Proteomic Profiling Skin Mucus of European Eel Anguilla anguilla Infected with Anguillid Herpesvirus
Abstract
1. Introduction
2. Results
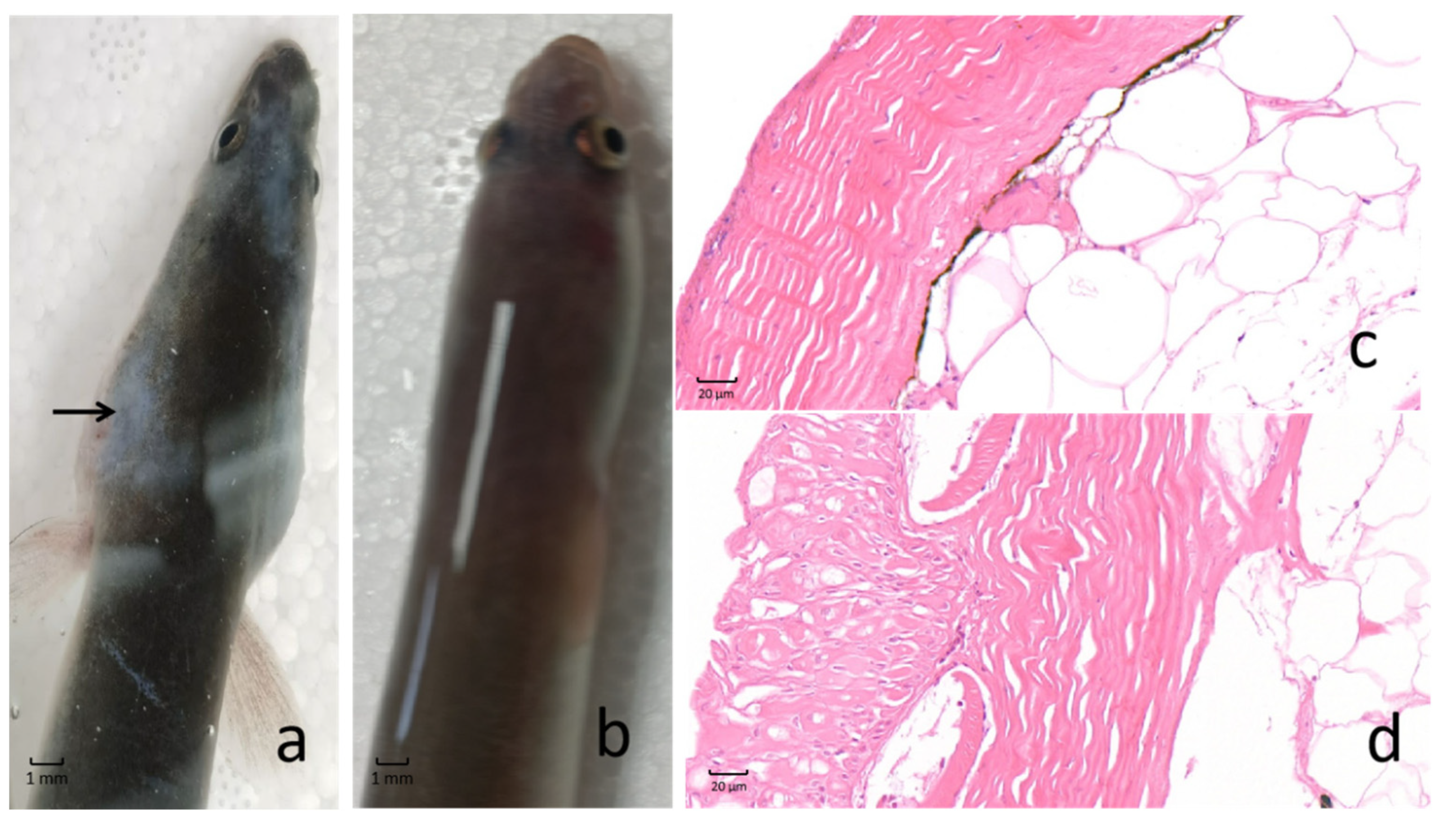
2.1. Morphological and Histopathological Changes in the Skin of AngHV-Infected Eels
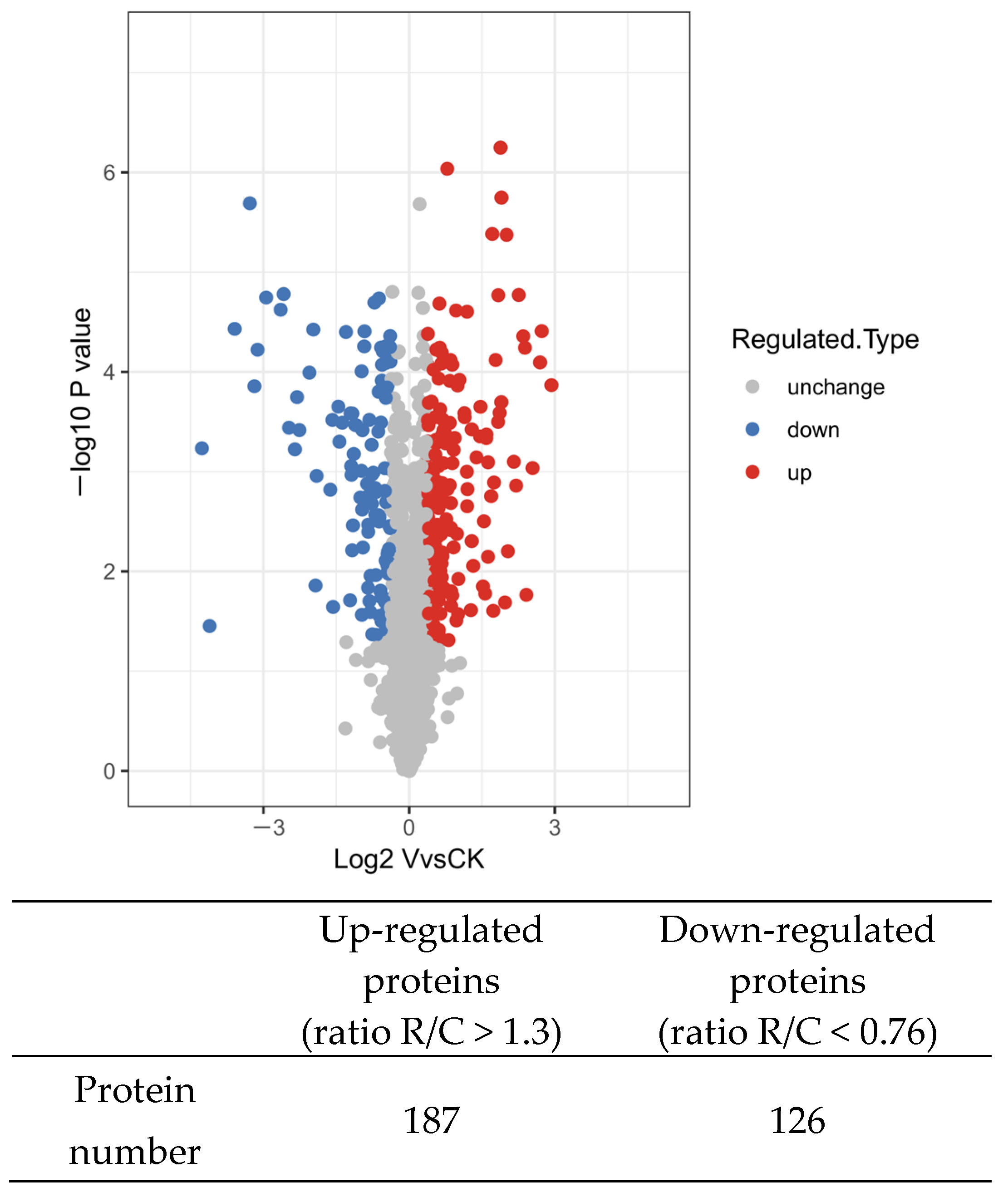
2.2. Protein Identification
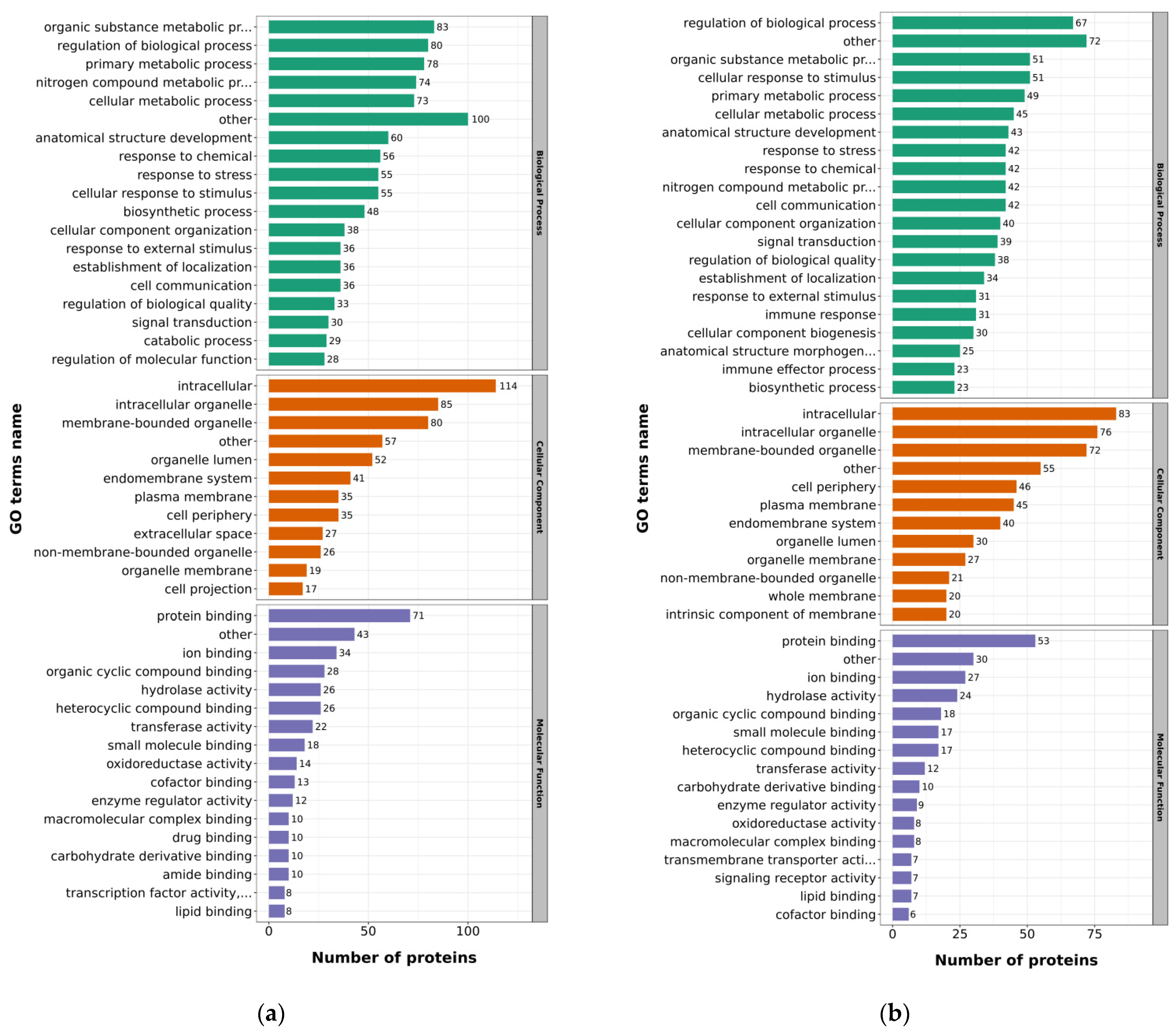
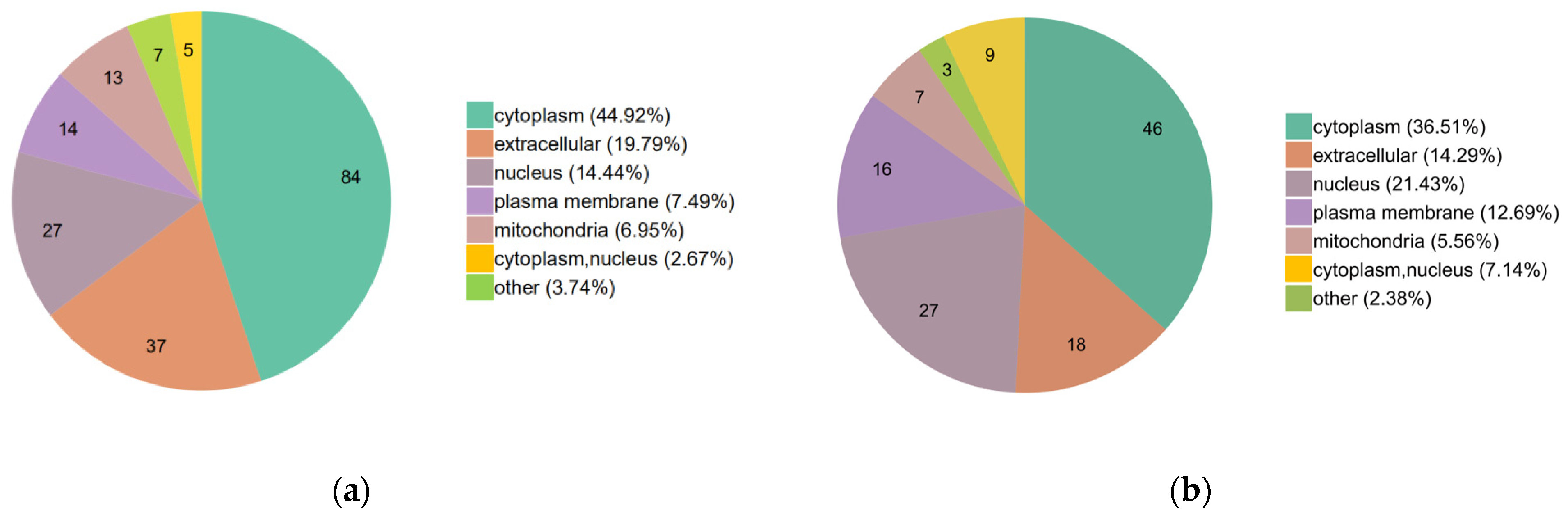
2.3. Functional Classification of the DEPs
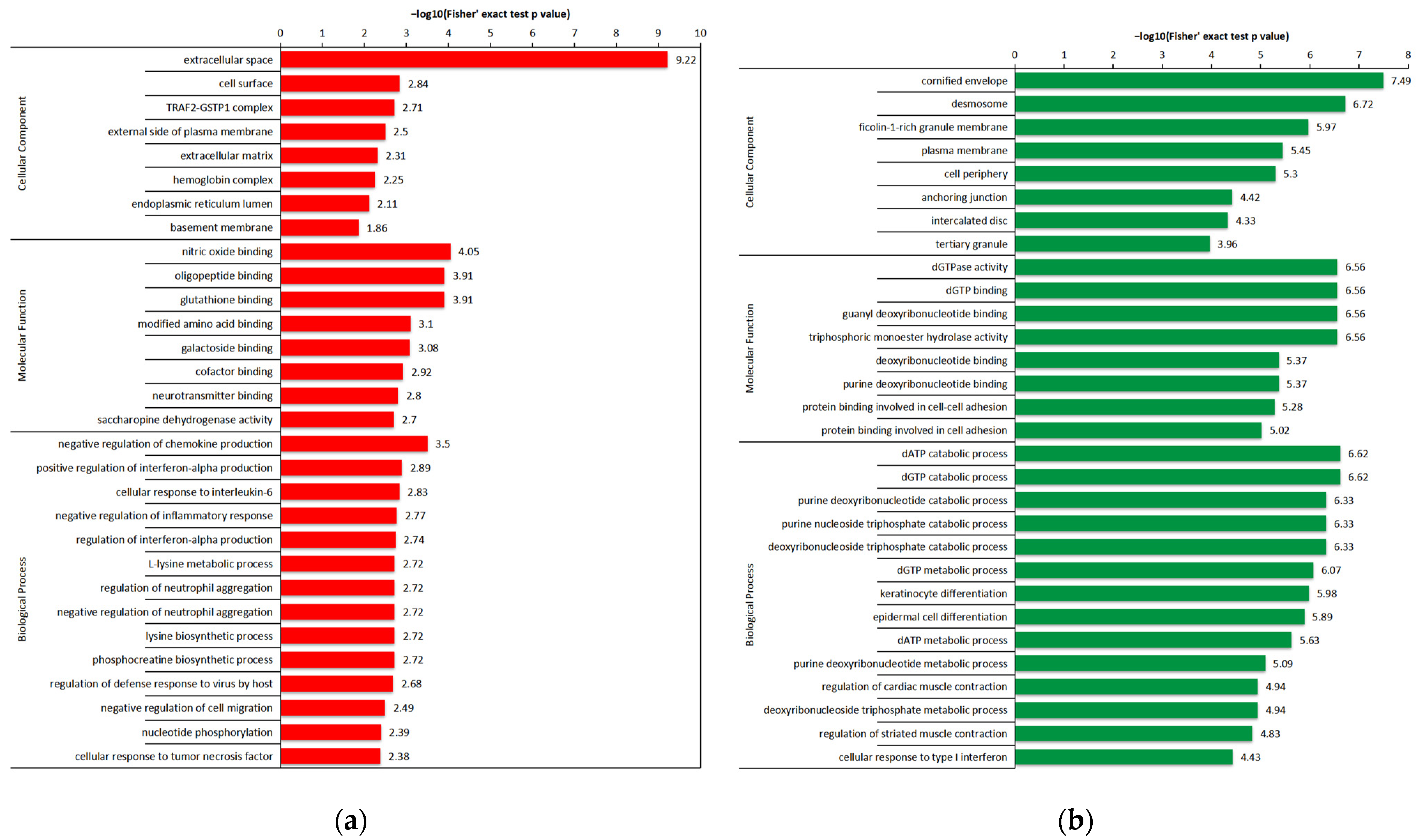
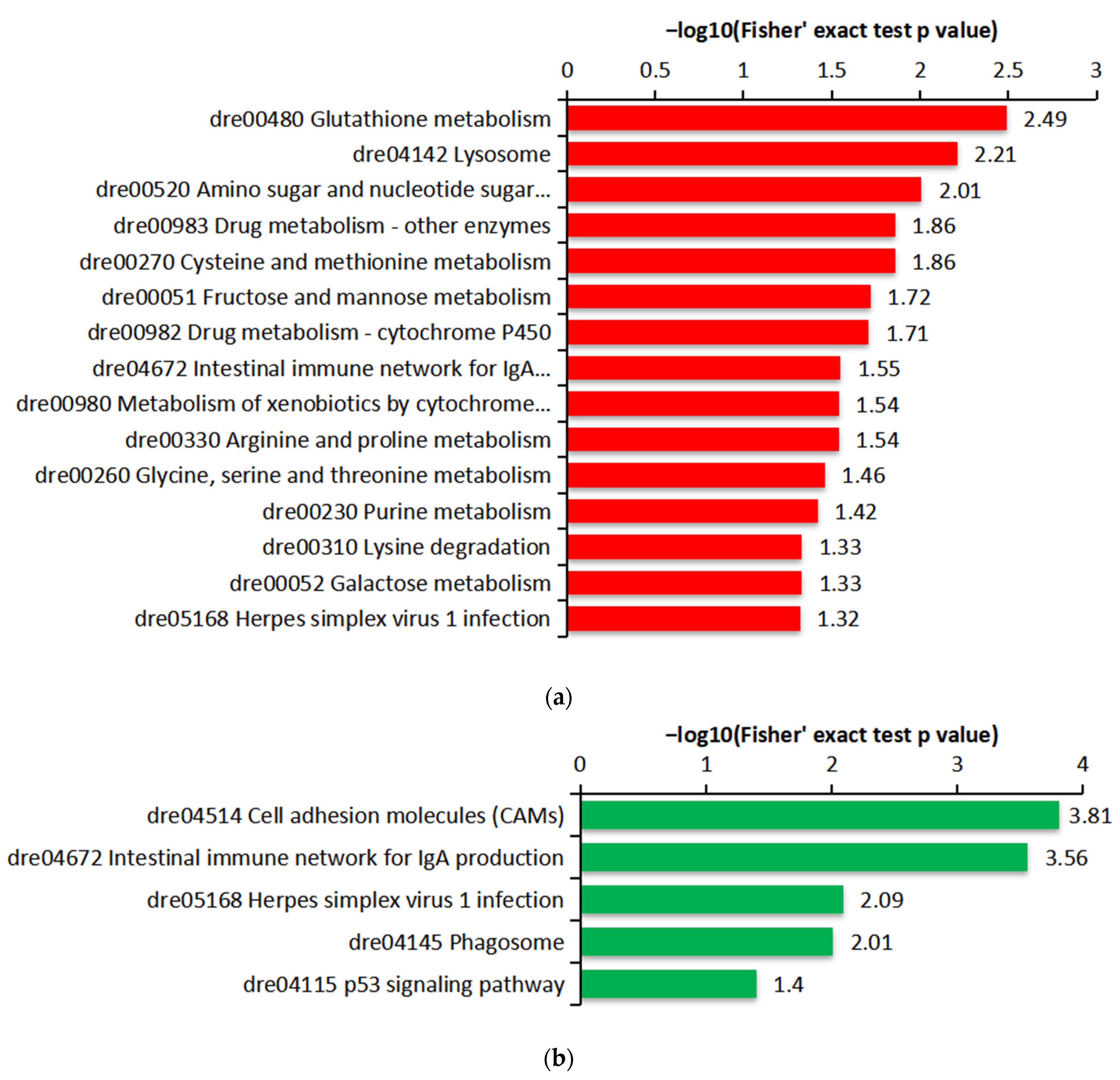
2.4. Functional Enrichment of the DEPs
2.5. Confirmation of DEPs by PRM Analysis
3. Discussion
4. Materials and Methods
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Van Beurden, S.J.; Gatherer, D.; Kerr, K.; Galbraith, J.; Herzyk, P.; Peeters, B.P.H.; Rottier, P.J.M.; Engelsma, M.Y.; Davison, A.J. Anguillid herpesvirus 1 transcriptome. J. Virol. 2012, 86, 10150–10161. [Google Scholar] [CrossRef]
- Bandín, I.; Souto, S.; Cutrín, J.M.; López-Vázquez, C.; Olveira, J.G.; Esteve, C.; Alcaide, E.; Dopazo, C.P. Presence of viruses in wild eels Anguilla anguilla L, from the Albufera Lake (Spain). J. Fish Dis. 2014, 37, 597–607. [Google Scholar]
- Sano, M.; Fukuda, H.; Sano, T. Isolation and characterization of a new herpesvirus from eel. In Pathology in Marine Science; Academic Press: Cambridge, MA, USA, 1990; pp. 15–31. [Google Scholar] [CrossRef]
- Ueno, Y.; Kitao, T.; Chen, S.N.; Al, E. Characterization of a herpes-like virus isolated from cultured Japanese eels in Taiwan. Fish Pathol. 1992, 27, 7–17. [Google Scholar]
