 0.36 (0.80) * | 1.94 ± 1.56 (1.42) *** | 0.95 ± 0.44 (0.93) * | |
| CFD | 1.35 ± 1.17 (0.87) | 1.71 ± 1.42 (1.08) * | 3.38 ± 2.93 (2.48) *** | 2.00 ± 1.42 (1.42) * | |
| CR1 | 0.83 ± 0.42 (0.78) | 0.53 ± 0.26 (0.47) ** | 1.30 ± 0.77 (1.33) | 1.53 ± 0.68 (1.53) *** | |
| Complementconcentration | Protein concentration (ng/mL) | ||||
| Mean ± SD (IQR) | C9 | 90,070.67 ± 22,913.75 (88,300) | 59,470.25 ± 22,633.56 (56,900) *** | 6130.59 ± 2099.12 (5730) *** | 4680.26 ± 1786.52 (4730) *** |
| CFD | 4153.29 ± 1170.17 (3923.83) | 3599.13 ± 1064.72 (3476.6) * | 3869.71 ± 1024.44 (3817.65) | 5038.22 ± 1344.68 (4732.5) * | |
| C2 | 26,629.12 ± 6388.90 (26,174.4) | 19,602.25 ± 5044.71 (19,233) *** | 19,343.79 ± 5003.17 (19,436.8) *** | 21,253.07 ± 4590.60 (21,145.8) *** | |
| Time of sample collection | Day 1 | Day 7 | Day 14 | Day 28 | |
|---|---|---|---|---|---|
| Groups | CMV (−) (n = 25) | ||||
| Number of individuals within the group | n = 21 | n = 10 | n = 10 | n = 11 | |
| Chemokine expression | Gene relative expression level | ||||
| Mean ± SD (IQR) | CXCL8 | 0.49 ± 0.40 (0.35) | 1.77 ± 1.06 (2.22) *** | 1.67 ± 1.97 (0.76) *** | 0.85 ± 0.61 (0.83) ** |
| CCL2 | 0.63 ± 0.83 (0.36) | 0.32 ± 0.12 (0.27) | 2.05 ± 2.43 (0.44) | 0.85 ± 0.71 (0.58) | |
| CCR1 | 0.91 ± 0.49 (0.78) | 0.82 ± 0.28 (0.68) * | 1.24 ± 0.48 (1.14) | 1.01 ± 0.34 (0.92) | |
| Chemokine concentration | Protein concentration (pg/mL) | ||||
| Mean ± SD (IQR) | CXCL8 | 4.47 ± 2,90 (3.93) | 3.03 ± 2.32 (1.97) *** | 3.78 ± 3.09 (2.87) | 3.38 ± 2.21 (2.64) |
| CXCL10 | 195.20 ± 132.24 (199.5) | 63.24 ± 31.96 (62.34) *** | 46.60 ± 38.58 (30.03) *** | 29.23 ± 7.47 (31.23) *** | |
| CCL2 | 255.20 ± 126.41 (243) | 191.52 ± 91.80 (202.5) * | 234.31 ± 133.63 (200) | 260.14 ± 89.04 (236) | |
| CCL3 | 287.33 ± 139.65 (284.5) *** | 144.60 ± 72.02 (119) *** | 134,54 ± 77,57 (125) *** | 83.18 ± 42.47 (80.01) *** | |
| Complement expression | Gene relative expression level | ||||
| Mean ± SD (IQR) | C4BPA | 0.28 ± 0.29 (0.18) | 0.98 ± 1.44 (0.49) | 0.41 ± 0.39 (0.43) | 0.75 ± 0.56 (0.65) |
| C5AR1 | 0.59 ± 0.34 (0.59) | 0.69 ± 0.54 (0.74) | 0.94 ± 0.82 (0.92) | 1.10 ± 0.62 (1.18) | |
| CFD | 0.89 ± 0.64 (0.82) | 0.89 ± 0.57 (0.84) | 1.63 ± 1.00 (1.26) | 1.84 ± 0.93 (1.55) | |
| CR1 | 0.77 ± 0.42 (0.67) | 0.43 ± 0.16 (0.38) | 0.56 ± 0.22 (0.55) | 2.07 ± 0.79 (2.01) * | |
| Complement concentration | Protein concentration (ng/mL) | ||||
| Mean ± SD (IQR) | C9 | 89,665 ± 20,673.83 (86,700) | 56,705 ± 20,082.18 (54,875) * | 4158.75 ± 1272.43 (4577.5) *** | 3480 ± 837.71 (3235) *** |
| CFD | 4074.60 ± 1041.90 (3750.7) | 3965.14 ± 1257.85 (3843.28) * | 4075.83 ± 851.52 (4157.25) | 4743.12 ± 1160.92 (4324) * | |
| C2 | 27,041.32 ± 4977.20 (28,002.1) | 19,133.4 ± 3607.63 (19,404.8) *** | 19,675.76 ± 3839.96 (18381) *** | 21,176.18 ± 3482.16 (21,065.4) *** | |
| Severity | CMV IgG Status | Frequency |
|---|---|---|
| ICU | Negative | 1 |
| Non-ICU | Negative | 24 |
| ICU | Positive | 51 |
| Non-ICU | Positive | 134 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pius-Sadowska, E.; Niedźwiedź, A.; Kulig, P.; Baumert, B.; Sobuś, A.; Rogińska, D.; Łuczkowska, K.; Ulańczyk, Z.; Wnęk, S.; Karolak, I.; et al. CXCL8, CCL2, and CMV Seropositivity as New Prognostic Factors for a Severe COVID-19 Course. Int. J. Mol. Sci. 2022, 23, 11338. https://doi.org/10.3390/ijms231911338
Pius-Sadowska E, Niedźwiedź A, Kulig P, Baumert B, Sobuś A, Rogińska D, Łuczkowska K, Ulańczyk Z, Wnęk S, Karolak I, et al. CXCL8, CCL2, and CMV Seropositivity as New Prognostic Factors for a Severe COVID-19 Course. International Journal of Molecular Sciences. 2022; 23(19):11338. https://doi.org/10.3390/ijms231911338
Chicago/Turabian StylePius-Sadowska, Ewa, Anna Niedźwiedź, Piotr Kulig, Bartłomiej Baumert, Anna Sobuś, Dorota Rogińska, Karolina Łuczkowska, Zofia Ulańczyk, Szymon Wnęk, Igor Karolak, and et al. 2022. "CXCL8, CCL2, and CMV Seropositivity as New Prognostic Factors for a Severe COVID-19 Course" International Journal of Molecular Sciences 23, no. 19: 11338. https://doi.org/10.3390/ijms231911338
APA StylePius-Sadowska, E., Niedźwiedź, A., Kulig, P., Baumert, B., Sobuś, A., Rogińska, D., Łuczkowska, K., Ulańczyk, Z., Wnęk, S., Karolak, I., Paczkowska, E., Kotfis, K., Kawa, M., Stecewicz, I., Zawodny, P., & Machaliński, B. (2022). CXCL8, CCL2, and CMV Seropositivity as New Prognostic Factors for a Severe COVID-19 Course. International Journal of Molecular Sciences, 23(19), 11338. https://doi.org/10.3390/ijms231911338