- Davidse, A.; Haenen, O.L.M.; Dijkstra, S.G.; Al, E. First isolation of herpesvirus of eel (Herpesvirus anguillae) in diseased European eel (Anguilla anguilla L.) in Europe. Bull. Eur. Assoc. Fish Pat. 1999, 19, 137–141. [Google Scholar]
- Lee, N.S.; Kobayashi, J.; Miyazaki, T. Gill filament necrosis in farmed Japanese eels, Anguilla japonica (Temminck & Schlegel), infected with Herpesvirus anguillae. J. Fish Dis. 1999, 22, 457–463. [Google Scholar]
- Haenen, O.L.M.; Dijkstra, S.G.; van Tulden, P.W.; Davidse, A.; van Nieuwstadt, A.P.; Wagenaar, F.; Wellenberg, G.J. Herpesvirus anguillae (HVA) isolations from disease outbreaks in cultured European eel, Anguilla anguilla in The Netherlands since 1996. Bull. Eur. Assoc. Fish Pathol. 2002, 22, 247–257. [Google Scholar]
- Jakob, E.; Neuhaus, H.; Steinhagen, D.; Luckhardt, B.; Hanel, R. Monitoring of Herpesvirus anguillae (HVA) infections in European eel, Anguilla anguilla (L.), in northern Germany. J. Fish Dis. 2009, 32, 557–561. [Google Scholar] [CrossRef]
- Van Nieuwstadt, A.P.; Dijkstra, S.G.; Haenen, O. Persistence of herpesvirus of eel Herpesvirus anguillae in farmed European eel Anguilla anguilla. Dis. Aquat. Organ. 2001, 45, 103–107. [Google Scholar] [CrossRef]
- Chang, P.H.; Pan, Y.H.; Wu, C.M.; Kuo, S.T.; Chung, H.Y. Isolation and molecular characterization of herpesvirus from cultured European eels (Anguilla anguilla) in Taiwan. Dis. Aquat. Organ. 2002, 50, 111–118. [Google Scholar] [CrossRef]
- Van Beurden, S.J.; Bossers, A.; Voorbergen-Laarman, M.H.A.; Haenen, O.L.M.; Peters, S.; Abma-Henkens, M.H.C.; Peeters, B.P.H.; Rottier, P.J.M.; Engelsma, M.Y. Complete genome sequence and taxonomic position of anguillid herpesvirus 1. J. Gen. Virol. 2010, 91, 880–887. [Google Scholar] [CrossRef]
- Yang, J.X.; Chen, Q.; Li, Y.Y.; Song, T.Y.; Ge, J.Q. Biological and physicochemical characteristics of Anguillid herpesvirus. J. Fish. China 2020, 44, 1435–1440. [Google Scholar] [CrossRef]
- Ge, J.Q.; Yang, J.X.; Li, Y.J.; Chen, Q.; Lin, T.L. Polymerase chain reaction for the detection of Herpesvirus anguillae in eel viral disease samples. Fujian J. Agric. Sci. 2012, 27, 961–964. [Google Scholar] [CrossRef]
- Ge, J.Q.; Yang, J.X.; Gong, H.; Lin, T.L. Isolation and identification of a herpesvirus from cultured European eels Anguilla anguilla in China. J. Fish. China 2014, 38, 1579–1583. [Google Scholar]
- Chen, Q.; Li, Y.Y.; Yang, J.X.; Song, T.Y.; Ge, J.Q. Pathogenicity of Anguillid herpesvirus to Anguilla anguilla. J. Fish. China 2021, 45, 940–947. [Google Scholar] [CrossRef]
- Hu, B.; Li, X.; Huo, Y.; Yu, Y.; Zhang, Q.; Chen, G.; Zhang, Y.; Fraser, N.W.; Wu, D.; Zhou, J. Cellular responses to HSV-1 infection are linked to specific types of alterations in the host transcriptome. Sci. Rep. 2016, 6, 28075. [Google Scholar] [CrossRef]
- Sano, T.; Morita, N.; Shima, N.; Akimoto, M. Herpesvirus cyprini: Lethality and oncogenicity. J. Fish Dis. 1991, 14, 533–543. [Google Scholar] [CrossRef]
- Davidovich, M.; Dishon, A.; Ilouze, M.; Kotler, M. Susceptibility of cyprinid cultured cells to cyprinid herpesvirus 3. Arch. Virol. 2007, 152, 1541–1546. [Google Scholar] [CrossRef]
- Cain, K.D.; Jones, D.R.; Raison, R.L. Characterisation of mucosal and systemic immune responses in rainbow trout (Oncorhynchus mykiss) using surface plasmon resonance. Fish Shellfish Immun. 2000, 10, 651–666. [Google Scholar] [CrossRef]
- Hellio, C.; Pons, A.M.; Beaupoil, C.; Bourgougnon, N.; Gal, Y.L. Antibacterial, antifungal and cytotoxic activities of extracts from fish epidermis and epidermal mucus. Int. J. Antimicrob. Agents 2002, 20, 214–219. [Google Scholar] [CrossRef]
- Lin, S.H.; Ellis, A.E.; Davidson, G.A.; Secombes, C.J. Migratory, respiratory burst and mitogenic responses of leucocytes isolated from the gills of rainbow trout (Oncorhynchus mykiss). Fish Shellfish Immun. 1999, 9, 211–226. [Google Scholar] [CrossRef]
- Dos Santos, N.M.S.; Taverne-Thiele, J.J.; Barnes, A.C.; van Muiswinkel, W.B.; Ellis, A.E.; Rombout, J.H.W.M. The gill is a major organ for antibody secreting cell production following direct immersion of sea bass (Dicentrarchus labrax, L.) in a Photobacterium damselae ssp. piscicida bacterin: An ontogenetic study. Fish Shellfish Immun. 2001, 11, 65–74. [Google Scholar] [CrossRef]
- Lin, S.H.; Davidson, G.A.; Secombes, C.J.; Ellis, A.E. Use of a lipid-emulsion carrier for immunisation of dabž/Limanda limanda by bath and oral routes: An assessment of systemic and mucosal antibody responses. Aquaculture 2000, 181, 11–24. [Google Scholar]
- Zhao, C.; Wang, F.; Wang, P.; Ding, H.; Huang, X.; Shi, Z. Early second-trimester plasma protein profiling using multiplexed isobaric tandem mass tag (TMT) labeling predicts gestational diabetes mellitus. Acta Diabetol. 2015, 52, 1103–1112. [Google Scholar] [CrossRef]
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 340–350. [Google Scholar] [CrossRef]
- Lü, A.; Hu, X.; Wang, Y.; Shen, X.; Li, X.; Zhu, A.; Tian, J.; Ming, Q.; Feng, Z. iTRAQ analysis of gill proteins from the zebrafish (Danio rerio) infected with Aeromonas hydrophila. Fish Shellfish Immun. 2014, 36, 229–239. [Google Scholar] [CrossRef]
- Long, M.; Zhao, J.; Li, T.; Tafalla, C.; Zhang, Q.; Wang, X.; Gong, X.; Shen, Z.; Li, A. Transcriptomic and proteomic analyses of splenic immune mechanisms of rainbow trout (Oncorhynchus mykiss) infected by Aeromonas salmonicida subsp. salmonicida. J. Proteom. 2015, 122, 41–54. [Google Scholar] [CrossRef]
- Wang, L.; Shao, C.; Xu, W.; Zhou, Q.; Wang, N.; Chen, S. Proteome profiling reveals immune responses in Japanese flounder (Paralichthys olivaceus) infected with Edwardsiella tarda by iTRAQ analysis. Fish Shellfish Immunol. 2017, 36, 229–239. [Google Scholar]
- Liu, M.; Wu, T.; Li, S.; Wei, P.; Yan, Y.; Gu, W.; Wang, W.; Meng, Q. Combined transcriptomic/proteomic analysis of crucian carp Carassius auratus gibelio in cyprinid herpesvirus 2 infection. Fish Shellfish. Immunol. 2018, 82, 386–399. [Google Scholar]
- Rinaldo, R.C. Immune suppression by herpesviruses. Annu. Rev. Med. 1990, 41, 331–338. [Google Scholar] [CrossRef]
- Schulz, P.; Deperasińska, I.; Kaczorek-Łukowska, E.; Siwicki, A.K. Immunosuppressive influence of Anguillid herpesvirus-1 (AngHV-1) infection on cellular defense mechanisms in European eel (Anguilla anguilla). Pol. J. Vet. Sci. 2019, 22, 785–787. [Google Scholar] [CrossRef]
- Beutler, B.; Eidenschenk, C.; Crozat, K.; Imler, J.; Takeuchi, O.; Hoffmann, J.A.; Akira, S. Genetic analysis of resistance to viral infection. Nat. Rev. Immunol. 2007, 7, 753–766. [Google Scholar] [CrossRef]
- Chen, H.Y.; Liu, W.; Wu, S.Y.; Chiou, P.P.; Li, Y.H.; Chen, Y.C.; Lin, G.H.; Lu, M.W.; Wu, J.L. RIG-I specifically mediates group II type I IFN activation in nervous necrosis virus infected zebrafish cells. Fish Shellfish Immun. 2015, 20, 427–435. [Google Scholar]
- Hu, X.; Fu, X.; Li, N.; Dong, X.; Zhao, L.; Lan, J.; Ji, W.; Zhou, W.; Ai, T.; Wu, S.; et al. Transcriptomic analysis of Mandarin fish brain cells infected with infectious spleen and kidney necrosis virus with an emphasis on retinoic acid-inducible gene 1-like receptors and apoptosis pathways. Fish Shellfish Immun. 2015, 45, 619–629. [Google Scholar] [CrossRef]
- Zou, P.F.; Chang, M.X.; Li, Y.; Huan Zhang, S.; Fu, J.P.; Chen, S.N.; Nie, P. Higher antiviral response of RIG-I through enhancing RIG-I/MAVS-mediated signaling by its long insertion variant in zebrafish. Fish Shellfish Immun. 2015, 43, 13–24. [Google Scholar] [CrossRef]
- Skjesol, A.; Hansen, T.; Shi, C.Y.; Thim, H.L.; Jorgensen, J.B. Structural and functional studies of STAT1 from Atlantic salmon (Salmo salar). BMC Immunol. 2010, 11, 17. [Google Scholar] [CrossRef]
- Sen, G.C. Viruses and interferons. Annu. Rev. Microbiol. 2001, 55, 255. [Google Scholar]
- Langevin, C.; Aleksejeva, E.; Passoni, G.; Palha, N.; Levraud, J.; Boudinot, P. The Antiviral Innate Immune Response in Fish: Evolution and Conservation of the IFN System. J. Mol. Biol. 2013, 425, 4904–4920. [Google Scholar] [CrossRef]
- Jin, B.Q. Cellular and Molecular Immunology; Science China Press: Beijing, China, 2001. [Google Scholar]
- Herberg, J.A.; Sgouros, J.; Jones, T.; Copeman, J.; Humphray, S.J.; Sheer, D.; Cresswell, P.; Beck, S.; Trowsdale, J. Genomic analysis of the Tapasin gene, located close to the TAP loci in the MHC. Eur. J. Immunol. 1998, 28, 459–467. [Google Scholar] [CrossRef]
- Peaper, D.R.; Cresswell, P.; Wearsch, P.A. Tapasin and ERp57 form a stable disulfide-linked dimer within the MHC class I peptide-loading complex. EMBO J. 2005, 24, 3613–3623. [Google Scholar] [CrossRef]

| Total Spectrum | Matched Spectrum | Peptides | Unique Peptides | Identified Proteins | Quantified Proteins |
|---|---|---|---|---|---|
| 298,207 | 44,795 (15.0%) | 20,633 | 16,815 | 3486 | 2935 |
| Protein Accession | Protein Name | Protein Description | Fold Change (AngHV/CK) | |
|---|---|---|---|---|
| PRM | TMT | |||
| transcript_14379 | STAT1 | signal transducer and activator of transcription 1 | 1.888 ± 0.021 | 1.915 ± 0.040 |
| transcript_28497 | CRISP-3 | Cysteine-rich secretory protein 2 | 1.847 ± 0.085 | 1.698 ± 0.028 |
| transcript_25607 | serpin B | serpin B | 1.695 ± 0.066 | 1.788 ± 0.024 |
| transcript_15352 | SAP | saposin | 1.406 ± 0.092 | 1.312 ± 0.029 |
| transcript_15360 | CD68 antigen | CD68 antigen | 1.372 ± 0.007 | 1.603 ± 0.006 |
| transcript_25331 | cathepsin B | cathepsin B [EC:3.4.22.1] | 1.354 ± 0.070 | 1.325 ± 0.036 |
| transcript_29917 | HBA | hemoglobin subunit alpha | 1.352 ± 0.132 | 1.585 ± 0.091 |
| transcript_29975 | HBE | hemoglobin subunit epsilon | 1.191 ± 0.126 | 1.567 ± 0.079 |
| transcript_1706 | DSG2 | desmoglein 2 | 0.683 ± 0.024 | 0.767 ± 0.003 |
| transcript_23496 | CAV-1 | caveolin 1 | 0.668 ± 0.041 | 0.758 ± 0.013 |
| transcript_27261 | ASC | apoptosis-associated speck-like protein containing a CARD | 0.622 ± 0.009 | 0.707 ± 0.012 |
| transcript_7859 | MTHFD1 | methylenetetrahydrofolate dehydrogenase (NADP+)/methenyltetrahydrofolate cyclohydrolase/formyltetrahydrofolate synthetase [EC:1.5.1.5 3.5.4.9 6.3.4.3] | 0.551 ± 0.017 | 0.765 ± 0.022 |
| transcript_2511 | plakophilin 1 | plakophilin 1 | 0.511 ± 0.028 | 0.712 ± 0.006 |
| transcript_8206 | CTNNβ1 | catenin beta 1 | 0.451 ± 0.007 | 0.678 ± 0.007 |
| transcript_3978 | ADA | adenosine deaminase [EC:3.5.4.4] | 0.364 ± 0.014 | 0.463 ± 0.007 |
| transcript_1362 | desmocollin 2 | desmocollin 2 | 0.306 ± 0.024 | 0.527 ± 0.007 |
| transcript_893 | DSP | desmoplakin | 0.276 ± 0.010 | 0.052 ± 0.002 |
| transcript_9314 | SAMHD1 | deoxynucleoside triphosphate triphosphohydrolase SAMHD1 | 0.027 ± 0.004 | 0.130 ± 0.006 |
| transcript_24094 | MHC II | major histocompatibility complex, class II | 0.007 ± 0.001 | 0.445 ± 0.015 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.-Y.; Yang, J.-X.; Chen, X.; Chen, Q.; Song, T.-Y.; Ge, J.-Q. Proteomic Profiling Skin Mucus of European Eel Anguilla anguilla Infected with Anguillid Herpesvirus. Int. J. Mol. Sci. 2022, 23, 11283. https://doi.org/10.3390/ijms231911283
Li Y-Y, Yang J-X, Chen X, Chen Q, Song T-Y, Ge J-Q. Proteomic Profiling Skin Mucus of European Eel Anguilla anguilla Infected with Anguillid Herpesvirus. International Journal of Molecular Sciences. 2022; 23(19):11283. https://doi.org/10.3390/ijms231911283
Chicago/Turabian StyleLi, Ying-Ying, Jin-Xian Yang, Xi Chen, Qiang Chen, Tie-Ying Song, and Jun-Qing Ge. 2022. "Proteomic Profiling Skin Mucus of European Eel Anguilla anguilla Infected with Anguillid Herpesvirus" International Journal of Molecular Sciences 23, no. 19: 11283. https://doi.org/10.3390/ijms231911283
APA StyleLi, Y.-Y., Yang, J.-X., Chen, X., Chen, Q., Song, T.-Y., & Ge, J.-Q. (2022). Proteomic Profiling Skin Mucus of European Eel Anguilla anguilla Infected with Anguillid Herpesvirus. International Journal of Molecular Sciences, 23(19), 11283. https://doi.org/10.3390/ijms231911283





